Abstract
Background
Genome-wide association studies have so far identified 56 loci associated with risk of coronary artery disease (CAD). Many CAD loci show pleiotropy; that is, they are also associated with other diseases or traits.Objectives
This study sought to systematically test if genetic variants identified for non-CAD diseases/traits also associate with CAD and to undertake a comprehensive analysis of the extent of pleiotropy of all CAD loci.Methods
In discovery analyses involving 42,335 CAD cases and 78,240 control subjects we tested the association of 29,383 common (minor allele frequency >5%) single nucleotide polymorphisms available on the exome array, which included a substantial proportion of known or suspected single nucleotide polymorphisms associated with common diseases or traits as of 2011. Suggestive association signals were replicated in an additional 30,533 cases and 42,530 control subjects. To evaluate pleiotropy, we tested CAD loci for association with cardiovascular risk factors (lipid traits, blood pressure phenotypes, body mass index, diabetes, and smoking behavior), as well as with other diseases/traits through interrogation of currently available genome-wide association study catalogs.Results
We identified 6 new loci associated with CAD at genome-wide significance: on 2q37 (KCNJ13-GIGYF2), 6p21 (C2), 11p15 (MRVI1-CTR9), 12q13 (LRP1), 12q24 (SCARB1), and 16q13 (CETP). Risk allele frequencies ranged from 0.15 to 0.86, and odds ratio per copy of the risk allele ranged from 1.04 to 1.09. Of 62 new and known CAD loci, 24 (38.7%) showed statistical association with a traditional cardiovascular risk factor, with some showing multiple associations, and 29 (47%) showed associations at p < 1 × 10-4 with a range of other diseases/traits.Conclusions
We identified 6 loci associated with CAD at genome-wide significance. Several CAD loci show substantial pleiotropy, which may help us understand the mechanisms by which these loci affect CAD risk.Free full text
Systematic Evaluation of Pleiotropy Identifies 6 Further Loci Associated With Coronary Artery Disease
Associated Data
Abstract
Background
Genome-wide association studies have so far identified 56 loci associated with risk of coronary artery disease (CAD). Many CAD loci show pleiotropy; that is, they are also associated with other diseases or traits.
Objectives
This study sought to systematically test if genetic variants identified for non-CAD diseases/traits also associate with CAD and to undertake a comprehensive analysis of the extent of pleiotropy of all CAD loci.
Methods
In discovery analyses involving 42,335 CAD cases and 78,240 control subjects we tested the association of 29,383 common (minor allele frequency >5%) single nucleotide polymorphisms available on the exome array, which included a substantial proportion of known or suspected single nucleotide polymorphisms associated with common diseases or traits as of 2011. Suggestive association signals were replicated in an additional 30,533 cases and 42,530 control subjects. To evaluate pleiotropy, we tested CAD loci for association with cardiovascular risk factors (lipid traits, blood pressure phenotypes, body mass index, diabetes, and smoking behavior), as well as with other diseases/traits through interrogation of currently available genome-wide association study catalogs.
Results
We identified 6 new loci associated with CAD at genome-wide significance: on 2q37 (KCNJ13-GIGYF2), 6p21 (C2), 11p15 (MRVI1-CTR9), 12q13 (LRP1), 12q24 (SCARB1), and 16q13 (CETP). Risk allele frequencies ranged from 0.15 to 0.86, and odds ratio per copy of the risk allele ranged from 1.04 to 1.09. Of 62 new and known CAD loci, 24 (38.7%) showed statistical association with a traditional cardiovascular risk factor, with some showing multiple associations, and 29 (47%) showed associations at p < 1 × 10−4 with a range of other diseases/traits.
Conclusions
We identified 6 loci associated with CAD at genome-wide significance. Several CAD loci show substantial pleiotropy, which may help us understand the mechanisms by which these loci affect CAD risk.
Central Illustration
Over the past decade, genome-wide association studies (GWAS) have identified several thousand robust associations (p < 5 × 10−8) for a range of human traits and diseases. For coronary artery disease (CAD), 56 such loci have been identified so far, explaining ~15% of the disease’s heritability 1, 2. Approximately one-third of the CAD loci also show association with a known or putative cardiovascular risk factor, particularly blood pressure and lipid traits (2). Furthermore, several loci show association with other diseases; for example, the CAD-associated variants in the chromosome 9p21 locus also associate with risk of stroke as well as abdominal, aortic, and intracranial aneurysms 3, 4. These observations suggest that a comprehensive analysis of variants associated with other diseases and traits might not only identify additional loci associated with risk of CAD, but also provide important insights into genetic mechanisms shared by different diseases.
Here, we leveraged the HumanExome BeadChip (Illumina, Inc., San Diego, California) to test the contribution of 29,393 common variants of single nucleotide polymorphisms (SNPs) (minor-allele frequency >5%) for association with CAD. The variants included the majority of reported trait-/disease-associated lead SNPs in the National Human Genome Research Institute GWAS catalogue as of August 2011, as well as a number of associations for complex diseases unpublished at that time, variants in the human leukocyte antigen (HLA) region, and a scaffold of approximately 5,000 SNPs placed on the array for identity by descent testing. The results of an analysis of rare (minor-allele frequency <5%) coding sequence (“exome”) variants on this array with CAD were recently reported (5).
We identified 6 new loci associated at genome-wide significance with CAD, annotated these, and undertook a detailed examination of the extent of pleiotropy of these loci as well the previously known CAD loci.
Methods
The study consisted of discovery and replication phases and has been described in more detail elsewhere (5). Briefly, the discovery cohort included 42,335 cases and 78,240 control subjects from 20 individual studies (Online Table 1); the replication cohort, which was separately assembled and ascertained to have no sample overlap with the discovery cohorts, included 30,533 cases and 42,530 control subjects from 8 studies (Online Table 2). With the exception of participants from 2 studies in the replication cohort who were of South Asian ancestry, all participants were of European ancestry (Online Table 2).
Samples were genotyped on the Illumina HumanExome BeadChip versions 1.0 or 1.1, or the Illumina OmniExome (which includes markers from the HumanExome BeadChip) arrays followed by quality control procedures as previously described (5).
Statistical analysis
In discovery samples that passed quality control procedures, we performed individual tests for association of the selected variants with CAD in each study separately, using logistic regression analysis with principal components of ancestry as covariates (5). We combined evidence across individual studies using an inverse-variance weighted fixed-effects meta-analysis. Heterogeneity was assessed by Cochran’s Q statistic (6). In the discovery phase, we defined suggestive novel association as a meta-analysis p value ≤1 × 10−6.
For variants with suggestive association, we performed association analysis in the replication studies (Online Appendix). We defined significant novel associations as those nominally significant (p < 0.05) in the replication study and with an overall (discovery and replication combined) p value <5 × 10−8.
Bioinformatics analysis
To identify any association between the novel loci and gene expression traits, we performed a systematic search of cis-expression quantitative trait loci (eQTL) (described in the Online Appendix). To identify candidate causal SNPs at the new loci, we annotated each of the lead variants as well as SNPs in high linkage disequilibrium (LD) (r2 > 0.8) on the basis of position, overlap with regulatory elements, and in silico SNP prioritization tools (Online Appendix).
For both the novel loci and all previously reported CAD loci 1, 2, we tested the association of the lead CAD-associated variant (or, if unavailable, a proxy) with traditional cardiovascular risk factors using publicly available GWAS meta-analyses datasets for systolic, diastolic, and pulse pressures 7, 8; low-density lipoprotein (LDL) cholesterol level; high-density lipoprotein (HDL) cholesterol level; triglycerides level 9, 10; type 2 diabetes mellitus (11); body mass index (BMI) (12); and smoking quantity (13). The maximum size of these datasets ranged from 41,150 to 339,224 individuals. For variants available on the exome array with a known genome-wide association with a risk factor, we also compared the magnitude of the reported association with the risk factor to the observed association with CAD in our analysis.
