Abstract
Free full text

The oppD Gene and Putative Peptidase Genes May Be Required for Virulence in Mycoplasma gallisepticum
Associated Data
ABSTRACT
Relatively few virulence genes have been identified in pathogenic mycoplasmas, so we used signature-tagged mutagenesis to identify mutants of the avian pathogen Mycoplasma gallisepticum with a reduced capacity to persist in vivo and compared the levels of virulence of selected mutants in experimentally infected chickens. Four mutants had insertions in one of the two incomplete oppABCDF operons, and a further three had insertions in distinct hypothetical genes, two containing peptidase motifs and one containing a member of a gene family. The three hypothetical gene mutants and the two with insertions in oppD1 were used to infect chickens, and all five were shown to have a reduced capacity to induce respiratory tract lesions. One oppD1 mutant and the MGA_1102 and MGA_1079 mutants had a greatly reduced capacity to persist in the respiratory tract and to induce systemic antibody responses against M. gallisepticum. The other oppD1 mutant and the MGA_0588 mutant had less capacity than the wild type to persist in the respiratory tract but did elicit systemic antibody responses. Although M. gallisepticum carries two incomplete opp operons, one of which has been acquired by horizontal gene transfer, our results suggest that one of the copies of oppD may be required for full expression of virulence. We have also shown that three hypothetical genes, two of which encode putative peptidases, may be required for full expression of virulence in M. gallisepticum. None of these genes has previously been shown to influence virulence in pathogenic mycoplasmas.
INTRODUCTION
Mycoplasma gallisepticum is the most important mycoplasmal pathogen in poultry (1, 2) and a particularly useful model of mycoplasmal pathogenesis, in part as a typical representative of the Pneumoniae phylogenic group, which includes the important human pathogen M. pneumoniae. There have been relatively few studies attempting to identify virulence genes in mycoplasmas, in part because of the limited availability of tools for such studies. Signature-tagged mutagenesis (STM) is a useful technique for identifying genes likely to be involved in virulence, as it enables the rapid comparison of populations of mutants used to inoculate animals with those recovered from these animals during the course of infection, allowing identification of mutants with a reduced capacity to persist in experimentally infected animals (3, 4). However, the examination of only pools of mutants may result in a failure to distinguish between a mutation in a gene that is essential in vivo and a mutation in a gene that reduces the capacity of the mutant to compete with other mutants in vivo. In addition, it is not possible to fully assess the virulence of individual mutants within populations. Therefore, STM needs to be complemented with direct comparisons of isolated mutants for their capacity to infect, persist, and cause disease.
In a previous study, we showed that several signature-tagged (ST) mutants of M. gallisepticum that were recovered infrequently from chickens inoculated with pools of mutants were significantly attenuated when assessed for their virulence in isolation (5). These mutants contained insertions in genes encoding the cytadhesin GapA and its accessory protein, CrmA. Three other virulence-associated determinants in M. gallisepticum, lpd, malF, and mslA, which has recently been shown to encode a novel oligonucleotide binding protein, have also been identified previously using transposon mutagenesis (5,–8). Two of these (malF and mslA) are members of ABC transporter operons.
The aims of the study reported here were to characterize further ST mutants that were recovered infrequently from chickens infected with a pool of mutants and to then examine and compare the levels of virulence of these mutants and their capacities to colonize the respiratory tract of experimentally infected chickens, with the aim of identifying additional genes that play a significant role in the pathogenesis of respiratory mycoplasmosis.
RESULTS
Identification of insertion sites in ST mutants.
The insertion points of the transposon in ST mutants examined in detail in this study are shown in Table 1. The insertion points in all the ST mutants in the pools are listed in Table 2. In most cases, the insertion site for a specific tagged transposon was the same in each of the three initial mutant pools. There were four ST mutants that had a tagged transposon inserted within the oppAB1C1D1F1 operon. Transposons were located within oppB1a in ST mutant 36-1 (and mutants 36-2 and 36-3) and within oppC1 in ST mutant 24-1 (and mutants 24-2 and 24-3). In ST mutant 20-1 (and mutants 20-2 and 20-3) and 26-1 (and mutants 26-2 and 26-3), the transposons were inserted at different sites in oppD1. No mutants with insertions in the other copy of the opp operon in M. gallisepticum (oppB2C2D2F2) were obtained. Three ST mutants, 03-1 (and mutants 03-2 and 03-3), 18-1 (and mutants 18-2 and 18-3), and 22-1 (and mutants 22-2 and 22-3), had the transposon inserted within genes encoding distinct hypothetical proteins.
