Abstract
Free full text

Exosome-Mediated Intercellular Communication between Hepatitis C Virus-Infected Hepatocytes and Hepatic Stellate Cells
ABSTRACT
Fibrogenic pathways in the liver are principally regulated by activation of hepatic stellate cells (HSC). Fibrosis is associated with chronic hepatitis C virus (HCV) infection, although the mechanism is poorly understood. HSC comprise the major population of nonparenchymal cells in the liver. Since HCV does not replicate in HSC, we hypothesized that exosomes secreted from HCV-infected hepatocytes activate HSC. Primary or immortalized human hepatic stellate (LX2) cells were exposed to exosomes derived from HCV-infected hepatocytes (HCV-exo), and the expression of fibrosis-related genes was examined. Our results demonstrated that HCV-exo internalized to HSC and increased the expression of profibrotic markers. Further analysis suggested that HCV-exo carry miR-19a and target SOCS3 in HSC, which in turn activates the STAT3-mediated transforming growth factor β (TGF-β) signaling pathway and enhances fibrosis marker genes. The higher expression of miR-19a in exosomes was also observed from HCV-infected hepatocytes and in sera of chronic HCV patients with fibrosis compared to healthy volunteers and non-HCV-related liver disease patients with fibrosis. Together, our results demonstrated that miR-19a carried through the exosomes from HCV-infected hepatocytes activates HSC by modulating the SOCS-STAT3 axis. Our results implicated a novel mechanism of exosome-mediated intercellular communication in the activation of HSC for liver fibrosis in HCV infection.
IMPORTANCE HCV-associated liver fibrosis is a critical step for end-stage liver disease progression. However, the molecular mechanisms for hepatic stellate-cell activation by HCV-infected hepatocytes are underexplored. Here, we provide a role for miR-19a carried through the exosomes in intercellular communication between HCV-infected hepatocytes and HSC in fibrogenic activation. Furthermore, we demonstrate the role of exosomal miR-19a in activation of the STAT3–TGF-β pathway in HSC. This study contributes to the understanding of intercellular communication in the pathogenesis of liver disease during HCV infection.
INTRODUCTION
Hepatitis C virus (HCV) is a positive-sense, single-stranded RNA virus with a genome size of 9.6 kb that encodes several structural and nonstructural proteins. About 170 million people are infected with HCV worldwide. Chronic HCV infection often results in the development of fibrosis, cirrhosis, and hepatocellular carcinoma (HCC). HCV-associated liver fibrosis is a key pathological process leading to liver cirrhosis and HCC (1, 2), although the underlying mechanisms of disease progression remain unclear. Hepatic stellate cells (HSC) are involved in the pathogenesis of liver fibrosis (3). Liver injury is characterized by a phenotypic and functional transformation of normally quiescent HSC into alpha-smooth muscle actin (α-SMA)-expressing myofibroblastic cells, which promote wound closure and produce collagen matrix that supports hepatocyte repopulation. Due to their anatomical position in the space of Dissé, between the fenestrated endothelium of the sinusoids and the hepatocytes, HSC are optimally located to interact with many cell types in the liver (4, 5). However, the molecular mechanisms associated with cross talk between exosomes derived from HCV-infected hepatocytes (HCV-exo) and HSC during pathogenesis remain unknown.
Exosomes contain a characteristic composition of proteins and express cell recognition molecules on their surfaces that facilitate their selective targeting of and uptake by recipient cells. Recent reports indicate that exosomes also harbor a variety of nucleic acids, including double-stranded DNA, mRNAs, microRNAs (miRNAs), and long noncoding RNAs (lncRNAs), which can be transferred to recipient cells and modulate their functions (6). These findings have increased interest in the role of exosomes in cell-to-cell communication and support the idea that exosomes might constitute an exquisite mechanism for local and systemic intercellular transfer, not only of proteins, but also of genetic information in the form of RNA. It is known that exosomes isolated from HCV-infected patients' sera contain HCV RNA and elevated levels of the proteins Ago2 and HSP90 and of viral receptor-mediated transmission to hepatocytes (7). Liver cells stimulated by type I interferon (IFN) secrete exosomes, which contain antiviral molecules and can attenuate hepatitis B virus (HBV) replication (8). Exosomes isolated from liver endothelial cells stimulated by either type I or III IFNs can also suppress viral replication in HCV-infected liver cells (9). Recently, we have also shown the role of exosomes in HCV release from infected hepatocytes (10).
In the present study, we investigated the exosome-mediated communication between HCV-infected hepatocytes and HSC. We found that exosomes derived from HCV-infected hepatocytes have the potential to activate HSC, as confirmed by activation of profibrotic markers. We demonstrated that miR-19a is shuttled from HCV-infected hepatocytes to HSC via exosomes and targets SOCS3 to induce STAT3-mediated transforming growth factor β1 (TGF-β1) gene expression. Moreover, we observed higher expression of serum miR-19a in samples from HCV-infected patients with fibrosis than in healthy counterparts. Thus, our study reveals one of the molecular mechanisms of HSC activation during HCV-associated liver pathogenesis.
RESULTS
Exosomes from HCV-infected hepatocytes are internalized into HSC for induction of fibrogenic makers.
HCV RNA can be found in exosomes, which can transmit HCV infection (7, 10, 11). We evaluated whether HCV RNA carried by HCV-exo can replicate in LX2 cells. We observed an initial peak of HCV RNA at 2 days post-exosome incubation (~8-fold); however, HCV RNA levels were lower at later time points (Fig. 1A). We also incubated LX2 cells with exosomes isolated from spiroepoxide-treated HCV-infected hepatocytes. Total RNA was isolated from LX2 cells for detection of viral RNA. HCV RNA in LX2 cells remained undetectable, suggesting the exosome-mediated transfer of HCV RNA (data not shown). Our result is also in agreement with earlier observations suggesting hepatic stellate cells do not support HCV infection and replication (12).
