Abstract
Free full text

PNAS Plus
Mass spectrometric identification of intermediates in the O2-driven [4Fe-4S] to [2Fe-2S] cluster conversion in FNR
Significance
The transcriptional regulator Fumarate and Nitrate Reduction is the master switch for the transition between anaerobic and aerobic respiration in many other bacteria. It fulfills this role by controlling the expression of >300 genes in response to O2. It senses O2 through a [4Fe-4S] cluster cofactor, which undergoes conversion to a [2Fe-2S] cluster on reaction with O2, leading to loss of DNA binding. By using time-resolved electrospray ionization mass spectrometry, we gained insight into the reaction through detection of cluster conversion intermediates and products, including a [3Fe-3S] cluster. The data also show that sulfide released from the cluster is oxidized during the conversion reaction and not after it. Our methodology has great potential for broad application to studies of cofactor reactivities.
Abstract
The iron-sulfur cluster containing protein Fumarate and Nitrate Reduction (FNR) is the master regulator for the switch between anaerobic and aerobic respiration in Escherichia coli and many other bacteria. The [4Fe-4S] cluster functions as the sensory module, undergoing reaction with O2 that leads to conversion to a [2Fe-2S] form with loss of high-affinity DNA binding. Here, we report studies of the FNR cluster conversion reaction using time-resolved electrospray ionization mass spectrometry. The data provide insight into the reaction, permitting the detection of cluster conversion intermediates and products, including a [3Fe-3S] cluster and persulfide-coordinated [2Fe-2S] clusters [[2Fe-2S](S)n, where n = 1 or 2]. Analysis of kinetic data revealed a branched mechanism in which cluster sulfide oxidation occurs in parallel with cluster conversion and not as a subsequent, secondary reaction to generate [2Fe-2S](S)n species. This methodology shows great potential for broad application to studies of protein cofactor–small molecule interactions.
A remarkable feature of bacteria is their ability to adapt to rapidly changing environments. Many bacterial species, including the model organism Escherichia coli, are capable of growth on a variety of substrates under varying oxygen tensions. In the absence of O2, alternative electron acceptors, such as fumarate or nitrate, are used to support growth (1). However, these alternative metabolic pathways are less efficient than aerobic respiration. Therefore, the ability to monitor the availability of O2 and respond by reprogramming gene expression is necessary for these bacteria to remain competitive.
In E. coli and many other bacteria, the O2-sensing Fumarate and Nitrate Reduction (FNR) protein is the master regulator of the switch between aerobic and anaerobic metabolism (2–5). The first crystal structure of dimeric holo-FNR, from Aliivibrio fischeri, was recently reported (6). AfFNR is the same length as and shares 84% sequence identity with E. coli FNR. Like other members of the cAMP receptor protein superfamily, FNR comprises two distinct domains, providing sensory and DNA binding functions linked by a dimer interface (Fig. 1A). The N-terminal sensory domain contains four essential Cys residues (Cys20, -23, -29, and -122) (7–9) that are capable of binding either a [4Fe-4S]2+ (Fig. 1B) or a [2Fe-2S]2+ cluster. The C-terminal DNA binding domain recognizes specific FNR binding sequences within target promoters. In the absence of O2, monomeric (~30 kDa) FNR acquires a [4Fe-4S]2+ cluster, triggering a conformational change at the dimerization interface that leads to the formation of homodimers (~60 kDa) and site-specific DNA binding (10, 11). Kiley and coworkers (8, 12) established that the [4Fe-4S]2+ EcFNR is converted into a [2Fe-2S]2+ form both in vivo and in vitro and that this results in a rearrangement of the dimer interface, leading to monomerization (10).

Crystal structure of FNR from A. fischeri. (A) AfFNR is a homodimer composed of monomeric subunits (blue and brown). Each subunit binds an all cysteine-ligated [4Fe-4S] cluster (shown as space filled) and contains an N-terminal sensory domain, a C-terminal DNA binding domain, and a large dimerization helix that forms the dimer interface. (B) The [4Fe-4S] cluster binding loop of AfFNR (Protein Bata Dank ID code 5E44). The location of Ser24 is shown relative to the positions of the cluster ligands (Cys20, -23, -29, and -122).