TABLE 1
Transposon insertion sites in the ST mutants examined in detail in this study
| ST mutant | Insertion site in genome (% of gene to insertion point) | Gene name | Function of disrupted gene |
|---|---|---|---|
| 03-1, -2, and -3 | 380999–381000 (16.0) | MGA_1102 | Hypothetical membrane protein, zinc peptidase-like motif |
| 18-1, -2, and -3 | 980912–980913 (25.1) | MGA_0588 | Hypothetical membrane protein, multiple paralogs |
| 20-1, -2, and -3 | 710237–710238 (7.9) | MGA_0220 | Oligopeptide transporter ATP-binding protein OppD1 |
| 22-1, -2, and -3 | 365386–365387 (22.3) | MGA_1079 | Hypothetical lipoprotein, trypsin-like peptidase motif |
| 24-1, -2, and -3 | 712236–712237 (67.8) | MGA_0221 | Oligopeptide transporter permease OppC1 |
| 26-1, -2, and -3 | 710587–719588 (34.2) | MGA_0220 | Oligopeptide transporter ATP-binding protein OppD1 |
| 36-1, -2, and -3 | 712897–712898 (45.4) | MGA_0223 | Oligopeptide transporter permease (OppB1a) |
TABLE 2
Classification of transposon insertion sites in all ST mutants in this study
| Gene category and ST mutanta | Transposon insertion site | Function |
|---|---|---|
| Cell envelope, membranes and lipoproteins | ||
    11-1, -2, and -3 11-1, -2, and -3 | MGA_0964 | VlhA 4.02 (hemagglutinin) |
    17-1 and -2 17-1 and -2 | MGA_0379 | VlhA 3.02 (hemagglutinin) |
    13-1, -2, and -3 13-1, -2, and -3 | No match in strain Rlow | Match to vlhA gene in draft Ap3AS sequence |
| Cellular processes, 01-1, -2, and -3 | MGA_1142 | OsmC-like stress-induced protein |
| Translation, protein synthesis, 19-1, -2, and -3 | MGA_0216 | Elongation factor P (EF-P) |
| Transport and binding proteins, ABC transport | ||
    20-1, -2, and -3 20-1, -2, and -3 | MGA_0220 | ATP-binding protein OppD1 |
    24-1, -2, and -3 24-1, -2, and -3 | MGA_0221 | Permease protein OppC1 |
    26-1, -2, and -3 26-1, -2, and -3 | MGA_0220 | ATP-binding protein OppD1 |
    36-1, -2, and -3 36-1, -2, and -3 | MGA_0223 | Permease protein OppB1a |
| Intergenic regions | ||
    10-1, -2, and -3 10-1, -2, and -3 | MGA_0537–MGA_0539 | HsdM endonuclease, hsd1 locus |
    16-1, -2, and -3 16-1, -2, and -3 | MGA_0395–MGA_0398 | vlhA 3.09, mal P |
    23-1, -2, and -3 23-1, -2, and -3 | MGA_0379–MGA_0380 | vlhA 4.04, vlhA 4.05 |
    27-1, -2, and -3 27-1, -2, and -3 | MGA_0226–MGA_0230 | Conserved hypothetical protein, oppF (OppF) |
    31-1, -2, and -3 31-1, -2, and -3 | MGA_0071–MGA_0073 | vlhA 1.05, putative transposase |
    38-1, -2, and -3 38-1, -2, and -3 | MGA_0518–MGA_0519 | Unique hypothetical protein, conserved hypothetical protein |
| Others | ||
    02-2 02-2 | MGA_0549 | Unique hypothetical protein |
    02-3 02-3 | MGA_0145 | Putative transposase |
    03-1, -2, and -3 03-1, -2, and -3 | MGA_1102 | Hypothetical membrane protein, zinc peptidase motif |
    04-3 04-3 | MGA_0073 | Putative transposase |
    07-1, -2, and -3 07-1, -2, and -3 | MGA_0554 | Hypothetical lipoprotein |
    09-1 and -2 09-1 and -2 | MGA_0662 | Unique hypothetical membrane protein |
    12-1, -2, and -3 12-1, -2, and -3 | 16S rRNA | 16S rRNA |
    14-1, -2, and -3 14-1, -2, and -3 | MGA_0549 | Hypothetical membrane protein unique to M. gallisepticum and M. synoviae |
    18-1, -2, and -3 18-1, -2, and -3 | MGA_0588 | Hypothetical membrane protein, multiple paralogs |
    21-1, -2, and -3 21-1, -2, and -3 | MGA_0554 | Hypothetical lipoprotein |
    22-1, -2, and -3 22-1, -2, and -3 | MGA_1079 | Hypothetical lipoprotein |
    32-1, -2, and -3 32-1, -2, and -3 | MGA_0817b | Unique hypothetical protein |
    35-1, -2, and -3 35-1, -2, and -3 | MGA_0758 | Putative hemoxygenase |
| Not determined | ||
    09-3 09-3 | Multiple insertions | |
    15-1, -2, and -3 15-1, -2, and -3 | Multiple insertions | |
    17-3 17-3 | Multiple insertions | |
    25-1, -2, and -3 25-1, -2, and -3 | Multiple insertions | |
    34-1, -2, and -3 34-1, -2, and -3 | Multiple insertions |
Initial screening. (i) Pathological and serological findings.
No anti-M. gallisepticum antibody was detected in the serum of any bird at the time of inoculation. More serum samples were anti-M. gallisepticum antibody positive at 4 weeks than at 2 weeks after infection, but no antibody response was detected in uninoculated in-contact birds in any group (Table 3). Fewer birds had air sac lesions at 4 weeks than at 2 weeks after inoculation, and more severe air sac lesions were generally observed at 2 weeks after infection (scores of 0.5 in group A, 1.0 to 2.0 in group B, and 0.5 to 2.5 in group C). The severity of the lesions did not correlate closely with the serology results, with some birds having high rapid serum agglutination (RSA) scores but no detectable air sac lesions.
TABLE 3
Serology, air sac lesions, and mutants targeted for further investigation detected in each group in initial and confirmatory screens
| Screen and group | No. of birds positive for the following/no. examined: | Mutant tag(s) detected (no. of times detected) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Antibody against M. gallisepticum | Air sac lesions | ||||||||
| Wk 0 | Wk 2 | Wk 4 | Control | Wk 2 | Wk 4 | Control | Air sacs | Trachea | |
| Initial screen | |||||||||
    Aa Aa | 0/30 | 7/9 | 8/9 | 0/8 | 2/10 | 0/9 | 0/8 | oppD1-20 (2)b | oppD1-20 (6), oppC1-24 (6), oppD1-26 (1) MGA_0588-18 (1) |
    B B | 0/31 | 6/10 | 9/11 | 0/10 | 2/10 | 1/11 | 0/10 | oppD1-20 (1) | oppD1-20 (6) |
    Cc Cc | 0/29 | 5/10 | 9/10 | 0/7 | 4/10 | 2/10 | 0/7 | oppD1-20 (7), MGA_0588-18 (1) | oppD1-20 (10), oppC2-24 (5), oppB1a-36 (1), MGA_0588-18 (1) |
| Confirmatory screend | |||||||||
    A A | 0/20 | 15/19 | 3/19 | MGA_1102-03 (1) | MGA_1102-03 (2) | ||||
    B B | 0/20 | 14/19 | 1/19 | MGA_1102-03 (1), MGA_1079-22 (2) | |||||
(ii) Identification of recoverable ST mutants.
The rates of recovery of ST mutants from birds at 14 and 28 days after inoculation are summarized in Tables 3 and and4.4. The oppD1 ST mutant 20 (mutants 20-1, -2, and -3) was the mutant most frequently recovered from the air sacs, as well as from the tracheas, and was isolated at a high frequency from all three groups. It was also recovered from the air sacs of one in-contact control bird in group A 4 weeks after inoculation. In contrast, oppD1 ST mutant 26 (mutants 26-1, -2, and -3) was recovered only from the trachea of one bird in group A. Six and five isolations of oppC1 ST mutant 24 (mutants 24-1 and -3) were made from the tracheas of birds in groups A and C, respectively, while only one isolation of the oppB1a ST mutant 36 (mutant 36-3) was made, and it was from the trachea of a bird in group C. MGA_0588 ST mutant 18 was reisolated only from the air sacs of one bird in group C (mutant 18-3) and from the trachea of one bird each in groups A and C (mutants 18-1 and -3, respectively). Sixteen ST mutants, including ST mutants 03 (mutants 03-1, -2, and -3; MGA_1102 interrupted) and 22 (mutants 22-1, -2, and -3; MGA_1079 interrupted), were unable to be recovered from any bird in any of the groups (Table 3).