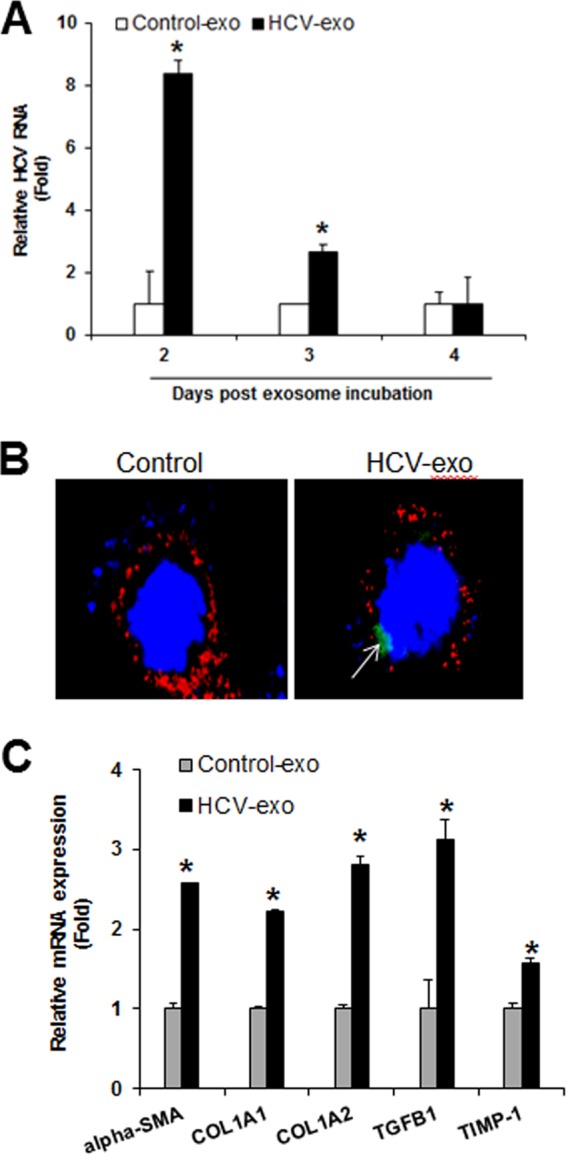
Exosomes derived from HCV-infected hepatocytes are internalized into human hepatic stellate (LX2) cells and induce upregulation of profibrogenic markers. (A) Exosomal uptake of HCV RNA by LX2 cells. LX2 cells were incubated with exosomes derived from control cells (Control-exo) or HCV-exo. RNA was extracted from control or HCV-exo-incubated LX2 cells at the indicated time points, and intracellular HCV RNA was analyzed by qRT-PCR. (B) Internalization of exosomes derived from HCV-infected hepatocytes into immortalized human hepatic stellate (LX2) cells was assessed by BODIPY labeling. Three hours after exosome incubation, LX2 cells were stained with Alexa Fluor 647-conjugated wheat germ agglutinin for 10 min at 37°C, washed, and fixed. The cell nuclei were stained with DAPI (blue) and observed under a confocal microscope. A representative image is shown. Exosomes are green (arrow), and the plasma membrane border stained with Alexa Fluor 647-conjugated wheat germ agglutinin is red. (C) Quantitative-PCR analyses for profibrogenic α-SMA, COL1A1 and COL1A2, TGF-β1, and TIMP-1 genes in control or HCV-exo-exposed LX2 cells. The values represent the results of three independent experiments and SD. Statistical significance was analyzed using the two-tailed Student t test. *, P < 0.05.
We next examined whether HCV-exo activate HSC. Exosome isolation and characterization were done (10) to examine whether HCV-exo is indeed internalized in LX2 cells. For this, we labeled HCV-infected hepatocytes with BODIPY and isolated exosomes. The BODIPY-labeled exosomes were incubated with LX2 cells and examined for exosomal uptake by immunofluorescence. Our results showed that HCV-exo were internalized in LX2 cells (Fig. 1B). To determine the effect of internalized exosomes on modulation of the HSC phenotype, we exposed LX2 cells to HCV-exo for 24 h and analyzed mRNA expression of profibrotic markers by quantitative reverse transcription (qRT) PCR. A significant increase of COL1A1, COL1A2, α-SMA, TGF-β1, and TIMP1 genes was noted compared to exosomes from mock-infected cells (Fig. 1C). Similar results were obtained with exosomes isolated with the HCV full-length genome containing replicons (data not shown).
Exosomes from HCV-infected hepatocytes induce the activation of primary human hepatic stellate cells.
Next, we extended our findings by examining primary human hepatic stellate cells. Primary HSC exposed to HCV-exo displayed significant upregulation of the profibrotic markers COL1A1/A2/3A1/4A1, TGF-β1, TIMP1, CTGF, matrix metalloproteinase 2 (MMP-2), and α-SMA (Fig. 2A and andB).B). Immunofluorescence analysis for α-SMA in exosome-treated cells showed a characteristic myofibroblast-like pattern of activated HSC and increased α-SMA expression (Fig. 2C). Further, an increase in α-SMA expression at the protein level in exosome-treated primary HSC was observed (Fig. 2D). The results suggest exosomes from HCV-infected hepatocytes promote primary human hepatic stellate cell activation similarly to immortalized LX-2 cells.
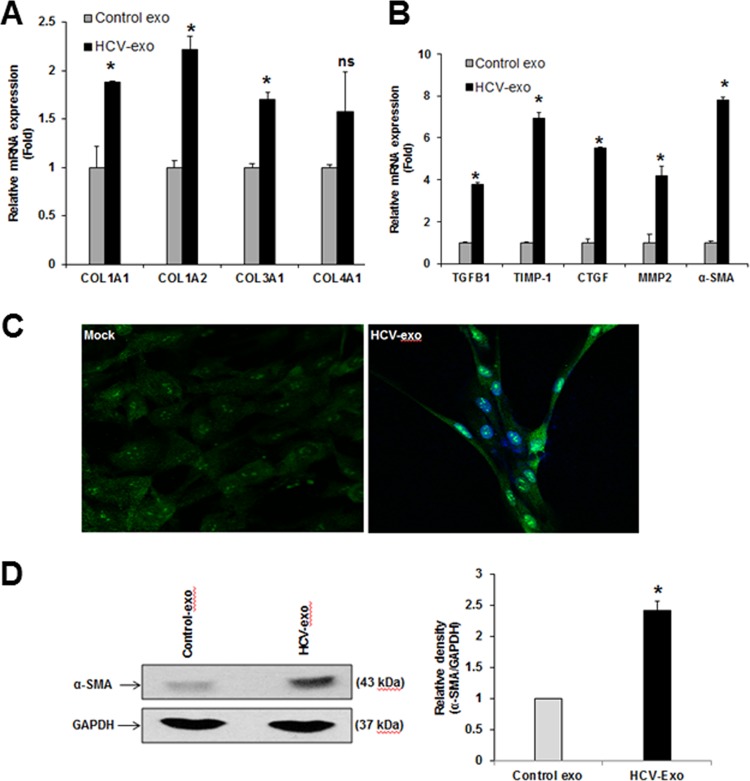
Exosomes derived from HCV-infected hepatocytes induce upregulation of profibrogenic markers in primary human hepatic stellate cells. (A and B) qRT-PCR analysis was performed for COL1A1 and COL1A2, COL3A1, COL4A1, TGF-β1, TIMP-1, CTGF, MMP-2, and α-SMA from RNA of primary HSC incubated with HCV-exo. (C) Immunofluorescence analysis of α-SMA protein (green) in control or HCV-exo-exposed primary HSC showing the phenotypic alteration to activated myofibroblast-like cells. Cell nuclei were stained with DAPI (blue). (D) Western blot analysis of α-SMA in primary HSC exposed to HCV-exo. (Right) Quantification of the α-SMA protein level was normalized with GAPDH. The values and SD represent the results of three independent experiments. Statistical significance was analyzed using the two-tailed Student t test. *, P < 0.05; ns, not significant.