To identify any associations with other diseases or traits, we searched version 2 of the GRASP (Genome-Wide Repository of Associations between SNPs and Phenotypes) database (14) and the National Human Genome Research Institute-European Bioinformatics Institute GWAS catalog (15), plus we collected all associations below 1 × 10−4. For all associations, we identified the lead variant for that trait or disease and calculated pairwise LD with the lead CAD-associated variant using the SNAP web server (16).
Results
In the discovery cohort, 28 variants not located in a known CAD locus (defined as ±300 kb from the published lead SNP) showed association with CAD at a p value <1 × 10−6 (Online Table 3). No marked heterogeneity was observed, justifying the use of a fixed-effects model. We then tested these 28 variants for replication, and 6 variants showed both a nominally significant (p < 0.05) association in the replication cohort and a combined discovery and replication meta-analyses p value exceeding the threshold for genome-wide significance (p < 5 × 10−8) (Table 1). As typical for GWAS findings, the risk alleles were common (allele frequencies ranging from 15% to 86%), and the risk increase per allele was modest (ranging from 4% to 9%) (Table 1).
Table 1
Novel Loci Significantly Associated With CAD
| Lead Variant | Locus | Locus Name | A1/2 (Freq) | Discovery | Replication | Meta-Analysis | Known Associations With Lead Variant | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases/Control Subjects | OR (95% CI) | p Value | Cases/Control Subjects | OR (95% CI) | p Value | OR (CI 95%) | p Value | |||||
| rs1801251 | 2q37 | KCNJ13-GIGYF2 | A/G (0.35) | 42,332/78,229 | 1.06 (1.04–1.08) | 1.46 × 10−8 | 30,528/42,521 | 1.03 (1.01–1.06) | 0.007 | 1.05 (1.03–1.06) | 1.48 × 10−9 | — |
| rs3130683 | 6p21 | C2 | T/C (0.86) | 39,494/72,267 | 1.09 (1.06–1.13) | 7.87 × 10−8 | 30,450/42,485 | 1.09 (1.05–1.14) | 2.97 × 10−5 | 1.09 (1.07–1.12) | 1.04 × 10−11 | — |
| rs11042937 | 11p15 | MRVI1-CTR9 | T/G (0.49) | 42,335/78,234 | 1.05 (1.03–1.07) | 3.21 × 10−8 | 30,533/42,527 | 1.03 (1.00–1.05) | 0.019 | 1.04 (1.03–1.06) | 1.18 × 10−8 | Bipolar disorder and schizophrenia |
| rs11172113 | 12q13 | LRP1 | C/T (0.41) | 42,335/78,234 | 1.06 (1.04–1.08) | 1.78 × 10−8 | 28,503/36,433 | 1.06 (1.03–1.08) | 1.16 × 10−6 | 1.06 (1.04–1.07) | 9.25 × 10−14 | Migraine, pulmonary function |
| rs11057830 | 12q24 | SCARB1 | A/G (0.15) | 42,331/78,237 | 1.09 (1.06–1.11) | 3.69 × 10−10 | 20,395/30,592 | 1.07 (1.03–1.11) | 0.0003 | 1.08 (1.06–1.10) | 4.61 × 10−13 | Vitamin E level |
| rs1800775 | 16q13 | CETP | C/A (0.51) | 38,810/62,756 | 1.06 (1.04–1.08) | 2.21 × 10−8 | 22,445/32,148 | 1.03 (1.00–1.05) | 0.032 | 1.04 (1.03–1.06) | 9.83 × 10−9 | HDL cholesterol |
A1/2 = allele 1/allele 2; CAD = coronary artery disease; CI = confidence interval; freq = frequency of allele 1; HDL = high-density lipoprotein; OR = odds ratio for disease for carriers of allele 1.
Annotation of novel loci
Forest and regional association plots for the 6 novel loci are shown in Online Figures 1 and 2, respectively. Interrogation of the 1000 Genomes Project phase 1 EUR data using Haploreg (BROAD Institute, Massachusetts Institute of Technology and Harvard, Boston, Massachusetts) (17) showed that the number of SNPs in high LD (r2 > 0.8) with the lead variant varied between 1 (LRP1 locus and CETP locus) and 111 (KCNJ13-GIGYF2 locus) (Online Table 4). Apart from the lead variant at the KCNJ13-GIGYF2 locus, which is a nonsynonymous SNP, none of the other loci had a variant affecting protein sequence in high LD with the lead variant.
Notable cis-eQTL findings for the new loci are shown in Online Table 5 and functional annotation of the lead variant and variants in high LD appear in Online Figure 3. The main findings from these analyses are discussed here locus by locus.
16q13
The lead variant, rs1800775, also known as −629C>A, is in the promoter of the cholesteryl ester transfer protein (CETP) gene, which mediates the transfer of cholesteryl esters from HDL cholesterol to other lipoproteins and was placed on the array because of its association with plasma HDL cholesterol level 9, 10. The risk (C) allele is associated with lower HDL cholesterol and modest increases in plasma LDL cholesterol and triglycerides levels 9, 10. Previous studies have shown that rs1800775 is itself functional in that the C allele disrupts binding of the Sp1 transcription factor resulting in increased promoter activity (18). This is in agreement with our annotation, which predicts this to be more likely to be a functional SNP than the only other SNP in high LD, rs3816117 (Online Figure 3). Consistent with this, we also found associations between rs1800775 and CETP expression (r2 of 0.77) with the best eSNP (i.e., the lead SNP for the eQTL) in monocytes and liver (Online Table 5), and previous studies have shown that the variant is also associated with plasma CETP level 19, 20.
12q24
The lead variant, rs11057830, and all 8 variants in high LD are located in a region of approximately 10 kb in intron 1 of SCARB1, which encodes SR-B1, a receptor for HDL cholesterol. Other variants at this locus have been associated with HDL cholesterol level 9, 10. However, these HDL cholesterol variants are not in high LD with the CAD-associated variants identified here, which only have a modest association with plasma HDL cholesterol level (Online Table 6), but a stronger association with plasma LDL cholesterol and triglycerides levels (Table 2). rs11957830 was included on the array because of an association of the A allele (CAD risk-associated allele) with higher levels of vitamin E (Table 3) (21). Variants in high LD with the CAD risk allele at rs11057830 have also been associated with increased lipoprotein-associated phospholipase A2 (Lp-PLA2) activity (22). Analysis of eQTL identified an association between rs11057841 (r2 = 0.92 with the lead variant), and expression of SCARB1 in the intestine (Online Table 5). Functional annotation of the locus did not identify a strong candidate causal SNP, but rs10846744 (r2 = 0.94 with the lead variant) overlaps a deoxyribonuclease I hypersensitivity peak in a region bound by several transcription factors (Online Figure 3).