TABLE 4
All mutants detected in each experimental group in initial and confirmatory screensa
| Screen and group | Signature tags detected (no. of times detected) | |
|---|---|---|
| Air sacs | Trachea | |
| Initial screen | ||
    Ab Ab | 16-1 (1),c 20-1 (2)c | 07-1 (3), 11-1 (1), 12-1 (4), 14-1 (1), 18-1 (1), 19-1 (2), 20-1 (6), 21-1 (1), 24-1 (6), 26-1 (1), 27-1 (2), 31-1 (1), 32-1 (1), 35-1 (1), 38-1 (1) |
    B B | 12-2 (1), 20-2 (1) | 07-2 (1), 11-2 (1), 12-2 (3), 14-2 (1), 16-2 (1), 20-2 (6), 27-2 (1) |
    Cd Cd | 11-2 (1), 12-2 (1), 16-2 (3), 18-2 (1), 20-2 (7), 27-2 (1) | 01-3 (1), 02-3 (8), 11-3 (1), 12-3 (5), 13-3 (1), 14-3 (1), 16-3 (3), 18-3 (1), 20-3 (10), 24-3 (5), 27-3 (6), 36-3 (1), 38-3 (2) |
| Confirmatory screene | ||
    A A | 03-1 (1), 09-1 (1), 10-1 (2), 15-1 (1), 23-1 (2) | 02-2 (4), 03-1 (2), 04-3 (1), 09-1 (3), 10-1 (10), 15-1 (1), 17-2 (12), 23-1 (2), 25-1 (1), 34-1 (1) |
    B B | 09-3 (2), 17-3 (1), 23-1 (2) | 02-2 (1), 03-1 (1), 09-3 (14), 10-1 (5), 15-1 (2), 17-3 (10), 23-1 (15), 25-1 (3), 34-1 (2) |
Confirmatory screening. (i) Pathological and serological assessments.
No anti-M. gallisepticum antibody was detected prior to inoculation in any bird (Table 3). Severe air sac lesions (lesion score, 2.5) were seen in one bird, and mild lesions (lesion scores, 0.5 and 1.0) were seen in another two chickens in group A. One bird in group B had mild air sac lesions (lesion score, 1.0). At 2 weeks after inoculation, 15/19 chickens in group A and 14/19 birds in group B had detectable antibody against M. gallisepticum (Table 3).
(ii) Reisolation of ST mutants.
Birds in group A were infected with a pool containing MGA_1102 ST mutant 03-1, and a pool that contained MGA_1079 ST mutant 22-1 was used to inoculate the birds in group B. A total of 11 ST mutants, including ST mutant 03-1, were reisolated from 16 chickens in group A, and 10 mutants, including ST mutants 22-1 and 03-1, were reisolated from 18 birds in group B (Table 3).
Infectivity and virulence analysis of selected ST mutants. (i) Clinical signs and postmortem examination.
The prevalence and severity of lesions in chickens infected with individual mutants are shown in Table 5. Air sac lesions were not seen in the uninfected control birds (group 1) or in birds exposed to aerosols of MGA_1079 ST mutant 22-1 (group 5). Mild lesions (lesion score, 0.25) were observed in one bird inoculated with MGA_1102 ST mutant 03-1 (group 2). Of the 20 birds infected with oppD1 ST mutant 26-1 (group 6), four had mild lesions (lesion scores, 0.5 to 1.0), while lesions were seen only in the abdominal air sacs of 5 birds exposed to MGA_0588 ST mutant 18-1 (group 3). Six of 20 birds in the group infected with oppD1 ST mutant 20-1 (group 4) had mild to severe lesions (lesion scores, 0.5 to 2.5), while mild to severe lesions (lesion scores, 0.5 to 3.0) were seen in 11/18 birds infected with the virulent Ap3AS strain (group 7).
TABLE 5
Serology, air sac lesion scores, and reisolation rates in infectivity and virulence study
| Group | Inoculum | No. of birds positive for antibody against M. gallisepticum/no. examineda | No. of birds positive/no. examineda,b | ||||
|---|---|---|---|---|---|---|---|
| Air sac lesions | Agar culture | Broth culturec | |||||
| Air sacs | Trachea | Air sacs | Trachea | ||||
| 1 | Medium | 0/20A | 0/20A (0–0)d | 0/20A | 0/20A | 0/20A | 0/20A |
| 2 | Mutant 03-1 | 1/20A | 1/20A,B (0–0.25) | 0/20A | 2/20A,B | 0/20A | 5/20B,C |
| 3 | Mutant 18-1 | 20/20B | 5/20B (0.5–3.5) | 5/20B,C | 17/20C | 7/20B | 19/20D |
| 4 | Mutant 20-1 | 20/20B | 6/20B (0.5–4.0) | 7/20C | 20/20C | 8/20B | 20/20D |
| 5 | Mutant 22-1 | 0/19A | 0/19A (0–0) | 0/19A | 1/19A,B | 0/19A | 3/19A,B |
| 6 | Mutant 26-1 | 0/20A | 4/20A,B (0.5–1.0) | 1/20A,B | 6/20B | 1/20A | 10/20C |
| 7 | Wild-type Ap3AS | 18/18B | 11/18C (0.5–8.5) | 9/18C | 18/18C | 10/18B | 15/18D |
(ii) Antibody responses and reisolation of ST mutants.
No anti-M. gallisepticum antibody was detected at the time of infection in the serum of any birds (Table 5). Two weeks after exposure, antibody responses were not detected in any of the birds in group 5 (MGA_1079 mutant 22-1 infected) or 6 (oppD1 mutant 26-1 infected), while a response was detectable in only one bird in group 2 (MGA_1102 mutant 03-1 infected). In contrast, strong antibody responses against M. gallisepticum were detectable in all the birds in groups 3 (MGA_0588 mutant 18-1 infected) and 4 (oppD1 mutant 20-1 infected). The uninfected control birds (group 1) did not develop any antibody against M. gallisepticum over the course of the experiment, while all birds infected with the wild-type Ap3AS strain (group 7) had strong antibody responses against M. gallisepticum.