MicroRNAs are present in exosomes derived from HCV-infected hepatocytes.
We next investigated the molecules that activate the HSC. MicroRNAs are carried through the exosomes and play a major role in cellular function. We examined the miRNA profile in HCV-exo. Clustering analysis revealed that exosomes from HCV-infected cells expressed distinct patterns of miRNAs compared with exosomes from the mock-treated cells (Fig. 3A). Selected miRNAs—miR-19a, miR-20a, miR-92, and miR-195 (Fig. 3A)—were validated by qRT-PCR and normalized with spiked-in Caenorhabditis elegans miR-39 (cel-miR-39). HCV-exo were significantly enriched in miR-19a, miR-20a, and miR-195 (Fig. 3B).
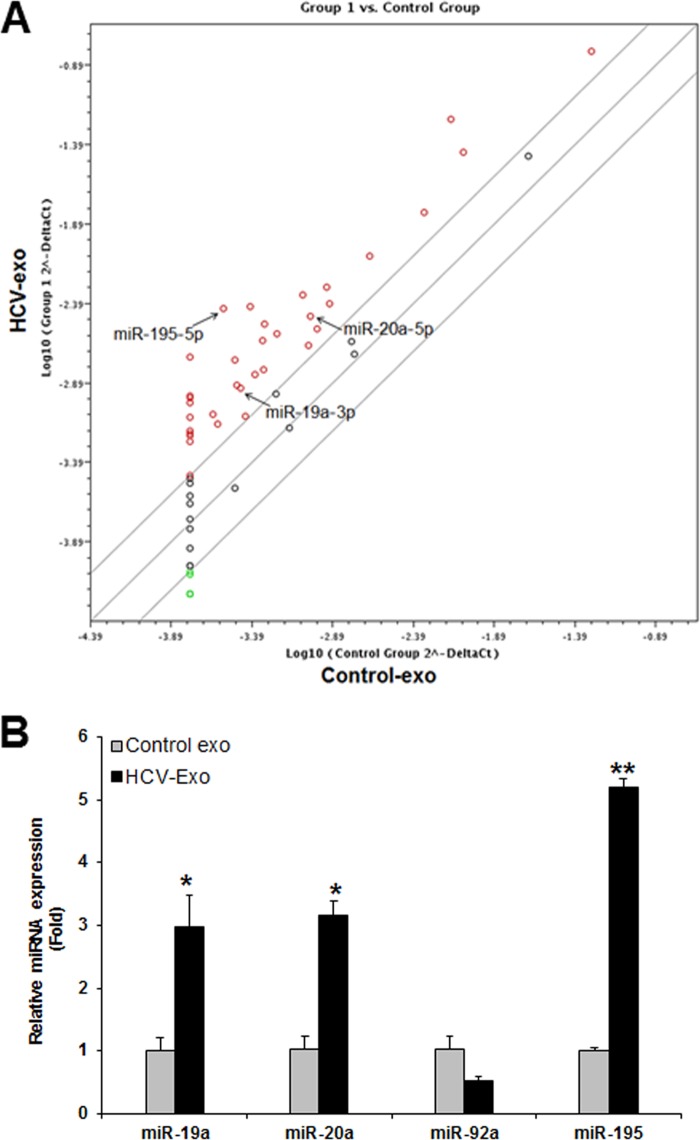
MicroRNA expression profiling of exosomes derived from HCV-infected hepatocytes. (A) miRNA expression profiling of exosomes isolated from control or HCV-infected hepatocytes using miScript miRNA PCR Array. The array data were analyzed using free Web-based software (http://pcrdataanalysis.sabiosciences.com/mirna/arrayanalysis.php). (B) Validation of differentially expressed miRNAs in HCV-exo by qRT-PCR. *, P < 0.05; **, P < 0.01. The error bars indicate SD.
We validated our in vitro findings in a small cohort of sera from healthy volunteers and non-HCV-infected and HCV-infected samples by determining the level of circulatory miR-19a. We observed that miR-19a expression was significantly upregulated in HCV-infected fibrotic patients compared to healthy volunteers and non-HCV-related liver disease patients with similar fibrosis grades (Fig. 4A). Higher expression of miR-19a in sera from late-fibrosis patients from cross-sectional patient samples was also observed (Fig. 4B). When we examined circulatory miRNA levels of miR-195 in sera of HCV-infected fibrotic patients, we did not observe a significant modulation in HCV fibrotic patients compared to healthy volunteers. We decided to characterize the function of miR-19a in HCV-mediated fibrosis.
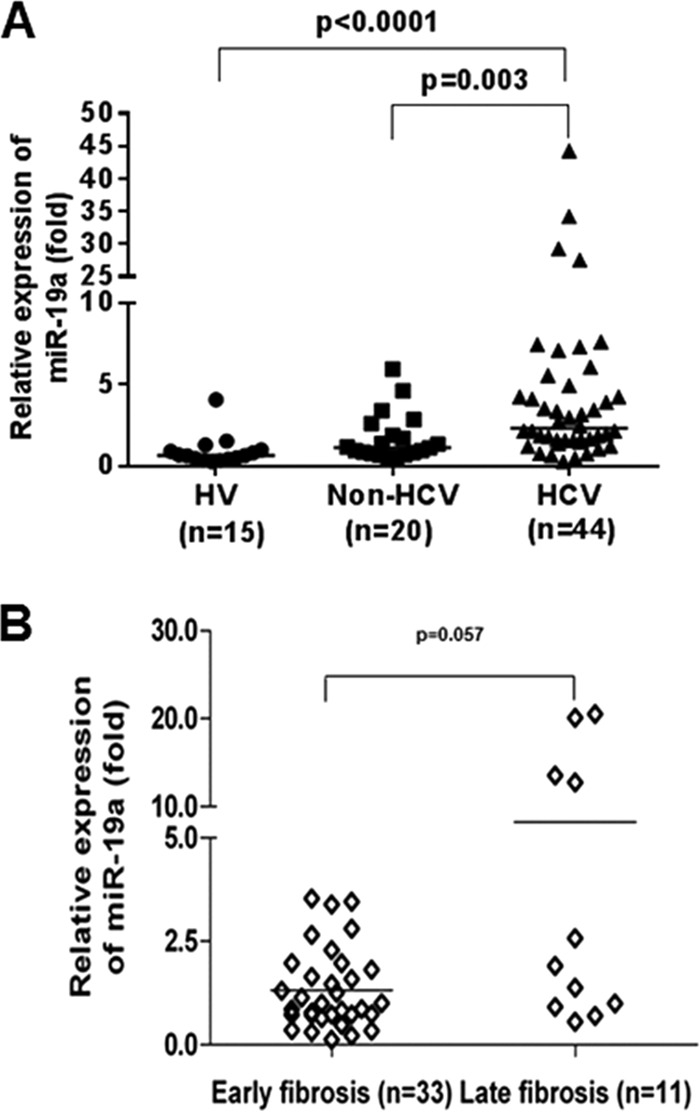
Upregulation of serum miR-19a levels in HCV-infected patients with liver fibrosis. (A) Scatter plots of serum miR-19a levels in healthy volunteers (HV) (n = 15), non-HCV-associated liver fibrosis patients (non-HCV) (n = 20), and HCV-infected fibrosis patients (HCV) (n = 44). (B) Increased expression of serum miR-19a levels in HCV-infected fibrosis patients from the early fibrosis stage (F0 to F2) (n = 33) to late-stage fibrosis (F3 and F4) (n = 11). The horizontal lines indicate the median values per group. Fold regulation is expressed as real-time quantitative (RQ) values based on the 2−ΔΔCT method. A Mann-Whitney U test (nonparametric) was used to determine statistical significance.