Table 2
Significant Associations of CAD Variants With Selected CV Risk Factors
| Locus | Locus Name | Lead Variant | Trait | Effect | p Value |
|---|---|---|---|---|---|
| New Loci | |||||
| 6p21 | C2 | rs3130683 | T2D | 1.12† | 2.7 × 10−5 |
| SCARB1 | rs11057830 | LDL | 0.006 | 2.6 × 10−5 | |
| TG | 0.022 | 8.3 × 10−5 | |||
| 16q13 | CETP | rs1800775 | LDL | 0.041 | 8.5 × 10−24 |
| HDL | −0.202 | 3.3 × 10−644 | |||
| TG | 0.04 | 1.3 × 10−26 | |||
| Known Loci | |||||
|---|---|---|---|---|---|
| 1p32 | PCSK9 | rs11206510 | LDL | 0.083 | 2.4 × 10−53 |
| 1p13 | SORT1 | rs602633 | LDL | 0.159 | 1.5 × 10−261 |
| HDL | −0.033 | 3.5 × 10−14 | |||
| 2p24 | APOB | rs515135 | LDL | 0.139 | 1.1 × 10−178 |
| 2p21 | ABCG5-ABCG8 | rs6544713 | LDL | 0.081 | 4.84 × 10−83 |
| 4q32 | GUCY1A3 | rs7692387 | DBP | 0.326 | 3.4 × 10−5 |
| 5q31 | SLC22A4-SLC22A5 | rs273909 | LDL | 0.022 | 2.3 × 10−5 |
| 2q33 | WDR12 | rs6725887 | LDL | −0.026 | 1.3 × 10−5 |
| 6q25 | LPA | rs3798220 | LDL | 0.158 | 6.1 × 10−11 |
| rs2048327 | LDL | 0.019 | 1.3 × 10−6 | ||
| 7q32 | ZC3HC1 | rs11556924 | DBP | NA | 1.8 × 10−5 |
| HDL | −0.018 | 1.3 × 10−5 | |||
| 7q36 | NOS3 | rs3918226 | SBP | 0.96 | 1.1 × 10−6 |
| DBP | 0.81 | 2.2 × 10−9 | |||
| 8p21 | LPL | rs264 | HDL | −0.098 | 8 × 10−77 |
| TG | 0.093 | 2.4 × 10−84 | |||
| 8q24 | TRIB1 | rs2954029 | LDL | 0.056 | 2.1 × 10−50 |
| HDL | −0.04 | 2.7 × 10−29 | |||
| TG | 0.076 | 1 × 10−107 | |||
| 9q34 | ABO | rs579459 | LDL | 0.067 | 2.4 × 10−44 |
| 10q24 | CYP17A1-CNNM2-NT5C2 | rs12413409 | SBP | 1.034 | 2 × 10−9 |
| DBP | 0.483 | 3.4 × 10−5 | |||
| PP | 0.56 | 5.7 × 10−8 | |||
| BMI | −0.03 | 2.2 × 10−8 | |||
| 11q23 | ZNF259-APOA5-APOA1 | rs964184 | LDL | 0.086 | 2 × 10−26 |
| HDL | −0.107 | 6.1 × 10−48 | |||
| TG | 0.234 | 6.6 × 10−244 | |||
| 12q24 | SH2B3 | rs3184504 | LDL | −0.027 | 4.2 × 10−12 |
| HDL | −0.026 | 4.1 × 10−12 | |||
| SBP | 0.598 | 2 × 10−9 | |||
| DBP | 0.483 | 8.8 × 10−6 | |||
| BMI | −0.131 | 9.4 × 10−6 | |||
| 15q26 | FURIN-FES | rs17514846 | SBP | 0.509 | 1.2 × 10−5 |
| 17p13 | SMG6 | rs2281727 | BMI | 0.015 | 3.64 × 10−6 |
| 18q21 | PMAIP1-MC4R | rs663129 | HDL | −0.026 | 5.5 × 10−9 |
| BMI | 0.056 | 8.8 × 10−53 | |||
| 19p13 | LDLR | rs1122608 | LDL | 0.074 | 8.5 × 10−57 |
| 19q13 | APOE-APOC1 | rs2075650 | LDL | 0.177 | 1.7 × 10−214 |
| HDL | −0.055 | 9.7 × 10−26 | |||
| TG | 0.044 | 2.3 × 10−21 | |||
| BMI | −0.026 | 1.3 × 10−8 | |||
| rs445925 | LDL | 0.363 | 6.6 × 10−397 | ||
| HDL | −0.051 | 1.9 × 10−10 | |||
| TG | 0.101 | 3.6 × 10−39 | |||
BMI = body mass index; CAD = coronary artery disease; CV = cardiovascular; DBP = diastolic blood pressure; HDL = high-density lipoprotein; LDL = low-density lipoprotein cholesterol level; T2D = type 2 diabetes mellitus; TG = triglycerides.
 Effects are either absolute beta estimates of the association of the CAD risk allele on the trait (with a positive association indicating a higher value of the trait per copy of the risk allele) or log odds ratio for diabetes mellitus, †per copy of the risk allele. This table only includes associations that passed Bonferroni correction.
Effects are either absolute beta estimates of the association of the CAD risk allele on the trait (with a positive association indicating a higher value of the trait per copy of the risk allele) or log odds ratio for diabetes mellitus, †per copy of the risk allele. This table only includes associations that passed Bonferroni correction.Table 3
Association of CAD Loci With Other Diseases or Traits
| Locus | Locus Name | Disease or Trait | Disease or Trait Lead SNP | p Value | Direction | r2 (CAD Lead and Disease or Trait Lead) |
|---|---|---|---|---|---|---|
| New Loci | ||||||
| 6p21 | C2 | Systemic lupus erythematosus | rs3130342 | 9.3 × 10−7 | + | 0.87 |
| Primary biliary cirrhosis | rs3134954 | 1 × 10−5 | + | 0.86 | ||
| Multiple sclerosis | rs3134954 | 3.2 × 10−9 | − | 0.86 | ||
| 11p15 | MRVI1-CTR9 | Bipolar disorder and schizophrenia | rs2018368 | 1 × 10−6 | + | 0.96 |
| 12q13 | LRP1 | Migraine | rs11172113 | 4.3 × 10−9 | − | Same SNP |
| Lung function (FEV1/FVC) | rs11172113 | 1.2 × 10−8 | + | Same SNP | ||
| Cervical artery dissection | rs11172113 | 3 × 10−7 | − | Same SNP | ||
| 12q24 | SCARB1 | Lp-PLA2 activity | rs11057841 | 6.1 × 10−14 | + | 0.92 |
| Circulating vitamin E levels | rs11057830 | 8.2 × 10−9 | + | Same SNP | ||
| Lp-PLA2 mass | rs10846744 | 6.8 × 10−6 | + | 0.94 | ||
| Known Loci | ||||||
|---|---|---|---|---|---|---|
| 1p32 | SORT1 | Lp-PLA2 activity | rs7528419 | 1.3 × 10−17 | + | 0.9 |
| Metabolic syndrome domains (Atherogenic dyslipidemia–PC1) | rs12740374 | 8 × 10−16 | + | Same SNP | ||
| Lp-PLA2 mass | rs7528419 | 7.1 × 10−5 | + | 0.9 | ||
| 1q21 | IL6R | C-reactive protein | rs4845625 | 4.2 × 10−7 | + | Same SNP |
| 2p21 | ABCG5-ABCG8 | Serum phytosterol | rs4245791 | 2.2 × 10−70 | + | 1 |
| 2p11 | VAMP5-VAMP8-GGCX | Prostate cancer | rs10187424 | 2.7 × 10−15 | − | 0.87 |
| 2q33 | WDR12 | Cerebral white matter hyperintensities burden | rs6705330 | 5.7 × 10−5 | + | 1 |
| 3q22 | MRAS | Coronary artery calcification | rs2306374 | 2.7 × 10−5 | + | 1 |
| 4q12 | REST-NOA1 | Height | rs17081935 | 6.7 × 10−17 | + | 0.95 |
| 4q31 | EDNRA | Carotid intima media thickness | rs1878406 | 7 × 10−12 | + | Same SNP |
| Intracranial aneurysm | rs6842241 | 2.4 × 10−9 | + | 0.94 | ||
| 6p24 | PHACTR1 | Coronary artery calcification | rs9349379 | 4 × 10−22 | + | Same SNP |
| Cervical artery dissection | rs9349379 | 1 × 10−11 | − | Same SNP | ||
| Migraine | rs9349379 | 5 × 10−8 | − | Same SNP | ||