M. gallisepticum was not isolated on mycoplasma agar (MA) plates inoculated with swab specimens of the air sacs of any birds in group 2 (MGA_1102 mutant 03-1 infected) or 5 (MGA_1079 mutant 22-1 infected), but it was isolated from the tracheas of two birds in group 2 (MGA_1102 mutant 03-1 infected) and one bird in group 5 (MGA_1079 mutant 22-1 infected). In group 6 (oppD1 mutant 26-1 infected), M. gallisepticum was isolated from the air sacs of one bird and the tracheas of six birds. M. gallisepticum was also isolated from the tracheas of 17/20 birds in group 3 (MGA_0588 mutant 18-1 infected) and of all the birds in group 4 (oppD1 ST mutant 20-1 infected) and also from swab specimens of the air sacs of 5/20 birds in group 3 and 7/20 birds in group 4. In the positive-control group (group 7), M. gallisepticum was isolated from the air sacs of 10/18 birds and the tracheas of 18/18 birds (Table 5).
M. gallisepticum was recovered in mycoplasma broth (MB) inoculated with swab specimens of the air sacs of one bird in group 6 (oppD1 mutant 26-1 infected), seven birds in group 3 (MGA_0588 mutant 18-1 infected), and eight birds in group 4 (oppD1 mutant 20-1 infected). M. gallisepticum was recovered from the tracheas of birds in all six of the groups exposed to ST mutants, with the number of colonized birds ranging from 3 in group 5 (MGA_1079 mutant 22-1 infected) to 20 in group 4 (oppD1 mutant 20-1 infected). The identity of the recovered M. gallisepticum ST mutants was confirmed using the unique signature tags that they carried (Table 5).
(iii) Tracheal lesions and mucosal thicknesses.
Median tracheal lesion scores are shown in Table 6. The scores of birds in all the mutant-infected groups differed significantly from those of birds in the wild-type-infected group (group 7) (P < 0.0001). There was no significant difference between the lesions scores for the uninfected group (group 1) and those for the mutant-infected groups, with the exception of those for the upper tracheas of the birds infected with MGA_1102 mutant 03-1. The upper tracheal lesion scores of the birds infected with MGA_1102 mutant 03-1 (group 2) did not differ from those of birds in groups 3, 5, and 6 but did differ significantly from those of birds in group 4 (oppD1 mutant 20-1 infected).
TABLE 6
Tracheal lesion scores and mucosal thicknesses in birds in infectivity and virulence study
| Group | Inoculum | Median tracheal lesion score (minimum, maximum)a | Mean tracheal mucosal thickness ± SD (μm)a | ||||
|---|---|---|---|---|---|---|---|
| Upper | Middle | Lower | Upper | Middle | Lower | ||
| 1 | Medium | 0.25 (0, 0.5)A | 0.25 (0, 0.5)A | 0 (0, 0.25)A | 50 ± 11A | 41 ± 8A | 37 ± 6A |
| 2 | Mutant 03-1 | 1 (0.25, 3)B | 0.375 (0, 1.5)A | 0.25 (0, 1.5)A | 89 ± 45B | 54 ± 12A | 46 ± 22A |
| 3 | Mutant 18-1 | 0.25 (0, 10)A,B | 0.25 (0, 1.5)A | 0.25 (0, 1.5)A | 62 ± 15A,B | 53 ± 15A | 49 ± 15A,B |
| 4 | Mutant 20-1 | 0.25 (0, 1.5)A | 0.125 (0, 1.5)A | 0.125 (0, 1.0)A | 56 ± 22A | 49 ± 23A | 51 ± 19A |
| 5 | Mutant 22-1 | 0.5 (0, 1)A,B | 0.25 (0, 1)A | 0 (0, 0.25)A | 64 ± 13A,B | 49 ± 8A | 39 ± 8A |
| 6 | Mutant 26-1 | 0.5 (0, 2)A,B | 0.25 (0, 1)A | 0 (0, 2)A | 64 ± 24A,B | 49 ± 14A | 48 ± 22A |
| 7 | Wild-type Ap3AS | 1.5 (1, 3)C | 1.50 (0.5, 3)B | 1.50 (0.25, 3)B | 168 ± 70C | 153 ± 84B | 120 ± 56B |
Mean tracheal mucosal thicknesses are shown in Table 6. The mean tracheal mucosal thicknesses of the wild-type-infected birds were significantly different from those of both the uninfected and the mutant-infected birds (P < 0.0001). The mean mucosal thicknesses in the middle and lower tracheas did not differ significantly between the uninfected group and any of the mutant-infected groups (groups 2 to 6). However, they were significantly different between the mutant-infected groups (groups 2 to 6) and the wild-type-infected group (group 7). While there was no significant difference between the tracheal mucosal thicknesses in the upper tracheas of the uninfected controls and birds in groups 3, 4, 5, and 6, the mean upper tracheal mucosal thickness in the group infected with MGA_1102 mutant 03-1 was significantly greater than that in the uninfected birds and birds infected with oppD1 mutant 20-1. In summary, the wild-type-infected controls had significantly more severe tracheal lesions, as assessed by histological lesion score or mucosal thickness, than any of the groups infected with the ST mutants. Group 2 (MGA_1102 mutant 03-1 infected) was the most severely affected among the groups infected with the ST mutants.
SDS-PAGE.
Total cell proteins of the M. gallisepticum strains used in this study were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie brilliant blue (Fig. 1). There were no detectable differences in the protein profiles of ST mutants 03-1 (MGA_1102 interrupted), 20-1 (oppD1 interrupted), 22-1 (MGA_1079 interrupted), 24-1 (oppC1 interrupted), and 36-1 (oppB1a interrupted) and wild-type strain Ap3AS. However, one protein band of about 40 kDa was absent in MGA_0588 mutant 18-1, while oppD1 mutant 26-1 contained an additional band of about 33 kDa compared to the banding patterns found for the other mutants and wild-type strain Ap3AS.