Exosomes from HCV-infected hepatocytes mediate the profibrogenic effect on hepatic stellate cells by shuttling miR-19a.
To gain insight into the exosome-mediated uptake of miR-19a in HSC, we incubated LX2 cells with control cells or HCV-exo at different time points and analyzed miR-19a expression by qRT-PCR. A significant upregulation of miR-19a in LX2 cells was observed within 2 to 3 h (Fig. 5A). Similarly, primary human hepatic stellate cells also showed internalization of exosomal miR-19a (Fig. 5B). To visualize the transfer of exosomal miR-19a into HSC, Cy3-labeled miR-19a or control miR (control) was transfected into Rep2a cells, and exosomes were isolated. When LX2 cells were incubated with labeled miR-19a-containing HCV-exo, red fluorescence of Cy3-miR-19a was detected in the cytoplasm of LX2 cells exposed to exosomes (Fig. 5C).
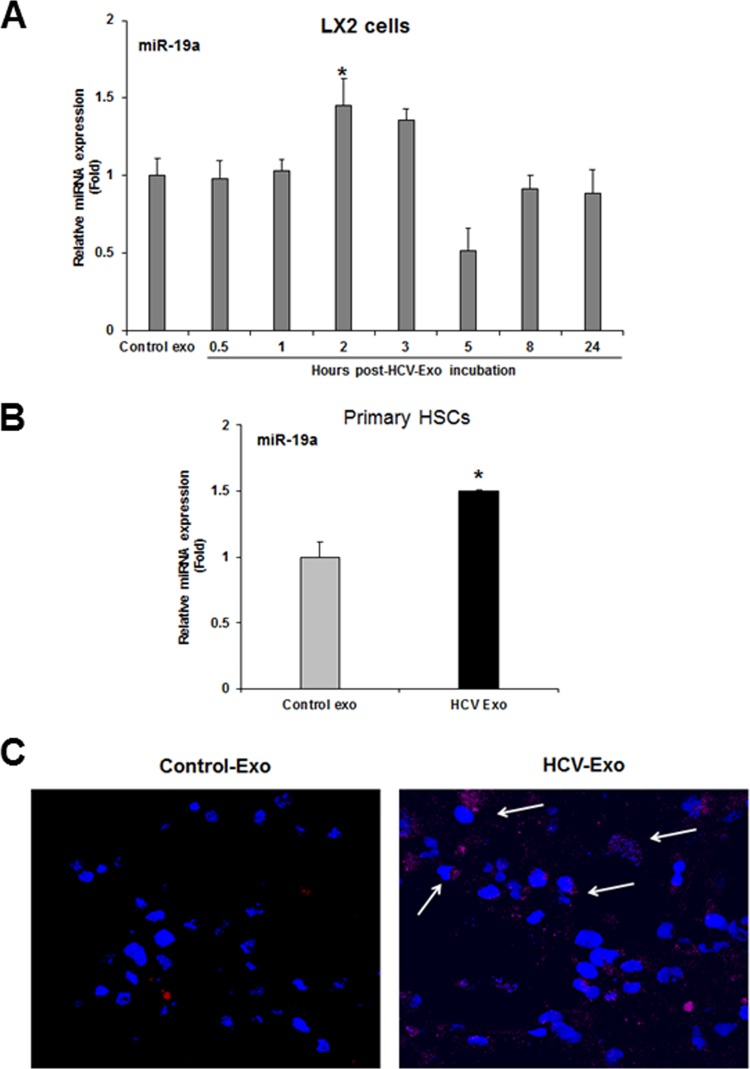
Uptake of exosomal miR-19a by human hepatic stellate (LX2) cells. (A) Time course analysis of exosomal miR-19a uptake in LX2 cells. LX2 cells were exposed to control or HCV-exo at the indicated time points and harvested for cellular RNA and qRT-PCR analysis of miR-19a. (B) Primary human HSC exposed to control or HCV-exo and processed to determine miR-19a expression as described for panel A. (C) Exosomes were isolated from HCV-infected cells transfected with Cy3-labeled control or Cy3-labeled miR-19a mimic and added to LX2 cells. Uptake of Cy3-labeled miR-19a was assessed by confocal microscopy. The arrows indicate cytoplasmic Cy3-labeled miR-19a (red) in LX2 cells. Nuclei were stained with DAPI (blue). *, P < 0.05. The error bars indicate SD.
To further verify the exosome-mediated miR-19a uptake in HSC, we depleted endogenous levels of miR-19a by transfecting miR-19a antagonist in LX2 cells and incubating for 30 h (Fig. 6A). We exposed the miR-19a-depleted LX2 cells to HCV-exo 30 h post-antagonist transfection and observed upregulation of miR-19a similar to that of exosome-exposed cells (Fig. 6B and andC).C). Together, our data suggested exosome-mediated shuttling of miR-19a into HSC. Given the key role of Rab27 in exosome release (13), we transfected Rep2a cells with Rab27a small interfering RNA (siRNA), and exosomes were isolated. HCV-infected cells were also treated with 10 or 20 μM spiroepoxide (exosome release inhibitor). Exosomes were isolated and incubated with LX2 cells for 3 h. As expected, HSC exposed to exosomes isolated from spiroepoxide-treated cells showed concentration-dependent inhibition of miR-19a uptake (Fig. 6D). Similarly, Rab27a knockdown cells showed no sign of miR-19a uptake (Fig. 6D). These results further confirmed the exosome-mediated uptake of miR-19a by HSC.