| Pulse wave velocity | rs7750679 | 5.4 × 10−5 | + | 1 | ||
| 6q25 | LPA | Lipoprotein (a) | rs3798220 | 1.6 × 10−49 | + | Same SNP |
| Colorectal cancer | rs7758229 | 5.6 × 10−9 | − | 0.85 | ||
| 7p21 | HDAC9 | Stroke (large vessel stroke) | rs11984041 | 1.9 × 10−11 | + | 1 |
| 7q22 | 7q22 | Endometriosis | rs10953541 | 3.2 × 10−5 | + | Same SNP |
| 8q24 | TRIB1 | Metabolic syndrome domains (Atherogenic dyslipidemia–PC1) | rs2954021 | 1.2 × 10−11 | + | Same SNP |
| Adiponectin levels | rs2954021 | 1.8 × 10−5 | + | Same SNP | ||
| Serum creatinine | rs2954021 | 2.3 × 10−5 | + | Same SNP | ||
| 9p21 | CDKN2BAS1 | Coronary artery calcification | rs1333049 | 3.3 × 10−24 | + | 0.97 |
| Abdominal aortic aneurysm | rs2383207 | 1.9 × 10−8 | + | 0.91 | ||
| Ankle brachial index | rs10757269 | 2.7 × 10−9 | + | 0.9 | ||
| Stroke (large vessel stroke) | rs2383207 | 2.4 × 10−6 | + | 0.91 | ||
| Intracranial aneurysm | rs10733376 | 4 × 10−12 | + | 0.94 | ||
| 9q34 | ABO | Alkaline phosphatase in plasma | rs579459 | 3 × 10−123 | + | Same SNP |
| Activated partial thromboplastin time | rs579459 | 1.7 × 10−74 | + | Same SNP | ||
| Soluble P-selectin | rs579459 | 1.9 × 10−41 | + | Same SNP | ||
| Soluble E-selectin | rs579459 | 1.3 × 10−29 | + | Same SNP | ||
| Plasma carcinoembryonic levels | rs579459 | 3 × 10−21 | + | Same SNP | ||
| Red blood cell count | rs579459 | 9.3 × 10−18 | + | Same SNP | ||
| Hemoglobin | rs579459 | 1.4 × 10−15 | + | Same SNP | ||
| Hematocrit | rs579459 | 7.6 × 10−14 | + | Same SNP | ||
| Interleukin-6 levels | rs579459 | 3.6 × 10−13 | + | Same SNP | ||
| Circulating galectin-3 levels | rs579459 | 1.9 × 10−10 | + | Same SNP | ||
| Factor XIII antigen | rs579459 | 2.3 × 10−8 | + | Same SNP | ||
| von Willebrand factor | rs651007 | 1 × 10−161 | + | 1 | ||
| Venous thromboembolism | rs495828 | 2 × 10−17 | + | 1 | ||
| Serum alkaline phosphatase levels | rs651007 | 1 × 10−56 | + | 1 | ||
| Ferritin levels | rs651007 | 1 × 10−8 | + | 1 | ||
| Soluble ICAM-1 | rs507666 | 3 × 10−91 | + | 0.83 | ||
| 10q24 | CYP17A1-CNNM2-NT5C2 | Intracranial aneurysm | rs12413409 | 1.2 × 10−9 | + | Same SNP |
| Schizophrenia | rs11191580 | 1.7 × 10−9 | + | 1 | ||
| Autism spectrum disorder | rs11191454 | 1.4 × 10−8 | + | 1 | ||
| Parkinson's disease | rs17115100 | 7.4 × 10−8 | + | 0.9 | ||
| 11q23 | ZNF259-APOA5-APOA1 | Metabolic syndrome domains (Atherogenic dyslipidemia–PC2) | rs964184 | 1.8 × 10−12 | + | Same SNP |
| Vitamin E levels | rs964184 | 7.8 × 10−12 | + | Same SNP | ||
| Lp-PLA2 activity | rs964184 | 8.4 × 10−11 | + | Same SNP | ||
| Metabolic syndrome domains (Atherogenic dyslipidemia–PC1) | rs964184 | 1.2 × 10−10 | + | Same SNP | ||
| 12q24 | SH2B3 | Selective immunoglobulin A deficiency | rs3184504 | 5.6 × 10−31 | + | Same SNP |
| Type 1 diabetes | rs3184504 | 2.8 × 10−27 | + | Same SNP | ||
| Celiac disease | rs3184504 | 5.4 × 10−21 | + | Same SNP | ||
| Hemoglobin | rs3184504 | 4.3 × 10−19 | + | Same SNP | ||
| Celiac disease | rs3184504 | 5.4 × 10−21 | + | Same SNP | ||
| Blood eosinophil count | rs3184504 | 6.5 × 10−19 | + | Same SNP | ||
| Generalized vitiligo | rs3184504 | 2.5 × 10−17 | + | 0.97 | ||
| Soluble ICAM-1 | rs3184504 | 2.9 × 10−17 | + | Same SNP | ||
| Hematocrit | rs3184504 | 7.9 × 10−16 | + | Same SNP | ||
| Hypothyroidism | rs3184504 | 2.6 × 10−12 | + | Same SNP | ||
| Rheumatoid arthritis and celiac disease | rs3184504 | 1.4 × 10−11 | + | Same SNP | ||
| Primary sclerosing cholangitis | rs3184504 | 5.9 × 10−11 | + | Same SNP | ||
| Serum urate | rs3184504 | 2.6 × 10−10 | + | Same SNP | ||
| Juvenile idiopathic arthritis | rs3184504 | 2.6 × 10−9 | + | 0.9 | ||
| Lymphocyte count | rs3184504 | 1.1 × 10−8 | + | Same SNP | ||
| Plasma beta-2 microglobulin levels | rs3184504 | 3.1 × 10−8 | + | Same SNP | ||
| White blood cell count | rs3184504 | 6.3 × 10−6 | + | Same SNP | ||
| Rheumatoid arthritis | rs3184504 | 6 × 10−6 | + | Same SNP | ||
| Tetralogy of Fallot | rs11065987 | 4.6 × 10−8 | + | 0.91 | ||
| 13q34 | COL4A1-COL4A2 | Coronary artery calcification | rs3809346 | 8.6 × 10−7 | + | 0.97 |
| 15q22 | SMAD3 | Crohn's disease | rs17293632 | 2.7 × 10−19 | − | 0.9 |
| Inflammatory bowel disease | rs17293632 | 6 × 10−16 | − | 0.9 | ||
| Ulcerative colitis | rs17293632 | 9.5 × 10−6 | − | 0.9 | ||
| Self-reported allergy | rs17228058 | 1.2 × 10−8 | − | 0.9 | ||
| 15q25 | ADAMTS7 | Coronary artery calcification | rs3825807 | 6.5 × 10−6 | + | Same SNP |
| 17p13 | SMG6 | Aortic root size | rs10852932 | 2.3 × 10−11 | + | 0.96 |
| 17q21 | UBE2Z | Height | rs318095 | 1.5 × 10−16 | − | 1 |
| Breast size (bra cup size in women) | rs12603969 | 3 × 10−5 | − | 1 | ||
| 18q21 | PMAIP1-MC4R | Obesity | rs17782313 | 4.8 × 10−15 | + | 0.86 |
| Height | rs11152213 | 6.9 × 10−13 | + | 0.91 | ||
| Antipsychotic drug induced weight gain | rs12967878 | 3.6 × 10−7 | + | 0.9 | ||
| 19q13 | APOE-APOC1 | Common carotid artery intima-media thickness | rs445925 | 1.7 × 10−8 | + | Same SNP |
| Lp-PLA2 activity | rs445925 | 3.3 × 10−10 | + | Same SNP | ||
| Metabolic syndrome domains (Atherogenic dyslipidemia–PC1) | rs445925 | 1.3 × 10−35 | + | Same SNP | ||
| Alzheimer disease | rs2075650 | 1 × 10−295 | + | Same SNP | ||
| Longevity | rs2075650 | 3.4 × 10−17 | − | Same SNP | ||
| Lp-PLA2 activity | rs2075650 | 8.1 × 10−15 | + | Same SNP | ||
| Age-related macular degeneration | rs2075650 | 8.4 × 10−8 | − | Same SNP | ||
| C-reactive protein | rs2075650 | 4.2 × 10−8 | + | Same SNP | ||
| Cognitive decline | rs2075650 | 2 × 10−8 | + | Same SNP | ||
+ indicates increase in disease or trait with the CAD risk allele.