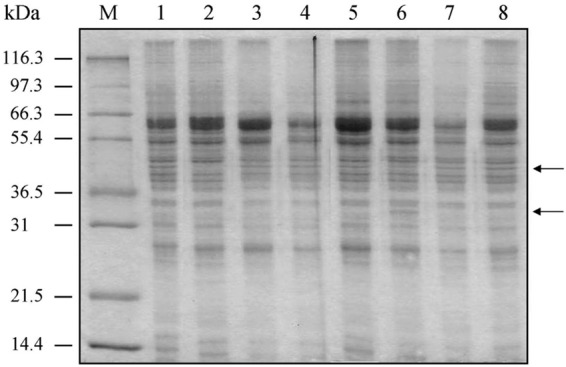
Protein profiles of ST mutants. Total cell proteins were separated in a 12.5% polyacrylamide gel together with molecular mass standards and then stained with Coomassie brilliant blue. The positions of the main differences in the protein profiles (in lanes 2 and 6) are indicated by arrows on the right. Lane 1, ST mutant 03-1 (MGA_1102 interrupted); lane 2, ST mutant 18-1 (MGA_0588 interrupted); lane 3, oppD1 mutant 20-1; lane 4, ST mutant 22-1 (MGA_1079 interrupted); lane 5, oppC1 mutant 24-1; lane 6, oppD1 mutant 26-1; lane 7, oppB1a mutant 36-1; lane 8, wild-type strain Ap3AS; lane M, broad-range protein size markers (Mark12 wide-range protein standard; Novex). The sizes of these markers are indicated on the left.
Two protein bands, of 38 and 30 kDa, were more prominently detected in MGA_0588 mutant 18-1 than in wild-type strain Ap3AS by the use of sera from birds infected with parent strain Ap3AS (Fig. 2; see also Fig. S1 in the supplemental material). One protein of 43 kDa was not detected in oppD1 mutant 20-1 by the use of sera from birds infected with Ap3AS, and two proteins of 40 kDa and 43 kDa were more prominently detected in oppD1 mutant 26-1 than in wild-type strain Ap3AS, while proteins of 20 kDa were less prominent in oppD1 mutant 26-1 than in the Ap3AS strain (Fig. 2 and S1).
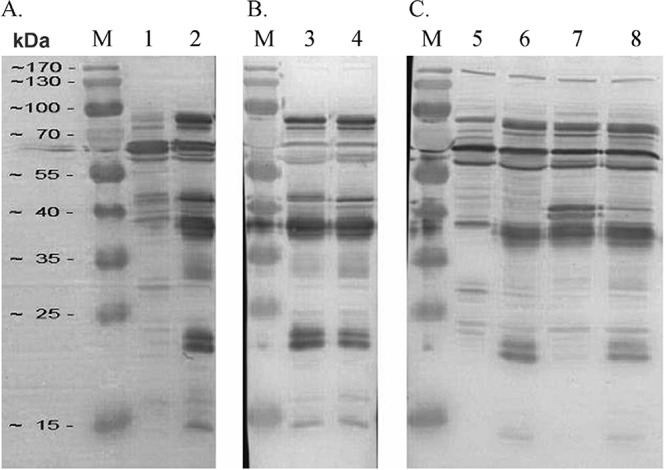
Western blot analysis of selected ST mutants. Whole-cell proteins of ST mutants and the parent strain Ap3AS were separated in 12.5% polyacrylamide gels and then transferred onto a PVDF membrane. The membranes were then probed with pooled sera from birds infected with Ap3AS diluted 1:2,000. Lanes 1 and 5, MGA_0588 ST mutant 18-1; lanes 3 and 6, oppD1 ST mutant 20-1; lanes 2, 4, and 8, Ap3AS; lane 7, oppD1 ST mutant 26-1; lanes M, the positions of the broad-range protein markers (PageRuler prestained protein ladder; Thermo Scientific).
DISCUSSION
ABC transporters are a family of multidomain membrane proteins that use energy from the hydrolysis of ATP to translocate solutes across cellular membranes (9, 10). The four domains within ABC transporters form a conserved core structure composed of two transmembrane domains (TMDs) and two nucleotide-binding domains (NBDs) (11,–14).
There are two adjacent copies of the opp ABC transporter operon (oppB1C1D1F1 and oppAB2C2D2F2) in M. gallisepticum strain Rlow (15). OppB and OppC are predicted to be transmembrane proteins, while oppD and oppF are predicted to encode ATP-binding proteins. The oppB1 gene contains a frameshift mutation, oppF2 appears to have been truncated, and there is only one gene encoding OppA, the substrate-binding lipoprotein (16), in M. gallisepticum. Thus, it appears that M. gallisepticum may contain two complementary partial copies of the opp operon, with only a single full-length copy of oppA, oppB, and oppF but two full-length copies of oppCD.
One of these opp operons (MGA_0237, MGA_0235, MGA_0234, MGA_0232, and MGA_0230, or oppAB2C2D2F2) is most similar to the opp operon of M. synoviae strain 53 (GenBank accession number AE017245), suggesting that the operon has been relatively recently acquired from M. synoviae or a closely related species within the Hominis phylogenic group (which includes M. hyopneumoniae, M. synoviae, and M. pulmonis) (Fig. 3). Phylogenetic analysis of these genes in a number of fully sequenced genomes suggested that this operon was duplicated in a member of the Hominis group and that these duplicated operons then diverged, with one subsequently being transferred horizontally to an ancestor of M. gallisepticum and Ureaplasma urealyticum. Ureaplasma urealyticum appears to have lost its original copy of the operon and retained only the horizontally transferred copy, while M. gallisepticum has retained most of both copies of the operon (Fig. 3). Studies in M. hominis have shown that OppA is a cytadhesin and also that it binds oligopeptides, suggesting that this operon encodes a transporter responsible for the import of oligopeptides (16,–18). It is notable that duplication of the oppABCDF operon by horizontal gene transfer has also occurred in the ruminant pathogens Mycoplasma agalactiae and Mycoplasma bovis, although this horizontal gene transfer event was clearly independent of that in M. gallisepticum (19,–21).