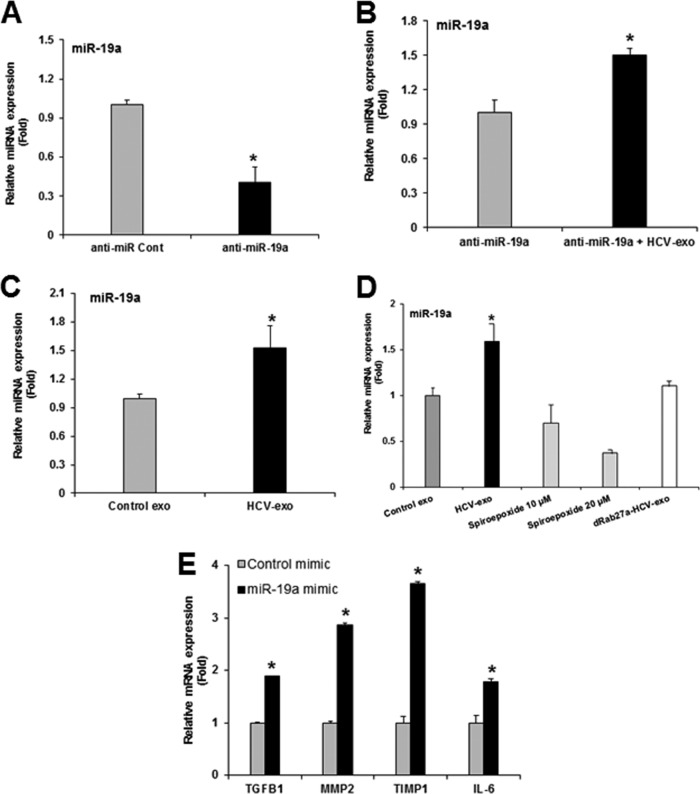
Inhibition of exosome release or exosome biogenesis affects miR-19a delivery in LX2 cells. (A) Depletion of endogenous miR-19a after antagonist transfection was confirmed by miR-19a-specific qRT-PCR. (B) The endogenous miR-19a level was depleted by transfecting LX2 cells with anti-miR-19a, and the cells were left untreated or exposed to HCV-exo. Expression of miR-19a was measured by qRT-PCR and normalized with endogenous U6. (C) LX2 cells were exposed to control or HCV-exo isolated from HCV-infected cells. The cells showed upregulation of the miR-19a level by qRT-PCR. (D) Exosomes isolated from HCV-infected cells with or without treatment with 10 and 20 μM spiroepoxide (exosome release inhibitor) and transfected with Rab27a siRNA. LX2 cells were exposed to the respective exosome preparations for 3 h, and miR-19a was analyzed by qRT-PCR. (E) Hepatic stellate (LX2) cells were transfected with miR-19a-3p mimic or control miR (25 nM each), and RNA was isolated. qRT-PCR analysis was performed using control and miR-19a-transfected cellular RNA with gene-specific probes and normalized with endogenous marker 18S RNA. The values represent the results of three independent experiments and SD. Statistical significance was analyzed using the two-tailed Student t test: *, P < 0.05.
In order to evaluate the profibrogenic role of miR-19a-3p encapsulated in HCV-exo, we transfected LX2 cells with control or miR-19a-3p mimic and analyzed the expression of profibrotic genes. LX2 cells transfected with miR-19a-3p mimic showed significant upregulation of TGF-β1, MMP-2, TIMP1, and interleukin 6 (IL-6) genes (Fig. 6E).
miR-19a targets the SOCS3/STAT3 axis and plays a profibrogenic role in hepatic stellate cells.
We next investigated the mechanism of miR-19a-mediated induction of fibrosis markers. In silico analyses using databases (TargetScan and miRanda) suggested miR-19a targets the 3′ untranslated region (UTR) of SOCS3 (Fig. 7A). A recent report demonstrated that miR-19a targets SOCS3 and modulates JAK-STAT signaling in juvenile idiopathic arthritis (14). We first determined SOCS3 expression from primary HSC exposed to HCV-exo. A significant reduction of protein was observed compared to control exo-treated cells (Fig. 7B and andC).C). To further evaluate the miR-19a-mediated downregulation of SOCS3, we transfected LX2 cells or primary HSC with miR-19a-3p mimic or control-miR as a negative control. We observed a significant reduction of SOCS3 protein compared to the cells transfected with the negative-control mimic (Fig. 7D and andEE).
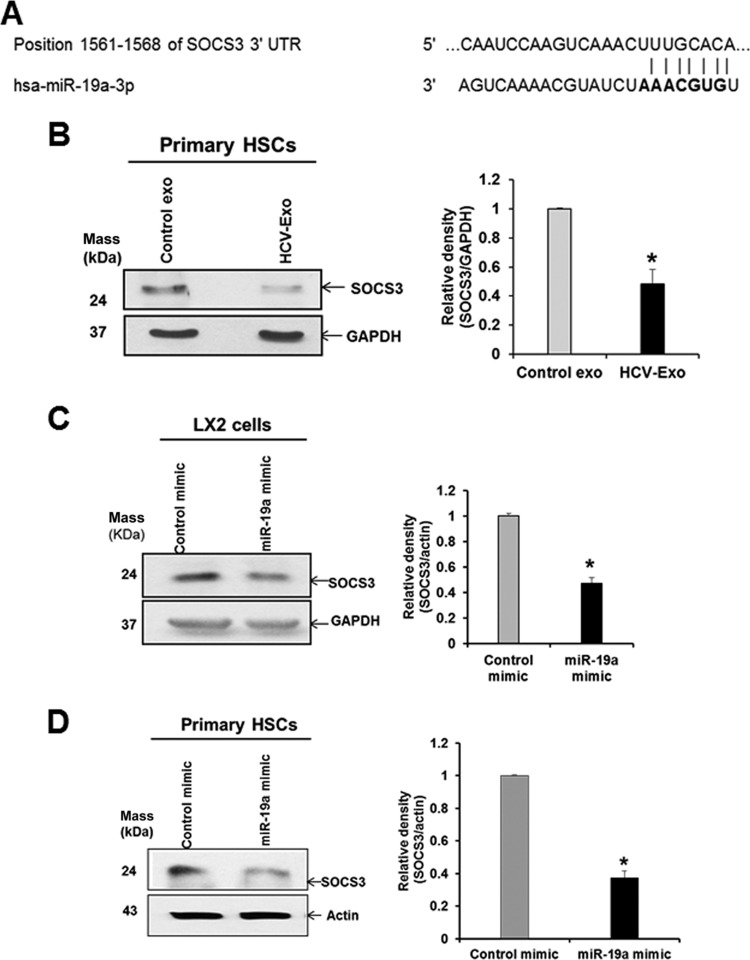
Verification of SOCS3 as a target of miR-19a. (A) Potential binding site for miR-19a on the 3′ UTR of SOCS3 mRNA. (B) Expression of SOCS3 protein was reduced in primary human HSC when exposed to HCV-exo compared with control exo-treated cells. (C and D) LX2 cells or primary human HSC were transfected with control or miR-19a mimic (25 nM), and SOCS3 expression was analyzed by Western blotting using specific antibodies. (B, C, and D, right) Quantification of the SOCS3 protein level was normalized with GAPDH or actin from three experiments, and the results of densitometric scanning are presented. *, P < 0.05. The error bars indicate SD.