CAD = coronary artery disease; FEV1 = forced expiratory volume 1; FVC = forced vital capacity; ICAM-1 = intercellular adhesion molecule 1; Lp-PLA2 = lipoprotein-associated phospholipase A2; PC = principal component; SNP = single nucleotide polymorphism.
12q13
The lead variant, rs11172113, is in intron 1 of LRP1 (LDL receptor–related protein-1) and only has 1 other adjacent SNP in high LD (Central Illustration, Online Table 4). The risk (C) allele of the lead variant has previously been associated with reduced risk of migraine (23), and there is an association of the alternate (T) allele with reduced lung function (24). There are also associations at this locus for abdominal aortic aneurysm (25) and triglyceride levels (10); however, these variants are in modest or low LD to the CAD-associated SNP (r2 of 0.54 and 0.07, respectively). The lead variant overlaps a region containing peaks in deoxyribonuclease I hypersensitivity in several cells and tissues, including aortic smooth muscle cells, within a predicted enhancer element (Online Figure 3). We found associations between the CAD risk allele at rs11172113 and reduced expression of LRP1 in atherosclerotic and nonatherosclerotic arterial wall, as well as eQTLs in omental and subcutaneous adipose tissue (Online Table 5).
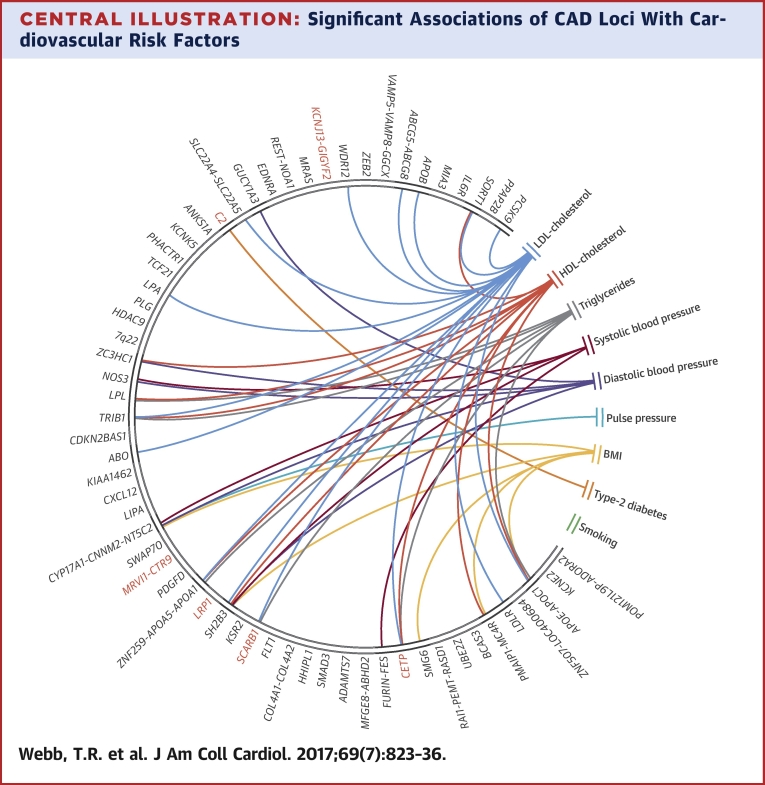
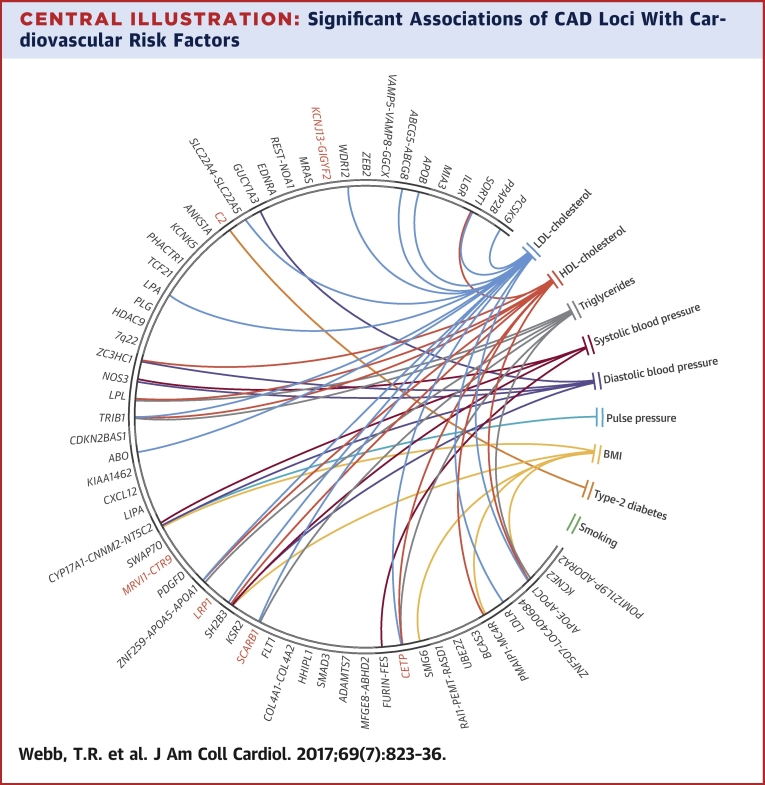
Significant Associations of CAD Loci With Cardiovascular Risk Factors
This chord diagram depicts associations that passed Bonferroni correction (Table 2). Connections indicate that single nucleotide polymorphisms at respective loci associate with both coronary artery disease (CAD) and the respective risk factor; they do not imply that the risk factor causally explains the association with CAD. Red indicates new CAD loci. BMI = body mass index; HDL = high-density lipoprotein; LDL = low-density lipoprotein.