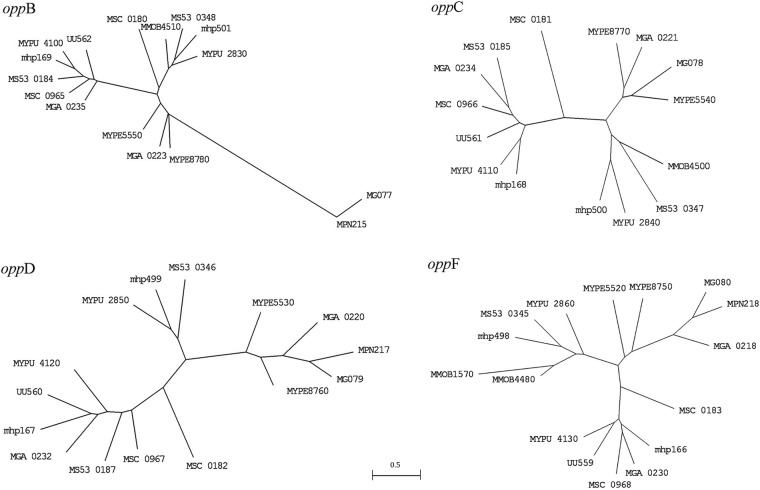
Phylogenetic analysis of the oppABCDF operons of Mollicutes. Trees were inferred using the DNAml program (35) and then drawn using the MEGA (version 3.1) program (36). Clustal W program (37)-aligned sequence data were analyzed using global rearrangement and randomized input orders to find the most likely tree. The optimal transition/transversion ratio was found to be 0.8 when the oppB genes were analyzed, so this ratio was used for analysis of all other genes. A single mutation rate was assumed. MG, Mycoplasma genitalium; MGA, Mycoplasma gallisepticum; MMOB, Mycoplasma mobile 163K; mhp, Mycoplasma hyopneumoniae; MPN, Mycoplasma pneumoniae; MS, Mycoplasma synoviae; MSC, Mycoplasma mycoides subsp. mycoides small colony type; MYPE, Mycoplasma penetrans; MYPU, Mycoplasma pulmonis; UU, Ureaplasma urealyticum/U. parvum.
In the studies presented here, four mutants had transposon insertions in genes in the same opp operon (in oppB1, oppC1, and oppD1), suggesting that mutations in some of the original opp genes may be tolerated (except possibly for the only full-length oppF, oppF1) but that the oppB2C2D2 genes may be required for survival in M. gallisepticum. The transposon was inserted into the oppD1 gene in mutants 20-1 (and mutants 20-2 and 20-3) and 26-1 (and mutants 26-2 and 26-3) but in different positions (Table 1). Mutant 20-1 induced seroconversion in more birds than mutant 26-1 and also colonized a greater proportion of birds, even though they were both much less pathogenic than the wild-type strain. The insertion site in mutant 20-1 was close to the start of the gene, while in mutant 26-1 the first third of the gene may be able to be expressed. It is possible that the two different oppD proteins form heterodimers in the transport complex and that, in the absence of expression of oppD1 in mutant 20-1, OppD2 could form functional homodimers. The partial copy of oppD1 in mutant 26-1 may retain a capacity to form a heterodimer, but one that is less functional, resulting in less efficient transportation and, thus, the greater reduction in colonization and virulence seen in this mutant. Further work will be needed to establish whether the two copies of oppD in the genome can partially complement each other and whether they form heterodimers.
In ST mutant 36-1 (and mutants 36-2 and 36-3), the transposon was inserted into the middle of the oppB1a gene (MGA_0223). As there is a frameshift mutation within the oppB1 gene, the two proteins predicted to be derived from it, OppB1a and OppB1b (MGA_0223 and MGA_0224, respectively), may not be fully functional, and thus, oppB2 may be the only fully functional copy in the genome. Although oppB1a mutant 36-1 was isolated very infrequently in the initial screening experiment, it was assessed only as a member of a pool, so the impact of mutation of the oppB1a gene on infectivity and virulence requires further investigation. ST mutant 24-1 (and mutants 24-2 and 24-3) had a transposon inserted into the oppC1 gene (MGA_0221) about two-thirds along the length of the gene, and this mutant could be frequently recovered from birds in the initial screening, suggesting either that the essential region of OppC may be within the first two-thirds of the gene or that oppC2 can complement the loss of oppC1.
ST mutant 18-1, with an insertion in one of multiple paralogous genes for a hypothetical protein (MGA_0588), was able to infect birds, but it was significantly less virulent than wild-type strain Ap3AS. This suggests that the paralog disrupted in this mutant may have a significant influence on virulence, although not on colonization. Mutant 22-1 (MGA_1079 interrupted) was relatively apathogenic and had a reduced capacity to colonize. ST mutant 03-1 (and mutants 03-2 and -3; MGA_1102 interrupted) was not recovered from any bird in the initial screen but was able to be reisolated in the confirmatory screen, albeit at a relatively low frequency, and was also able to spread, as it was recovered from birds in both groups. Although only one bird infected with this mutant developed detectable serum antibodies, it was the most virulent of all the mutants assessed in isolation, suggesting that mutants need to be assessed individually in hosts to examine the severity and the extent of the lesions that they induce if their virulence is to be fully assessed and that the capacity to induce seroconversion is not always a reliable indicator of virulence.
The oppD1 mutant 20-1, which caused mild air sac lesions and minor damage in the trachea, induced a detectable antimycoplasma antibody response. The site of insertion of the transposon would allow expression of only the N-terminal 7.9% of the protein. In contrast, less virulent oppD1 mutant 26-1 could express the N-terminal 34% of the protein and did not induce detectable antimycoplasma antibody. Western blotting detected differences between this mutant and wild-type strain Ap3AS, suggesting that the transposon insertion did result in changes in protein expression.
However, the protein profiles of the mutants did not provide clear evidence of the loss of expression of the proteins encoded by the genes that had been interrupted in each mutant, although some differences in the protein profiles were detected. The lack of a clear correlation between the site of insertion of the transposon and the changes in protein profiles may be attributable to the prevalence of posttranslational cleavage in mycoplasmas, which complicates the direct attribution of a protein band to a specific gene. In addition, assuming that oppD1 was expressed at sufficiently high levels to be detected in these assays, the presence of the product of the paralogous oppD2 gene would be expected to obscure detection of changes in the expression of oppD1. In other bacteria, signals imported through the Opp system can have diverse effects on the expression of other genes, so the changes detected in these mutants could reflect similar effects in M. gallisepticum. We would predict that the two oppD1 mutants might have different patterns of protein expression as a result of the different extents of their effects on the function of the Opp transport system.