Through a combination of visible absorbance and EPR spectroscopies, we identified an EPR active [3Fe-4S]1+ (S = 1/2) species as a transient intermediate in the cluster conversion process, indicating a two-step process (1 and 2) (13, 14):
In step 1, an Fe2+ ion is released to generate the [3Fe-4S]1+ intermediate. This release likely occurs after one-electron oxidation of the [4Fe-4S]2+ cluster to yield an unstable [4Fe-4S]3+ cluster that ejects Fe2+. Step 2 corresponds to the conversion of the [3Fe-4S]1+ species to the [2Fe-2S]2+ cluster. Although two sulfide ions are released from the cluster during the [4Fe-4S]2+ to [2Fe-2S]2+ cluster conversion (15), how this happens is unknown. They may be ejected into aqueous solution or undergo two-electron oxidation to form sulfane (S0), which subsequently reacts with Cys side chains (RS−) to form persulfides (RSS−). The latter can serve as ligands to the [2Fe-2S] cluster as recently shown (16, 17). Similarly, liquid chromatography mass spectrometry (LC-MS) experiments showed that multiple cysteine persulfides [RS(S)n, where n = 1–4] were formed during the FNR reaction time course (16). Step 2 may thus also be O2-dependent and, hence, more complex than initially envisaged (3 and 4):
or
A persulfide-coordinated [2Fe-2S] cluster may serve to retain sulfur that can be used for conversion back to the original [4Fe-4S] form. This process was shown on addition of a reductant (to reduce sulfane back to sulfide) plus Fe2+. This reversibility points to a pathway for iron-sulfur cluster repair that does not need to involve Fe-S biogenesis machinery (16). The complexity of the [3Fe-4S] to [2Fe-2S] conversion process means that it probably involves more than one reaction. Additional complexity arises depending on whether sulfide oxidation occurs simultaneously with iron release or after it.
Here we report time-resolved electrospray ionization mass spectrometry (ESI-MS) of E. coli FNR in response to its physiologically relevant analyte, O2. This methodology couples soft ESI and TOF detection with solution and ionization conditions under which proteins remain folded (18, 19). This experimental approach enables accurate mass detection of intact folded proteins and protein complexes and has already been used extensively to study protein–protein interactions and the interactions of proteins with other molecules (20, 21) as well as protein structural changes (19, 22). ESI-MS of metalloproteins, where the metal cofactor remains bound after vaporization/ionization, has also been shown to be a valuable technique for the study of metalloproteins (23–25), including iron-sulfur proteins (26, 27), but still remains relatively underexploited given its potential to provide significant insight into the chemistry taking place at metal cofactor active sites.
For this study, we have used the S24F variant of FNR, which undergoes the same cluster conversion reaction as the WT protein but at a significantly reduced rate (28), thus facilitating analysis by ESI-MS. The data provide an unusually detailed view of the cluster conversion process, revealing the formation of a [3Fe-3S] protein-bound intermediate and the oxidation of cluster sulfide during cluster conversion.
Results
Detection of [4Fe-4S] FNR by ESI-MS.
ESI-MS was used to detect [4Fe-4S] S24F FNR (hereafter referred to as FNR). The m/z spectrum (Fig. 2A) has three distinct regions corresponding to monomeric [4Fe-4S] FNR (1,000–2,500 m/z), monomeric apo-FNR containing multiple sulfur adducts (2,300–3,100 m/z), and dimeric [4Fe-4S] FNR (3,000–4,500 m/z). The deconvoluted spectrum of [4Fe-4S] FNR (Fig. S1A) revealed a major species at 29,898 Da caused by monomeric [4Fe-4S] FNR (predicted mass of 29,897 Da) (Table 1 shows a list of observed and predicted masses). A number of low-intensity, poorly resolved features were also observed to the higher mass side of [4Fe-4S] FNR, the clearest of which was at +40 Da. This peak was also present in LC-MS spectra, indicating that it arises from a small proportion of the protein that is covalently modified in some way (Fig. S2). In the lower-mass region, a minor peak was observed at 29,545 Da, corresponding to apo-FNR (predicted mass of 29,547 Da), with a 2-Da shift possibly caused by a single disulfide bond. More intense peaks were also observed at 29,609 and 29,673 Da, corresponding to two and four sulfur adducts of apo-FNR (+64 and +128 Da), respectively. Appreciable amounts of [2Fe-2S] FNR at 29,720 Da (predicted mass of 29,721 Da) were detected in some samples, indicative of a small amount of O2-mediated damage. Optimization of ionization conditions to favor the detection of the monomeric form of [4Fe-4S] FNR resulted in m/z and deconvoluted spectra (Fig. 2B and Fig. S1B) dominated by [4Fe-4S] FNR, with significantly less (<12%) of the lower mass forms of FNR.