11p15
The lead variant, rs11042937, at this locus lies in an intergenic region between MRVI1 (murine retrovirus integration site 1 homolog) encoding inositol-trisphosphate receptor-associated cyclic guanosine monophosphate kinase substrate, a mediator of smooth muscle tone and CTR9 that encodes a component of the PAF1 complex with some SNPs in high LD located within intron 1 of MRVI1 (Online Figure 3). The lead variant was included on the array because of a suggestive association with bipolar disorder and schizophrenia (26). There was no association of the locus with any cardiovascular risk factors, and we did not identify any eQTLs. Evidence for a regulatory function for either the lead variant or any of the SNPs in high LD was also weak (Online Figure 3).
6p21
The lead variant, rs3130683, lies in the HLA complex in intron 1 of C2, which encodes the complement C2 protein. There are just 14 SNPs in high LD with the lead variant (Online Table 4), but the CAD signal spans a region of approximately 300 kb including more than 20 genes (Online Figure 3). Apart from a single synonymous variant in HSPA1A (heat shock 70kDa protein 1A), the other high LD variants are noncoding with several of the variants showing evidence for regulatory functionality (Online Figure 3). Although there is a large number of eQTLs in the HLA region, most of these are variants with modest (r2 < 0.5) LD with the CAD-associated variants, and the only eQTL of note was with CYP21A2 (cytochrome P450, family 21, subfamily A, polypeptide 2) expression in whole blood (Online Table 5). rs3869109, another variant at the HLA locus approximately 700 kb away from the new lead variant, has been reported to be associated with CAD (27). In our discovery cohort, rs3869109 has a p value of association with CAD of 0.23.
2q37
The lead variant, rs1801251, was included on the array for identity by descent testing; rs1801251 causes a threonine to isoleucine amino acid change at position 95 in KCNJ13, an inwardly rectifying potassium channel protein. However, this is not predicted to be functionally important. There is extended linkage at this locus, with more than 100 SNPs in high LD and the lead variant in a region of ~170 kb also spanning GIGYF2 (GRB10 interacting GYF protein 2) (Online Figure 3). KCJN13 is located entirely within GIGYF2 and transcribed in the opposite direction. A number of the associated variants are in annotated regulatory regions, with the top scoring candidate by in silico prediction, rs11555646, lying in the 5′-UTR of GIGYF2 close to the initiating methionine (Online Figure 3). There was no association of the locus with any of the cardiovascular risk factors, but we found eQTLs for the lead variant or a variant in high LD for both GIGFY2 and KCNJ13 (Online Table 5).
CAD loci and pleiotropy
We undertook an updated analysis of the association of all 62 CAD loci (56 published and 6 novel in this report) with traditional cardiovascular risk factors (blood pressure traits, lipid traits, BMI, type 2 diabetes, and smoking). The full results are shown in Online Table 6, and the significant associations are summarized in Table 2. Of the 62 CAD loci, 24 (38.7%) showed a statistical association at a Bonferroni corrected p value <8.32 × 10−5 with a traditional cardiovascular risk factor with some loci showing multiple associations (Central Illustration). The largest number of associations were with lipid traits (14 with LDL cholesterol, 9 with HDL cholesterol, and 7 with triglycerides), followed by blood pressure traits (5 with diastolic blood pressure, 4 with systolic blood pressure, and 1 with pulse pressure), BMI (5 associations), and type 2 diabetes (1 association). Most associations were in the direction consistent from the epidemiological association of these risk factors with CAD, although a few displayed effects in the opposite direction (the risk variants at 2q33 and 12q24 are associated with reduced plasma LDL cholesterol, and those at 10q24, 12q24, and 19q13 are associated with lower BMI).
To inform the interpretation of these data, we conducted a complementary analysis for variants available on the array with a known genome-wide association with a risk factor; also, we compared the magnitude of the reported association with the risk factor to the observed association with CAD in our data. Except for LDL cholesterol and BMI, the correlations between the 2 effects were either weak or insignificant (Online Figure 4). In a separate analysis conducted in the 150,000 participants in UK Biobank with currently released genotype data, we confirmed that none of the CAD-associated variants showed a sex difference in allele frequency (data not shown).
We next analyzed the association of the 62 CAD loci with other diseases and traits. When restricted to variants with a high LD (r2 > 0.8) with the lead CAD variant, 29 of 62 (47%) loci showed an association with another disease/trait at a p value <1 × 10−4. Several loci showed multiple associations (Table 3). Although in most cases, the CAD-associated risk allele was also associated with an increased risk (or level) of the other disease or trait, this was not always the case. Furthermore, in some loci with multiple associations, the direction of association varied between diseases (Table 3).
Discussion
This large-scale meta-analysis of common variants, including many with prior evidence for association with another complex trait, resulted in the identification of 6 new CAD loci at genome-wide significance. We also showed that almost one-half of the CAD loci that have been identified to date demonstrate pleiotropy, an association with another disease or trait. The findings added to our understanding of the genetic basis of CAD and might provide clues to the mechanisms by which such loci affect CAD risk.
Our findings of a genome-wide association with CAD of a functional variant in the promoter of the CETP gene that is also associated with its expression and plasma activity 18, 19, 20 have added to previous evidence linking genetically determined increased activity of this gene with higher risk of CAD (20). There has been a longstanding interest in CETP inhibition as a therapeutic target, primarily because of the effect on plasma HDL cholesterol level. However, several CETP inhibitors have recently failed to improve cardiovascular outcomes in large randomized clinical trials 28, 29, 30 and, in 1 case, caused harm (28), despite markedly increasing plasma HDL cholesterol. Furthermore, Mendelian randomization studies have questioned the causal role of lower plasma HDL cholesterol in increasing CAD risk 31, 32. Although previous studies have shown that the CETP genetic variant we report here affects CETP activity, the precise mechanism(s) by which this variant modifies CAD risk remains uncertain.
A notable finding was the association with CAD of common variants located in the SCARB1 gene. Association of variants at the SCARB1 locus with CAD was also reported by the CARDIoGRAMplusC4D consortium, but this did not reach genome-wide significance (1). The gene encodes the canonical receptor, SR-BI, responsible for HDL cholesteryl ester uptake in hepatocytes and steroidogenic cells (33). Genetic modulation of SR-BI levels in mice is associated with marked changes in plasma HDL cholesterol (34). Consistent with this, a rare loss of function variant in which leucine replaces proline 376 (P376L) in SCARB1 was recently identified through sequencing of individuals with high plasma HDL cholesterol (35). Interestingly, despite having higher plasma HDL, 346L carriers had an increased risk of CAD, suggesting that the association of variation at this locus on CAD is not driven primarily through plasma HDL (35). Indeed, there is only a nominal association of the lead CAD variant at this locus (rs11057830) with plasma HDL cholesterol (Online Table 6). The variant is also modestly associated with plasma LDL cholesterol and serum triglycerides (Table 2). All 3 of these lipid associations are directionally consistent with epidemiological evidence linking them to CAD risk and could, in combination, explain the association of the locus with CAD. However, the lead variant is more strongly associated with Lp-PLA2 activity and mass (Table 3), which could provide an alternative explanation for its association with CAD. Irrespective of the mechanism, our findings, when combined with those of Zanoni et al. (35), suggest that modulating SR-B1 may be therapeutically beneficial.
After adjusting for multiple testing, we found that slightly more than one-third of the CAD loci showed an association with traditional cardiovascular risk factors. Although the vast majority of associations were in the direction consistent with the epidemiological association of these risk factors with CAD, as noted in the previous text with respect to loci affecting the HDL cholesterol level, this should not be interpreted as implying that these loci affect CAD risk through an effect on the specific risk factor. Indeed, for variants available on the array with a known genome-wide association with these risk factors, we found a poor correlation between the magnitudes of their effect of the risk factor and their association with CAD in our dataset except for LDL cholesterol (Online Figure 4). Nonetheless, formal causal inference analyses, using Mendelian randomization, have implicated LDL cholesterol, triglyceride-rich lipoproteins, blood pressure, type 2 diabetes, and BMI as causally involved in CAD (36).