As it is possible that the mutants described here could have other mutations, in addition to the transposon insertion, complementation would be required to definitively establish the role of the genes interrupted by the transposon in virulence in M. gallisepticum. Unfortunately, stable extrachromosomal expression of introduced genes, which would be needed to perform experimental infection studies, is difficult to achieve with our current tools for genetic manipulation of M. gallisepticum. In addition, the oppD insertions may have polar effects on the expression of other genes in the operon. However, the attenuation of virulence, albeit to different levels, by two independent insertions in the oppD gene strongly suggests that this gene in particular does play a role in virulence. Furthermore, recent studies have shown that this operon can play a role in virulence in both Gram-negative and Gram-positive bacteria (22, 23).
Notably, genes MGA_1102 and MGA_1079, which, on the basis of the results of the studies described here, may be required for full pathogenicity in M. gallisepticum, appear to have been transferred horizontally from M. synoviae, as all their most closely related homologs are found in mycoplasmas in the Hominis phylogenic group. The protein encoded by MGA_1102 has a zinc peptidase-like motif at its carboxyl end, while the protein encoded by MGA_1079 has a trypsin-like peptidase motif at its carboxyl end.
Thus, in the work presented here, we used STM to screen pools of mutants of M. gallisepticum and identified a number of genes that may have a role in establishing infection in the natural host of this pathogen and in pathogenicity. Our results suggest that genes that appear to have been acquired by horizontal gene transfer from M. synoviae, MGA_1102 and MGA_1079, as well as the oppD2 gene, which has been complemented by horizontal gene transfer from M. synoviae, may play a significant role in the pathogenicity in this important pathogen, even though they are not essential for colonization and persistence.
MATERIALS AND METHODS
Bacterial strain and construction of mutants.
Virulent M. gallisepticum strain Ap3AS was originally isolated from the air sacs of a broiler chicken with airsacculitis (24). It was cultured in modified Frey's broth (MB) containing 10% swine serum (25), as described previously (26).
Plasmid pISM 2062.2, carrying the transposon Tn4001mod, was used to construct a signature-tagged (ST) library, as described previously (5, 27,–30). The transposon contained a gentamicin resistance gene, allowing selective culture. The presence of the different tags in the ST mutants was confirmed by PCR using the P2/P4 primer set (Table 7) (5). The insertion site of the transposon in each mutant was determined by genomic DNA sequencing using the IGstmGenmeF3 primer (Table 7) and comparing the sequence to that of the M. gallisepticum strain Rlow genome (15).
TABLE 7
Oligonucleotides used in this study
| Name | Use | 5′–3′ sequence (size [bp]) |
|---|---|---|
| P2 | Signature tag region PCR | ATCCTACAACCTCAAGCT (18) |
| P4 | Signature tag region PCR | ATCCCATTCTAACCAAGC (18) |
| STM13-KE-complement-1-Rev | Wild-type Ap3AS PCR | CCACAGAGAACTTGAAG (17) |
| STM13-KF-C | Wild-type Ap3AS PCR | TATAAACCTGGTACGG (16) |
| IGstmGenmeF3 | DNA sequencing and ST mutant PCR | GGACTGTTATATGGCCTTTTTGGATC (26) |
| STM03-B-Rev | ST mutant PCR | ACAGCTTGACGTTTTCCA (18) |
| STM18-Rev | ST mutant PCR | AGCAAAATTTCCACCCAAGA (20) |
| STM20-Rev | ST mutant PCR | GAGACATCAATCGCGGTTTT (20) |
| JZ-STM22 | ST mutant PCR | CTCAATTGAAGAATTATATGATG (23) |
| STM26-Rev | ST mutant PCR | CATCCCACCTGAAAACTCGT (20) |
Screening of ST mutants in infected birds.
All infection experiments were approved by the University of Melbourne Animal Ethics Committee. The experimental design has been described previously (5). Briefly, in the initial screening, 102 ST mutants labeled with 34 different signature tags were cultured and combined into three pools, each of which contained 34 different mutants with distinct tags (Tables 1 and and2),2), and each pool was used to infect 20 specific-pathogen-free (SPF) chickens by aerosol (31), with an additional 10 uninfected chickens being included with each group to assess the transmissibility of the mutants. Necropsies were conducted on 10 inoculated birds at 14 days after infection and on the remaining 20 birds at 28 days after infection. Gross air sac lesions were assessed for severity as described previously (32). Swab specimens of the trachea and air sacs of each bird were assessed by culture in mycoplasma broth and on mycoplasma agar, as described previously (5), and the signature tags present were amplified by PCR using the P2/P4 primer pair if the broth culture showed signs of growth of M. gallisepticum. The tags present were identified by dot blotting, as described previously (5). Blood samples were collected from all birds before inoculation and before euthanasia, and the serum was assessed for antibody against M. gallisepticum using a commercial agglutination antigen (Intervet International) in the rapid serum agglutination (RSA) test (33).
Dilutions of the ST mutants that were not reisolated in the initial screening experiment were subjected to confirmatory screening to further reduce the number of candidate ST mutants for definitive testing. The mutants were separated into two pools, with the mutants in each pool being distinguishable from each other by their distinct tags. Each pool contained a similar concentration of each mutant, and each pool was used to infect 20 birds, as described above. In-contact controls were not included, but all 40 birds were placed into the same positive-pressure fiberglass isolator so that each group could be used as an in-contact group for the other. Necropsies and sample collection were performed as described above at 14 days after inoculation. Dot blot hybridization and ST mutant-specific PCRs were performed to identify the ST mutants recovered.
Comparison of the levels of persistence and virulence of selected mutants.
Several mutants were detected infrequently in the second screening experiment, suggesting that the genes that were interrupted in these mutants may have a significant influence on virulence. Two different oppD1 ST mutants were recovered from birds at distinct rates, suggesting that the different insertion sites may have different impacts on persistence in vivo and potentially on pathogenicity. To assess this and to assess the persistence and virulence of three mutants with insertions in hypothetical genes, the pathogenicities and rates of colonization of five distinct mutants were compared. Seven groups of 20 4-week-old SPF chickens were housed separately. Group 1 was exposed to an aerosol of mycoplasma broth only, group 2 was exposed to MGA_1102 ST mutant 03-1, group 3 was exposed to MGA_0588 ST mutant 18-1, group 4 was exposed to oppD1 ST mutant 20-1, group 5 was exposed to MGA_1079 ST mutant 22-1, group 6 was exposed to oppD1 ST mutant 26-1, and the 7th group was exposed to wild-type strain Ap3AS. All birds were subjected to necropsies at 14 days after infection, with samples being collected as described above. Sections of the upper, middle, and lower trachea were also taken from each bird.