ESI-MS of [4Fe-4S] FNR and the effect of exposure to O2. (A) Full m/z spectrum for FNR. Charge states corresponding to monomeric [4Fe-4S] (black), persulfide adducts of apo (gray), and dimeric [4Fe-4S] (red) FNR species are shown. (B) Deconvoluted mass spectrum of [4Fe-4S] S24F FNR (black line) before and after exposure to dissolved atmospheric oxygen (18 min exposure; red line). This exposure results in the formation of a variety of protein-bound clusters, including [3Fe-4S], [3Fe-3S], and [2Fe-2S] forms. Persulfide adducts of [2Fe-2S] and apoprotein are also observed. (C) Deconvoluted spectrum of dimeric FNR showing the presence of the [4Fe-4S]/[4Fe-4S] form under anaerobic conditions and only low-intensity features caused by cluster conversion species after the addition of O2 [20 (gray line) and 80 min (red line)]. The longer exposure time resulted in the loss of all cluster-containing dimer species, and the appearance of a broad, poorly resolved feature centered on 59,320 Da, approximating the mass of FNR containing multiple sulfane sulfurs (S0n, nave ~ 8). (D) 3D plot showing the formation and decay of all monomeric FNR species during the O2 reaction time course. Spectra are representative of multiple repeat experiments.
Table 1.
Predicted and observed masses for cluster-bound forms of FNR
| FNR species | Predicted mass (Da)* | Average observed mass (Da)† | ΔMass (Da)‡ |
| Monomeric FNR | |||
 [4Fe-4S]2+ [4Fe-4S]2+ | 29,897 | 29,898 | +1 |
 [3Fe-4S]1+ [3Fe-4S]1+ | 29,842 | 29,843 | +1 |
 [3Fe-3S]3+ [3Fe-3S]3+ | 29,808 | 29,811 | +3 |
 [3Fe-3S]1+ [3Fe-3S]1+ | 29,810 | 29,811 | +1 |
 [2Fe-4S]2+ [2Fe-4S]2+ | 29,785 | 29,784 | −1 |
 [2Fe-3S]2+ [2Fe-3S]2+ | 29,753 | 29,752 | −1 |
 [2Fe-2S]2+ [2Fe-2S]2+ | 29,721 | 29,720 | −1 |
| Dimeric FNR | |||
 [4Fe-4S]2+/[4Fe-4S]2+ [4Fe-4S]2+/[4Fe-4S]2+ | 59,794 | 59,796 | +2 |
 [4Fe-4S]2+/[3Fe-4S]1+ [4Fe-4S]2+/[3Fe-4S]1+ | 59,739 | 59,740 | +1 |
 [3Fe-4S]1+/[3Fe-4S]1+ [3Fe-4S]1+/[3Fe-4S]1+ | 59,684 | 59,685 | +1 |
 [3Fe-3S]3+/[3Fe-4S]1+ [3Fe-3S]3+/[3Fe-4S]1+ | 59,650 | 59,653 | +3 |
 [3Fe-3S]1+/[3Fe-4S]1+ [3Fe-3S]1+/[3Fe-4S]1+ | 59,652 | 59,653 | +1 |
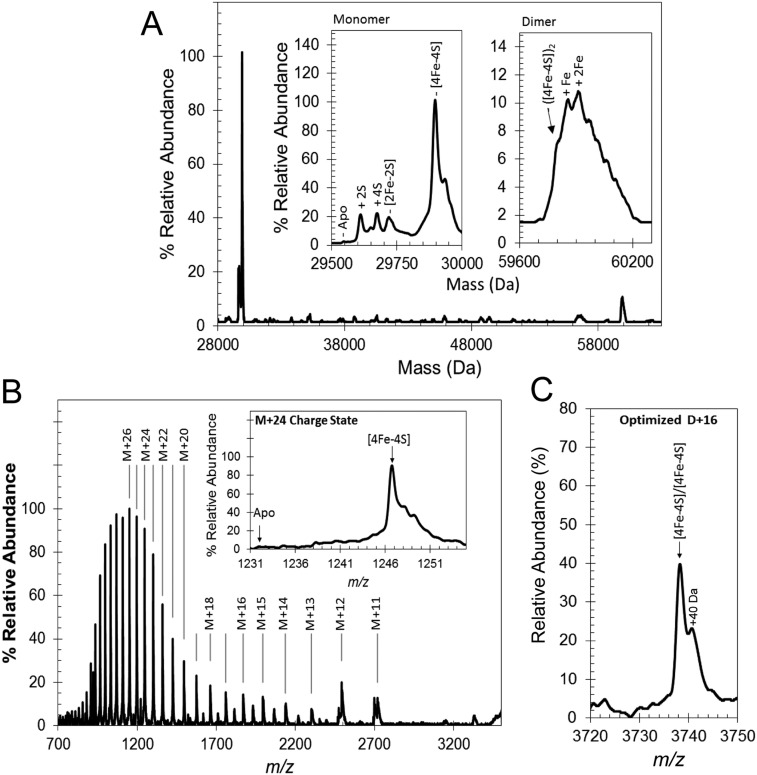