Almost one-half of the CAD loci showed a strong or suggestive association with other diseases or traits with, in many cases, the identical variant being the lead variant reported for the association with these other conditions (Table 3). Some of the associations with other traits—for example, coronary calcification (3q22, 6p24, 9p21, 13q34, and 15q25) or carotid intima-media thickness (4q31 and 19q13)—are not surprising, as these traits are known to be correlated with CAD. Others, such as risk of stroke (7p21 and 9p21), might reflect a shared etiology. However, the mechanism(s) behind most of the observed pleiotropy is not clear, although the findings could provide clues as to how the locus may affect CAD risk. As an example, 5 loci (12q24, 1p13, 6q25, 11q23, and 19q13) show strong associations with plasma activity and/or mass of Lp-PLA2. Lp-PLA2 is expressed in atherosclerotic plaques where studies have suggested a role in the production of proinflammatory and pro-apoptotic mediators, primarily through interaction with oxidized LDL 37, 38. A meta-analysis of prospective studies showed an independent and continuous relationship of plasma Lp-PLA2 with CAD risk (39). However, it should be noted that Mendelian randomization analyses have not supported a causal role of secreted Lp-PLA2 in coronary heart disease (40), and phase III trials of darapladib, an Lp-PLA2 inhibitor, have shown no benefit in patients with stable coronary heart disease (41) or acute coronary syndromes (42) when added to conventional treatments including statins.
Chronic inflammation plays a key role in both the pathogenesis of CAD and of inflammatory bowel disease. It is therefore interesting to note the association of the same locus at 15q22 with CAD as well as Crohn’s disease and ulcerative colitis (Table 3). Association of this locus with CAD at genome-wide significance was recently reported by the CARDIoGRAMplusC4D consortium (2) with the lead SNP (rs56062135) showing strong linkage disequilibrium (r2 = 0.9) with the lead SNP (rs17293632) associated with inflammatory bowel disease. Both rs56062135 and rs17293632 lie in a region of ~30 kb within the initial introns of the SMAD family member 3 gene (SMAD3), a signal transducer in the transforming growth factor–beta pathway. Indeed, rs17293632 was included on the exome array because of its known association with Crohn’s disease and showed a significant association with CAD in our combined dataset (p = 1.78 × 10−8). Farh et al. (43) interrogated ChIP-seq data from ENCODE and found allele-specific binding of the AP-1 transcription factor to the major (C) allele in heterozygous cell lines and suggested that the T allele of rs17293632 increases risk of Crohn’s disease by disrupting AP-1 regulation of SMAD3 expression. Interestingly, the direction of effect on CAD risk observed for this variant was in the opposite direction to that for inflammatory disorders, with the C allele being the risk allele. Recent analysis of this variant in arterial smooth muscle cells confirmed that the CAD risk allele preserves AP-1 transcription factor binding and increases expression of SMAD3 (44). Further investigation of the discordant effects of SMAD3 may shed light on the mechanisms of both diseases.
Study limitations
First, in our discovery study, we were only able to interrogate common variants associated with other diseases and traits that were known at the time of the creation of the exome array in late 2011 and, thus, included on the array. Conversely, our interrogation for pleiotropic associations of the new and known CAD has used the latest data available in the GWAS catalogs and other sources. Second, the common variants tested in our study conferred statistically robust yet quantitatively modest effects on both CAD and potentially related traits. Thus, we may have missed associations with other traits. However, if such traits were considered as intermediary steps in the etiology of CAD, exploration of our large GWAS sample sets and respective GWAS catalogs should have detected relevant associations. Third, our discovery analysis is largely on the basis of subjects with Western-European ancestry, and any association with CAD of the new loci in other populations needs further evaluation. Finally, although we used relatively stringent criteria (minimal r2 > 0.8 between the CAD SNP and the lead variant associated with the other disease/trait), the limited content of the exome array and the information available in the GWAS catalogs meant that we could not examine the extent of overlap in the loci in detail.
Conclusions
Through an analysis of selected variants associated with other disease traits, we reported the discovery of 6 further loci associated with CAD. Furthermore, in the most comprehensive analysis to date, we showed that several of the new and previously established loci demonstrated substantial pleiotropy, which may help our understanding of the mechanisms by which these loci affect CAD risk.
Footnotes
Drs. Akinsanya, Wu, Yin, and Reilly are employees of Merck Sharp & Dohme; and Dr. Vogt was an employee of Merck when aspects of this research was conducted, but is now retired from Merck. A cholesteryl ester transfer protein inhibitor, Anacetrapib (MK-0859), is currently undergoing clinical investigation in the REVEAL outcome trial sponsored by Merck Sharp & Dohme. Dr. Schick is an employee of Recombine. Dr. Dube has equity in DalCor Pharmaceuticals. Dr. McCarthy is a member of advisory boards for Pfizer and Novo Nordisk; has received honoraria from Pfizer, Novo Nordisk, and Eli Lilly; and has received research funding provided by Pfizer, Novo Nordisk, Eli Lilly, Servier, Sanofi-Aventis, Janssen, Roche, Boehringer-Ingelheim, Takeda, Merck, and AstraZeneca. Dr. Ferrieres has received grants from Merck Sharp & Dohme, Amgen, and Sanofi. Dr. Sattar has served as a consultant for Amgen and Sanofi. Dr. Butterworth has received grants from Pfizer and Merck. Dr. Danesh has served as a consultant for Takeda; has served on the Novartis Cardiovascular & Metabolic Advisory Board and International Cardiovascular and Metabolism Research and Development Portfolio Committee of Novartis; has served on the UK Atherosclerosis Advisory Board of Merck Sharp & Dohme; has served on the advisory board of Sanofi; has served on the Pfizer Population Research Advisory Panel; and has financial relationships with the British Heart Foundation, BUPA Foundation, diaDexus, European Research Council, European Union, Evelyn Trust, Fogarty International Centre, GlaxoSmithKline, Merck, National Heart, Lung, and Blood Institute, National Health Service Blood and Transplant, National Institute for Health Research, National Institute of Neurological Disorders and Stroke, Novartis, Pfizer, Roche, Sanofi, Takeda, The Wellcome Trust, UK Biobank, University of British Columbia, and UK Medical Research Council. Dr. Tardif has received research grants from Amarin, AstraZeneca, Merck, Pfizer, Eli Lilly, Sanofi, Servier, and DalCor; has received honoraria from Pfizer (to his institution), Servier, DalCor, and Sanofi (to his institution); and has received modest equity interest from DalCor. Dr. Kathiresan has financial/other relationships with Regeneron, Bayer, Catabasis, Merck, Celera, Genomics PLC, San Therapeutics, Novartis, Sanofi, AstraZeneca, Alnylam, Eli Lilly, Leerink Partners, and Noble Insights. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose. A full list of acknowledgments and funding sources is included in the Online Appendix. Drs. Webb, Erdmann, Strirrups, Stitziel, Samani, Schunkert, Deloukas, and Kathiresan contributed equally to this work.
Listen to this manuscript's audio summary by JACC Editor-in-Chief Dr. Valentin Fuster.
For supplemental tables and figures as well as a full list of acknowledgments and funding sources, please see the online version of this article.
Appendix
References
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.jacc.2016.11.056
Article citations
Identification of novel proteins for coronary artery disease by integrating GWAS data and human plasma proteomes.