The JAK-STAT pathway is under tight regulation by the induction of SOCS proteins. SOCS proteins silence the pathway by acting as pseudosubstrates that block JAK kinase ability, binding to the receptor to prevent STAT interaction and targeting proteins for proteasomal degradation (15). Since HCV-exo and miR-19a reduced SOCS3 in HSC, we examined downstream signaling for activation of pSTAT3. An enhanced pSTAT3 level in primary HSC exposed to HCV-exo was observed compared to control cells (Fig. 8A). Similar expression was observed in LX2 cells exposed to HCV-exo (data not shown). STAT3 regulates the expression of cyclin D1 (16), which is required for hepatocyte and HSC proliferation (17, 18). We examined the expression of cyclin D1 in HSC exposed to HCV-exo. Our results demonstrated that cyclin D1 was increased in HCV-exo-exposed cells compared to control exo-treated LX2 cells (Fig. 8B).
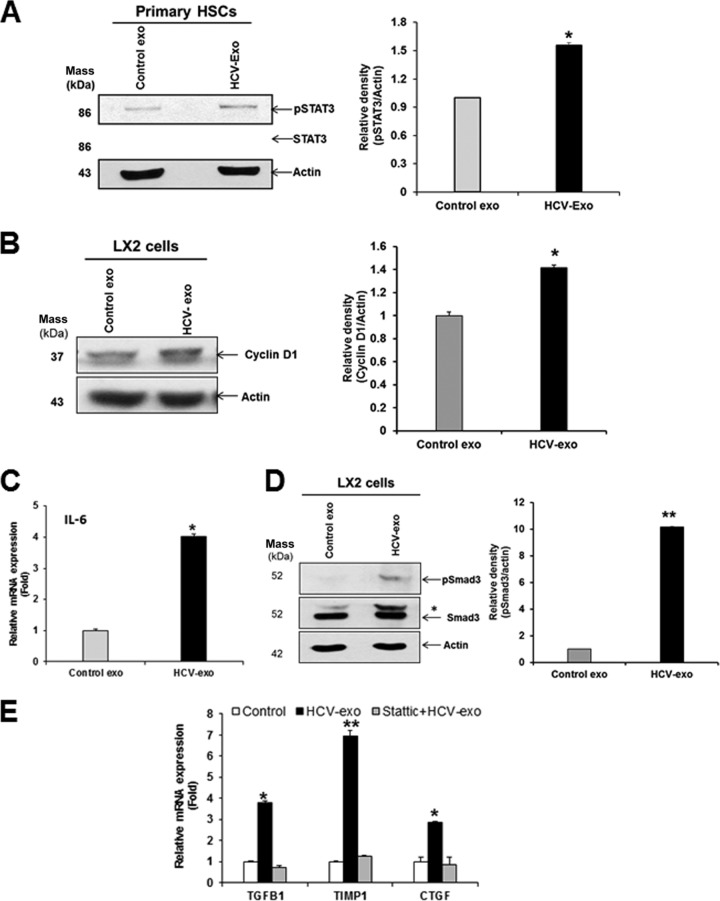
Exosomes from HCV-infected hepatocytes mediate the profibrogenic effect on hepatic stellate cells by activating the STAT3/TGF-β signaling axis. (A) Increased phosphorylation of STAT3 (pSTAT3) in primary human HSC exposed to HCV-exo compared to control exo-treated cells. (B) Cyclin D1 expression was increased in HCV-exo-exposed cells compared to control exo-treated cells. (C) Activation of STAT3 led to a concomitant increase in IL-6 mRNA expression levels in HCV-exo-exposed primary HSC. (D) Increased phosphorylation of Smad3 (pSmad3-Ser423/425) in HSC exposed to HCV-exo compared to control exo-treated cells. *, pSmad3 band detected with total Smad3 antibody. (E) LX2 cells were left untreated or treated with 5 μM Stattic, followed by exposure to HCV-exo for 24 h. TGFβ1, CTGF, and TIMP1 gene expression was analyzed by qRT-PCR as described for Fig. 6. *, P < 0.05; **, P < 0.01. The error bars indicate SD.
Next, we determined the expression of STAT3 target genes, IL-6 and TGF-β1 genes, in HSC exposed to HCV-exo. Activation of STAT3 led to a concomitant increase in IL-6 mRNA expression in HCV-exo-exposed primary HSC (Fig. 8C). We observed increased TGF-β1 gene expression in LX2 cells and primary HSC exposed to HCV-exo (Fig. 1B and and2B).2B). To functionally evaluate the TGF-β signaling in HSC exposed to HCV-exo, we determined phosphorylation-mediated activation of Smad3. HSC exposed to HCV-exo showed higher expression of pSmad3 than mock-treated cells (Fig. 8D). To determine the role of HCV-exo-mediated STAT3 signaling and induction of TGF-β1, LX2 cells were treated with the chemical inhibitor Stattic, which was shown to selectively inhibit activation of STAT3 by blocking tyrosine phosphorylation and dimerization (19). Stattic treatment attenuated HCV-exo-induced expression of TGF-β1, CTGF, and TIMP1 genes (Fig. 8E). Together, these data indicated that exosomes mediate enhancement in STAT3-mediated TGF-β signaling in HSC.
DISCUSSION
HCV infection is closely associated with liver fibrosis, cirrhosis, and HCC. However, the mechanism by which HCV induces liver fibrosis remains poorly understood. In this study, we demonstrated that (i) exosomes released from HCV-infected hepatocytes are efficiently internalized in HSC, (ii) exosomes internalized into HSC induce a phenotypic switch from quiescent to activated HSC, (iii) exosomes shuttle miRNAs into target cells, (iv) exosomes from HCV-infected cells carry miR-19a, and (v) miR-19a targets SOCS3 to activate the STAT3-mediated TGF-β signaling mechanism.
Exosomes are considered important mediators of cell-cell communication by delivering the information they carry to neighboring cells and through their role in liver pathology (20). In the liver, some exosomes that include miRNAs take part in the pathogenesis of liver fibrosis by modulating the epigenetic regulation of HSC (21). Exosome-mediated intercellular transfer of connective tissue growth factor (CCN2) by HSC is also implicated in fine-tuning of liver fibrosis (22). Conversely, exosomes derived from human umbilical cord mesenchymal cells attenuate liver fibrosis in mice by suppressing the Smad signaling pathway (23).
Activation of HSC during chronic liver injury triggers fibrogenic pathways in the cells, a process that results in increased expression of profibrogenic markers, like collagens, matrix metalloproteinases, and tissue inhibitor of MMPs. Upon hepatic injury HSC transdifferentiate into myofibroblast-like cells marked by expression of smooth muscle α-actin and increased proliferation and contractility (4, 5). Myofibroblastic HSC respond to and secrete a variety of profibrogenic cytokines, including connective tissue growth factor, tissue inhibitor of metalloproteinases, and TGF-β.