Native MS of [4Fe-4S] FNR. (A) Deconvoluted spectrum (28,000–60,300 Da) corresponding to the full m/z spectrum measured for FNR (shown in Fig. 2A). Insets are the monomeric (Left) and dimeric (Right) regions of the spectrum in more detail. (B) Full m/z spectrum optimized for passage of [4Fe-4S] monomeric charges states. Inset is the corresponding deconvoluted spectrum. (C) The m/z spectrum for the +16 charge state of dimeric [4Fe-4S] measured under optimized conditions for the passage of dimeric FNR.
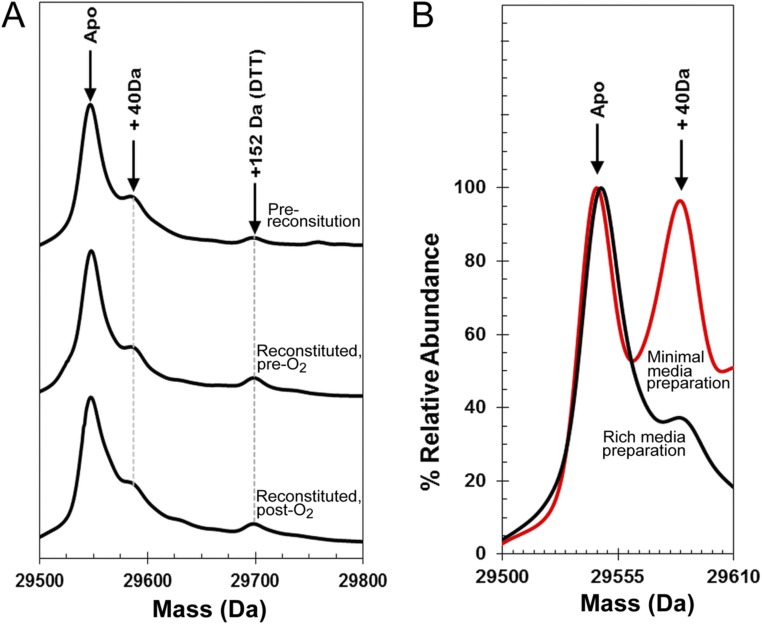
LC-MS of FNR. (A) Isolated FNR (prereconstitution), FNR after reconstitution of the [4Fe-4S] cluster, and reconstituted FNR after exposure to O2 as indicated. Each sample was treated with 20 mM DTT before measurement to remove any persulfides or other readily reducible (and removable) species. The data show that FNR samples contain a small amount of a covalently modified protein at +40 Da and that there is no significant formation of oxygen adducts of FNR after exposure to O2. (B) FNR protein samples purified from Escherichia coli cells grown in rich media (black spectrum) and minimal media (red spectrum). The data illustrate that the +40-Da form of the protein is significantly increased relative to the unmodified form when E. coli cells are grown under nutrient-limiting conditions. The nature of the modification is unknown.
Because [4Fe-4S] FNR is a dimer in solution, the observation of monomer [4Fe-4S] FNR suggests that the protein–protein interactions that mediate dimerization did not survive the ionization process. However, dimeric FNR was also detected in the m/z spectrum (Fig. 2A), and in the deconvoluted spectrum (Fig. S1A), a broad low-intensity peak centered on 59,906 Da was observed that corresponds to dimeric FNR containing iron-sulfur clusters. There is a shoulder at the predicted mass for the [4Fe-4S] FNR dimer (59,794 Da), but the more intense peaks correspond to various adduct species, including likely single (+56 Da) and double (+112 Da) iron adducts. Optimization of experimental conditions for the transmission of dimeric species (2,750–5,000 m/z) provided sharper, better resolved dimer charge states (Fig. S1C). The deconvoluted spectrum of dimeric [4Fe-4S] FNR (Fig. 2C) contained a well-resolved peak centered on 59,796 Da, indicative of dimeric [4Fe-4S] FNR; around the dimer peak were a number of poorly resolved features, with the principal high-mass species caused by one covalently modified subunit within the dimer at +40 Da. No peaks for dimeric [2Fe-2S] or apo-FNR species were observed, consistent with them being monomeric in solution.