Heliyon, 10(19):e38036, 19 Sep 2024
Cited by: 0 articles | PMID: 39386869 | PMCID: PMC11462259
Polygenic Risk Score Assessment for Coronary Artery Disease in Asian Indians.
J Cardiovasc Transl Res, 17(5):1086-1096, 24 Apr 2024
Cited by: 0 articles | PMID: 38658478
GENIUS-MAWII: for robust Mendelian randomization with many weak invalid instruments.
J R Stat Soc Series B Stat Methodol, 86(4):1045-1067, 14 Mar 2024
Cited by: 0 articles | PMID: 39279912
Multi-ancestry genetic analysis of gene regulation in coronary arteries prioritizes disease risk loci.
Cell Genom, 4(1):100465, 15 Dec 2023
Cited by: 1 article | PMID: 38190101 | PMCID: PMC10794848
Protein interaction networks in the vasculature prioritize genes and pathways underlying coronary artery disease.
Commun Biol, 7(1):87, 12 Jan 2024
Cited by: 1 article | PMID: 38216744 | PMCID: PMC10786878
Go to all (145) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe (Showing 7 of 7)
-
(2 citations)
PDBe - 7q22View structure
-
(1 citation)
PDBe - 4q31View structure
-
(1 citation)
PDBe - 6p24View structure
-
(1 citation)
PDBe - 3q22View structure
-
(1 citation)
PDBe - 6q25View structure
-
(1 citation)
PDBe - 1q21View structure
-
(1 citation)
PDBe - 2q33View structure
Show less
SNPs (Showing 71 of 71)
- (17 citations) dbSNP - rs3184504
- (12 citations) dbSNP - rs579459
- (7 citations) dbSNP - rs2075650
- (6 citations) dbSNP - rs11057830
- (6 citations) dbSNP - rs11172113
- (4 citations) dbSNP - rs445925
- (4 citations) dbSNP - rs964184
- (4 citations) dbSNP - rs17293632
- (3 citations) dbSNP - rs2954021
- (3 citations) dbSNP - rs651007
- (3 citations) dbSNP - rs1801251
- (3 citations) dbSNP - rs9349379
- (3 citations) dbSNP - rs1800775
- (3 citations) dbSNP - rs3130683
- (2 citations) dbSNP - rs12413409
- (2 citations) dbSNP - rs10846744
- (2 citations) dbSNP - rs2383207
- (2 citations) dbSNP - rs3798220
- (2 citations) dbSNP - rs3134954
- (2 citations) dbSNP - rs11042937
- (2 citations) dbSNP - rs7528419
- (1 citation) dbSNP - rs10757269
- (1 citation) dbSNP - rs10733376
- (1 citation) dbSNP - rs1122608
- (1 citation) dbSNP - rs3816117
- (1 citation) dbSNP - rs12740374
- (1 citation) dbSNP - rs17514846
- (1 citation) dbSNP - rs17115100
- (1 citation) dbSNP - rs6842241
- (1 citation) dbSNP - rs507666
- (1 citation) dbSNP - rs10852932
- (1 citation) dbSNP - rs11556924
- (1 citation) dbSNP - rs2048327
- (1 citation) dbSNP - rs495828
- (1 citation) dbSNP - rs515135
- (1 citation) dbSNP - rs663129
- (1 citation) dbSNP - rs2954029
- (1 citation) dbSNP - rs7692387
- (1 citation) dbSNP - rs11191580
- (1 citation) dbSNP - rs2306374
- (1 citation) dbSNP - rs17081935
- (1 citation) dbSNP - rs6725887
- (1 citation) dbSNP - rs264
- (1 citation) dbSNP - rs17782313
- (1 citation) dbSNP - rs11984041
- (1 citation) dbSNP - rs12603969
- (1 citation) dbSNP - rs11555646
- (1 citation) dbSNP - rs17228058
- (1 citation) dbSNP - rs3918226
- (1 citation) dbSNP - rs11191454
- (1 citation) dbSNP - rs4245791
- (1 citation) dbSNP - rs3130342
- (1 citation) dbSNP - rs6544713
- (1 citation) dbSNP - rs6705330
- (1 citation) dbSNP - rs1878406
- (1 citation) dbSNP - rs4845625
- (1 citation) dbSNP - rs3825807
- (1 citation) dbSNP - rs11057841
- (1 citation) dbSNP - rs11206510
- (1 citation) dbSNP - rs11065987
- (1 citation) dbSNP - rs2018368
- (1 citation) dbSNP - rs56062135
- (1 citation) dbSNP - rs10953541
- (1 citation) dbSNP - rs2281727
- (1 citation) dbSNP - rs10187424
- (1 citation) dbSNP - rs7750679
- (1 citation) dbSNP - rs11152213
- (1 citation) dbSNP - rs318095
- (1 citation) dbSNP - rs7758229
- (1 citation) dbSNP - rs602633
- (1 citation) dbSNP - rs1333049
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Partitioning the Pleiotropy Between Coronary Artery Disease and Body Mass Index Reveals the Importance of Low Frequency Variants and Central Nervous System-Specific Functional Elements.
Circ Genom Precis Med, 11(2):e002050, 01 Feb 2018
Cited by: 9 articles | PMID: 29444804
Genetics of type 2 diabetes and coronary artery disease and their associations with twelve cardiometabolic traits in the United Arab Emirates population.
Gene, 750:144722, 30 Apr 2020
Cited by: 6 articles | PMID: 32360841
Improved detection of common variants in coronary artery disease and blood pressure using a pleiotropy cFDR method.
Sci Rep, 9(1):10340, 17 Jul 2019
Cited by: 1 article | PMID: 31316127 | PMCID: PMC6637206
Shared genetic susceptibility to ischemic stroke and coronary artery disease: a genome-wide analysis of common variants.
Stroke, 45(1):24-36, 21 Nov 2013
Cited by: 209 articles | PMID: 24262325 | PMCID: PMC4112102
Review Free full text in Europe PMC
Funding
Funders who supported this work.
British Heart Foundation (3)
Genomics of Coronary Artery Disease
Panos Deloukas, Queen Mary, University of London
Grant ID: RG/14/5/30893
Systematic approaches to the evaluation of emerging coronary risk markers. Large-scale epidemiological analyses of existing data and stored biological samples (renewal)
John Danesh, University of Cambridge
Grant ID: RG/08/014/24067
Funding for the Leicester Cardiovascular Genomics Group
Professor Thompson Robinson, University of Leicester
Grant ID: SP/16/4/32697
Medical Research Council (5)
The Scottish eHealth Informatics Research Centre
Professor Andrew Morris, University of Dundee
Grant ID: MC_PC_13040
Large-scale integrative studies of risk factors in coronary heart disease: from discovery to application
John Danesh, University of Cambridge
Grant ID: MR/L003120/1
MICA: The Scottish eHealth Informatics Research Centre
Professor Jill Pell, University of Dundee
Grant ID: MR/K007017/1
UK Health Informatics Research Network
Professor Andrew Morris, University of Dundee
Grant ID: MR/M501633/1
UK Health Informatics Research Network
Professor Andrew Morris, University of Edinburgh
Grant ID: MR/M501633/2
NHLBI NIH HHS (4)
Grant ID: K08 HL114642
Grant ID: R01 HL125863
Grant ID: R01 HL127564
Grant ID: R35 HL135824
NIH HHS (2)
Grant ID: S10 OD018522
Grant ID: S10 OD020069
National Institute for Health Research (NIHR) (3)
Grant ID: NF-SI-0512-10165
Grant ID: NF-SI-0611-10170
Grant ID: NF-SI-0611-10099