Dot blot hybridization and specific PCRs were used to identify ST mutants reisolated in broth cultures. Because strain Ap3AS did not have a specific sequence tag, dot blot hybridization could not be used for cultures from the positive-control group. A pair of PCR primers, STM13-KE-complement-1-Rev and STM13-KF-C, which yielded a 400-bp product, were used to confirm the presence of Ap3AS (Table 7) (5). Sections of the trachea were processed and stained for histological examination and scored for mucosal lesions, and the mucosal thickness was measured (32). Statistical analyses were conducted using Kruskal-Wallis tests to compare median tracheal lesion scores, with the Dunn post hoc test being used to assess the significance of differences between pairs of groups, and an analysis of variance and Tukey's test were used to evaluate the mean tracheal mucosal thicknesses. Differences in the proportions of birds with lesions, of birds with serum antibody against M. gallisepticum, and of birds positive by culture for M. gallisepticum were assessed using Fisher's exact test (34). A probability of less than or equal to 0.05 was regarded as significant.
SDS-PAGE and Western blot analysis of ST mutants.
The cells in a 1-ml sample of a mid-log-phase culture of each of the ST mutants, as well as the Ap3AS strain, were collected by centrifugation at 16,000 × g for 5 min and then resuspended in 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) lysis buffer and incubated at 100°C for 5 min before rapid cooling on ice. Total cell proteins were separated in a 12.5% polyacrylamide gel together with molecular mass standards (Mark12 wide-range protein standard; Novex) and then stained with Coomassie brilliant blue.
Whole-cell proteins of MGA_0588ST mutant 18, oppD1 mutant 20, oppD1 mutant 26, and M. gallisepticum strain Ap3AS were subjected to SDS-PAGE, Western blotting, and immunostaining to detect differences in the proteins expressed in the mutants.
Total cell proteins of mutants with protein profiles that differed in Coomassie-stained gels were separated in a 10% polyacrylamide gel along with molecular mass standards (PageRuler prestained protein ladder; Thermo Scientific) and electrophoretically transferred onto a polyvinylidene difluoride (PVDF) membrane (Immobilon-P transfer membrane; Millipore), and antigen-free sites were blocked by overnight incubation in 5% skim milk (Devondale) in phosphate-buffered saline (PBS) containing 0.1% Tween 20 (PBS-T) at 4°C. The membrane was then washed three times (10 min each time) in PBS-T and then incubated in pooled sera collected from birds infected with Ap3AS (1:2,000 in PBS-T) at room temperature for 1 h with gentle rocking, again washed as described above, and then incubated for 1 h at room temperature in anti-chicken immunoglobulin-horseradish peroxidase conjugate (Dako) at a dilution of 1:2,000 in PBS-T. After again washing as described above, bound conjugate was detected using enhanced chemiluminescence (CD/DAB substrate kit; Thermo Scientific) following the manufacturer's recommendations, and the results were recorded digitally using a Bio-Rad molecular imager (Bio-Rad) according to the manufacturer's instructions.
ACKNOWLEDGMENTS
This work was partially supported by funding from the Australian Egg Corporation Ltd. and the Australian Poultry CRC.
We thank Chris Minion, Amir Hadji Noormohammadi, Cheryl Colson, and Tony Belfiore for their assistance with these studies.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00023-17.
REFERENCES
Articles from Infection and Immunity are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/iai.00023-17
Read article for free, from open access legal sources, via Unpaywall:
https://iai.asm.org/content/iai/85/6/e00023-17.full.pdf
Citations & impact
Impact metrics
Article citations
Unveiling genome plasticity and a novel phage in Mycoplasma felis: Genomic investigations of four feline isolates.
Microb Genom, 10(3), 01 Mar 2024
Cited by: 1 article | PMID: 38546735 | PMCID: PMC11004492
Virulence factors of Mycoplasma synoviae: Three genes influencing colonization, immunogenicity, and transmissibility.
Front Microbiol, 13:1042212, 25 Nov 2022
Cited by: 2 articles | PMID: 36532420 | PMCID: PMC9749132
Infection strategies of mycoplasmas: Unraveling the panoply of virulence factors.
Virulence, 12(1):788-817, 01 Dec 2021
Cited by: 22 articles | PMID: 33704021 | PMCID: PMC7954426
Review Free full text in Europe PMC
A Mycoplasma gallisepticum Glycerol ABC Transporter Involved in Pathogenicity.
Appl Environ Microbiol, 87(11):e03112-20, 11 May 2021
Cited by: 8 articles | PMID: 33741628 | PMCID: PMC8208142
Deletion of the oligopeptide transporter Lmo2193 decreases the virulence of Listeria monocytogenes.
J Vet Sci, 21(6):e88, 01 Nov 2020
Cited by: 5 articles | PMID: 33263235 | PMCID: PMC7710461
Go to all (8) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences
- (1 citation) ENA - AE017245
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Metabolite profiling of Mycoplasma gallisepticum mutants, combined with bioinformatic analysis, can reveal the likely functions of virulence-associated genes.
Vet Microbiol, 223:160-167, 02 Aug 2018
Cited by: 10 articles | PMID: 30173742
Safety and efficacy of a Mycoplasma gallisepticum oppD knockout mutant as a vaccine candidate.
Vaccine, 35(45):6248-6253, 21 Sep 2017
Cited by: 2 articles | PMID: 28941621
MalF is essential for persistence of Mycoplasma gallisepticum in vivo.
Microbiology (Reading), 159(pt 7):1459-1470, 08 May 2013
Cited by: 14 articles | PMID: 23657682
Haemagglutinins of pathogenic avian mycoplasmas.
Avian Pathol, 31(6):535-547, 01 Dec 2002
Cited by: 26 articles | PMID: 12593736
Review
Funding
Funders who supported this work.
Australian Egg Corporation Limited (1)
Grant ID: UM54-A
Australian Poultry Cooperative Research Centre (1)
Grant ID: 3.11
 a and
a and 