Our results demonstrated that LX2 cells or primary human hepatic stellate cells after exposure to HCV-exo showed a myofibroblast like appearance with increased expression of α-SMA and enhanced expression of profibrogenic markers. We have shown uptake of HCV RNA by HSC through exosomes. However, HCV RNA replication was not supported in HSC and does not activate HSC. Exosomal miRNA profiling suggested modulation of several miRNAs in exosomes isolated from HCV-infected hepatocytes. We observed that miR-19a was highly upregulated in chronic HCV patient sera with fibrosis compared to fibrosis specimens from other etiologies. We therefore focused our study on miR-19a, which was highly expressed in HCV-exosomes. Members of the miR-17-92 cluster have been implicated in the progression of liver fibrosis, as well as in cirrhosis and HCC (24). We have reported increased levels of miR-17-92 cluster members miR-20a and miR-92a during acute and chronic HCV infections, and significant upregulation of these miRNAs was observed in the sera of HCV-infected patients with liver fibrosis (25). By using a gain-of-function approach, we identified miR-19a as one of the key mediators of HSC activation. We observed that exosome-mediated shuttling of miR-19a in HSC downregulates SOCS3 expression. Since SOCS3 is an effective regulator of STAT3 signaling (15), our observation of subsequent STAT3 activation in exosome-exposed HSC implied the importance of this pathway in HSC activation.
STAT3 activation has been detected in several human diseases, and STAT3 signaling is involved in liver injury, steatosis, inflammation, regeneration, fibrosis, and hepatocarcinogenesis (26, 27). We have shown previously that HCV infection enhances STAT3 expression (28, 29). The activation of STAT3 in HSC by IL-6 or leptin stimulates HSC survival, proliferation, and activation (26). In chronic hepatitis B patients with advanced liver fibrosis, pSTAT3 expression in HSC was demonstrated and correlated with the fibrotic score and plasma IL-6 levels (30). The study also indicated that sorafenib and its derivative SC-1 could ameliorate liver fibrosis through STAT3 inhibition in HSC.
During liver fibrosis, TGF-β1 induces trans differentiation of quiescent HSC into myofibroblasts, which in turn produce other cytokines and matrix proteins (31). We observed higher TGF-β1 expression in primary human hepatic stellate cells exposed to HCV-exo than in mock-treated cells. STAT3-dependent TGF-β1 expression has been reported to play an important role in the activation and antiapoptotic effect of HSC in liver tissues of chronic hepatitis B patients and diethylinitrosamine-induced liver fibrosis in a rat model (32). Further, STAT3 is involved in modulating CTGF production upon TGF-β treatment in activated HSC (33). We also observed higher expression of CTGF (~5.5-fold) in primary human hepatic stellate cells exposed to HCV-exo, suggesting a role of the Stat3/TGF-β1 axis in activation of HSC. In addition, expression of TIMP-1, which plays an important role in promoting liver fibrosis (34, 35) and was suggested to be regulated by STAT3 (26), was upregulated. This is further supported by our STAT3 inhibition experiment, which led to reduced TGF-β1, CTGF, and TIMP1 gene expression in HCV-exo-exposed cells in the presence of inhibitor. Smad3 is an essential mediator of TGF-β signaling (36) and triggers phosphorylation of Smad2/3, which forms a complex with Smad4 and shuttles into the nucleus to regulate gene transcription (37). We also observed increased phosphorylation of Smad3 in HSC.
Taken together, our study demonstrated that exosomes from HCV-infected hepatocytes are internalized in hepatic stellate cells and changed to the activated state by inducing the expression of profibrotic markers. Furthermore, we have shown the exosomal miR-19a-mediated mechanism of HSC activation through depletion of SOCS3 and subsequent activation of the STAT3/TGF-β1/Smad3 signaling pathway. These data provide novel and advanced information for a better understanding of molecular and cellular mechanisms of HSC activation through the cross talk with HCV-infected hepatocytes.
MATERIALS AND METHODS
Cell culture, HCV infection, and transfection.
Immortalized human hepatocytes (IHH) and LX2 cells (immortalized human hepatic stellate cells) were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), 100 U/ml penicillin G, and 100 μg/ml streptomycin. Huh7.5 cells harboring an HCV genotype 2a genome length replicon with a Renilla luciferase reporter gene (Rep2a-Rluc; kindly provided by Hengli Tang) were cultured in DMEM containing 10% FBS with 1% antibiotics. Primary human hepatic stellate cells were procured from ScienCell and maintained following the manufacturer's instructions. For infection, Huh7.5 or IHH (3 × 105 cells) were incubated with HCV genotype 2a (clone JFH1) in a minimum volume of medium. After 8 h of virus adsorption on the hepatocytes, Dulbecco's modified Eagle's medium supplemented with 5% heat-inactivated fetal bovine serum was added as described previously (10). All the cells were maintained at 37°C in a humidified 5% CO2 atmosphere.
Transfection of miR-19a-3p miRNA mimic or anti-miR-19a-3p (Life Technologies) into HSC was performed using Interferin transfection reagent (Polyplus). Briefly, LX2 cells were plated at a density of 0.5 × 105 to 4 × 105 cells/well of a 12-well plate and transfected with miR-19a mimic or inhibitor. After 5 h of transfection, the medium was replaced with the standard cultured medium supplemented with 2% FBS. For inhibition of STAT3 signaling, primary HSC were treated with the selective STAT3 inhibitor Stattic (Santa Cruz) at 5 μM concentration with or without exosome treatment. Cells were harvested for RNA extraction and qRT-PCR after 24 h.
Exosome isolation.
Exosomes from cell culture supernatants were isolated and characterized as described previously (10). Briefly, virus-infected cells were washed twice with PBS and then supplemented with DMEM containing 2% exosome-depleted serum (System Biosciences) and incubated for 3 days. To inhibit exosome release, the cells were treated with N-SMase spiroepoxide inhibitor (Santa Cruz) as described previously (38). Culture supernatants were collected and processed for exosome isolation by the ExoQuick method (System Biosciences).
BODIPY labeling of exosomes and uptake.
BODIPY493/503 solution (1 mM; Molecular Probes) was added in the culture medium of Rep2a cells, and the cells were cultured for 1 h at 37°C in a CO2 incubator. The cells were washed three times with PBS, and fresh medium containing exosome-depleted FBS was added to the cells. After 24 h of incubation, the medium was collected and processed for exosome isolation. LX2 cells were seeded in 4-chambered slides (Nunc). LX2 cells were washed with serum-free medium, and the BODIPY-labeled exosomes were added to the LX2 cells for 3 h. The cells were washed with Hanks' balanced salt solution and labeled with Alexa Fluor 647-conjugated wheat germ agglutinin (Life Technologies) to stain the plasma membranes of the cells. The cells were fixed, stained with DAPI (4′,6-diamidino-2-phenylindole), and observed under confocal microscopy (Olympus FV1000).
RNA isolation and qRT-PCR.
Total cellular or exosomal RNA was isolated using TRIzol reagent (Invitrogen, CA). cDNA was synthesized by using the respective miRNA-specific or U6-specific primers with a TaqMan microRNA reverse transcription kit or random hexamers with Superscript III reverse transcriptase for gene expression studies. Real-time PCR was performed for quantitation by using TaqMan universal PCR master mix and 6-carboxyfluorescein (FAM)-minor groove binder (MGB) probes for miRNAs or for gene expression assays. U6 (assay number 001973) or 18S rRNA (assay number Hs03928985_g1) was used as an endogenous control for intracellular microRNA or gene expression, respectively, while synthetic spiked-in C. elegans miR-39 added to the exosomes prior to RNA extraction was used as an endogenous control for exosomal miRNA expression. The relative expression levels were normalized to the endogenous control by using the 2−ΔΔCT formula (ΔΔCT = ΔCT of the sample − ΔCT of the untreated control). RNA isolation from serum samples, miRNA profiling by miRNA Array, and exosomal HCV RNA determination were carried out as described previously (10, 25). For microRNA arrays, we isolated exosomes from cell supernatants from mock- and HCV-infected hepatocytes and performed exosomal miRNA profiling using miScript miRNA PCR Array and Qiagen miRNA Array Web-based software.
Western blotting and immunofluorescence.
Cell lysates were subjected to polyacrylamide gel electrophoresis and transferred onto a nitrocellulose membrane. The membranes were blocked with 5% nonfat dry milk and incubated with specific antibodies. The membrane was probed with antibodies to SOCS3, cyclin D1, phosphorylated STAT3 (pSTAT3), STAT3, pSmad3, Smad3 (Cell Signaling), and α-SMA (Santa Cruz). Proteins were detected by using enhanced chemiluminescence (ECL). The membrane was reprobed with actin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal control. The densitometric scanning of Western blots was performed with ImageJ software (NIH). For immunofluorescence, primary HSC were seeded in 4-well chamber slides and exposed to HCV-exo for 2 days. The cells were washed in phosphate-buffered saline (PBS), fixed with 3.7% formaldehyde for 20 min at room temperature, and blocked with 3% bovine serum albumin (BSA) for 1 h. The fixed cells were permeabilized with 0.2% Triton X-100 for 5 min at room temperature. The cells were incubated with α-SMA-specific mouse monoclonal antibody (Santa Cruz Biotechnology) overnight at 4°C, washed, and incubated with anti-mouse Ig conjugated with Alexa 488 antibody (Molecular Probes) for 1 h at room temperature. Finally, the cells were washed, and the nuclei were stained with DAPI and mounted for confocal microscopy (Olympus FV1000).
Shuttling assay for Cy3-labeled miRNA.
Control miR or miR-19a mimic (Invitrogen) was labeled with a Label It siRNA Tracker Cy3 kit that covalently attaches the Cy3 marker to siRNA (Mirus, Madison, WI, USA), following the manufacturer's instruction. This allowed tracking of fluorescence-labeled miR-19a. Huh 7.5 (Rep2a) cells harboring the HCV full-length genome were transfected with 10 nM control or Cy3-labeled miR-19a using Interferin transfection reagent (Polyplus). The cells were maintained for 24 h in medium containing exosome-depleted FBS. Exosomes were isolated and incubated on LX2 cells or primary human hepatic stellate cells for 4 h. The cells were washed three times with 1× PBS before fixation and processed for confocal imaging as discussed above.
Statistical analysis.
Results are presented as means and standard deviations (SD). Data were analyzed with Student's t test. A P value of < 0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We thank Scott Friedman for LX2 cells and Hengli Tang for Rep2a cells.
This work was supported by research grant R01 DK081817 from the National Institutes of Health.
REFERENCES
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.02225-16
Read article for free, from open access legal sources, via Unpaywall:
https://jvi.asm.org/content/jvi/91/6/e02225-16.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jvi.02225-16
Article citations
Novel insights into the role of immunomodulatory extracellular vesicles in the pathogenesis of liver fibrosis.
Biomark Res, 12(1):119, 12 Oct 2024
Cited by: 0 articles | PMID: 39396032 | PMCID: PMC11470730
Review Free full text in Europe PMC
Exosomes as Conduits: Facilitating Hepatitis B Virus-Independent Hepatitis D Virus Transmission and Propagation in Hepatocytes.
Viruses, 16(6):825, 22 May 2024
Cited by: 0 articles | PMID: 38932118 | PMCID: PMC11209184
Distinct serum exosomal miRNA profiles detected in acute and asymptomatic dengue infections: A community-based study in Baiyun District, Guangzhou.
Heliyon, 10(10):e31546, 18 May 2024
Cited by: 0 articles | PMID: 38807894 | PMCID: PMC11130723
Suppressor of cytokine signaling-3 expression and its regulation in relation to inflammation in Chronic Obstructive Pulmonary Disease.
Front Immunol, 15:1320077, 12 Mar 2024
Cited by: 0 articles | PMID: 38533493 | PMCID: PMC10963451
Extracellular Vesicles as Delivery Vehicles for Non-Coding RNAs: Potential Biomarkers for Chronic Liver Diseases.
Biomolecules, 14(3):277, 26 Feb 2024
Cited by: 4 articles | PMID: 38540698 | PMCID: PMC10967855
Review Free full text in Europe PMC
Go to all (111) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
New Mechanism of Hepatic Fibrogenesis: Hepatitis C Virus Infection Induces Transforming Growth Factor β1 Production through Glucose-Regulated Protein 94.
J Virol, 90(6):3044-3055, 30 Dec 2015
Cited by: 13 articles | PMID: 26719248 | PMCID: PMC4810663
Suppression of fibrogenic signaling in hepatic stellate cells by Twist1-dependent microRNA-214 expression: Role of exosomes in horizontal transfer of Twist1.
Am J Physiol Gastrointest Liver Physiol, 309(6):G491-9, 30 Jul 2015
Cited by: 87 articles | PMID: 26229009 | PMCID: PMC4572411
Exosomal Transmission of MicroRNA from HCV Replicating Cells Stimulates Transdifferentiation in Hepatic Stellate Cells.
Mol Ther Nucleic Acids, 14:483-497, 18 Jan 2019
Cited by: 48 articles | PMID: 30753992 | PMCID: PMC6369229
Mechanisms Underlying Hepatitis C Virus-Associated Hepatic Fibrosis.
Cells, 8(10):E1249, 14 Oct 2019
Cited by: 72 articles | PMID: 31615075 | PMCID: PMC6829586
Review Free full text in Europe PMC
Funding
Funders who supported this work.
HHS | National Institutes of Health (1)
Grant ID: DK081817
NIDDK NIH HHS (1)
Grant ID: R01 DK081817
 a,b,c
a,b,c