ESI-MS Reveals Mechanism of O2 Sensing.
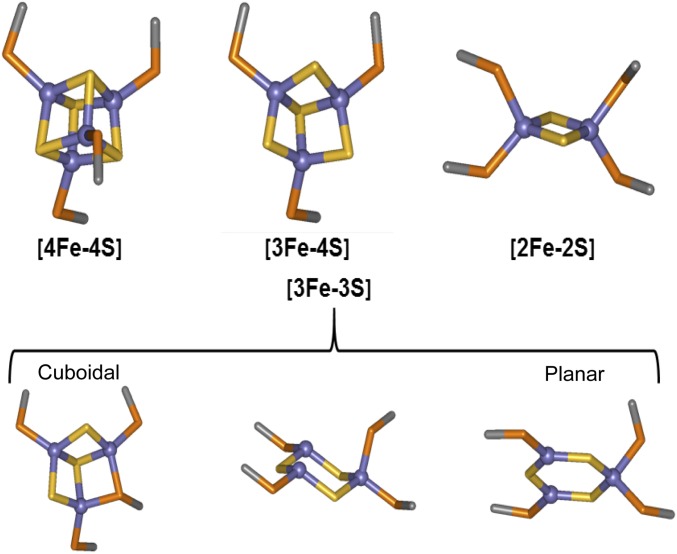
To study the mechanism of O2 sensing by FNR, optimized ionization conditions for detection of the monomeric rather than dimeric form of [4Fe-4S] FNR were used, so that intermediate species could be clearly and unambiguously identified (Fig. 2B). The addition of O2 (under pseudofirst-order reaction conditions where the [O2]:[4Fe-4S] ratio was ~10) resulted in the formation of a complex series of overlapping peaks (Fig. 2B). These peaks can be subdivided into two distinct groups corresponding to protein-bound cluster fragments (29,700–29,850 Da) and sulfur adducts of apo-FNR (29,550–29,700 Da). The cluster-bound fragments include the [2Fe-2S] end product of the cluster conversion reaction (at 29,720 Da) along with species with masses that correspond to [2Fe-3S] (29,752 Da) and [2Fe-4S] (29,784 Da) forms, which most likely represent singly and doubly persulfide-coordinated [2Fe-2S] clusters, respectively, as recently shown by resonance Raman (16) (3 and 4). In addition, a peak corresponding to a [3Fe-4S] form that is a well-characterized intermediate of the cluster conversion process (13, 14) was observed at 29,843 Da (Table 1). Each of these species has been previously observed in solution using various spectroscopic methods. The spectra also contained a peak at 29,811 Da consistent with the presence of a [3Fe-3S] cluster form (Table 1), which may represent a previously undetected intermediate in the conversion process, in which one of two sulfides that are eventually lost from the original [4Fe-4S] cluster has been ejected.
To investigate further these intermediates of cluster conversion, a 34S-substituted form of [4Fe-4S] FNR was generated (16) The deconvoluted spectrum contained a major peak at 29,905 Da, +8 Da relative to the predicted mass of natural abundance [4Fe-4S] FNR (Fig. 3A). The predicted increase in mass on substitution of all sulfides is +7.6 Da (calculated from natural isotope abundance). Exposure of the 34S-labeled [4Fe-4S] FNR sample to O2 resulted in cluster conversion as described above, except that the peaks corresponding to the cluster breakdown products were mass shifted as follows: [3Fe-4S] by +8 Da, [2Fe-2S] by +4 Da, [2Fe-3S] by +6 Da, and [2Fe-4S] by +8 Da (Fig. 3 A and B). The isotope substitution data also showed that the sulfur adducts of apo-FNR observed after reaction of [4Fe-4S] FNR with O2 are derived from the cluster (Fig. 3C) and that oxygen adducts of FNR are not formed (Fig. S2). Overall, the isotope substitution data provide unambiguous confirmation of the assignments for the cluster conversion intermediates and products.
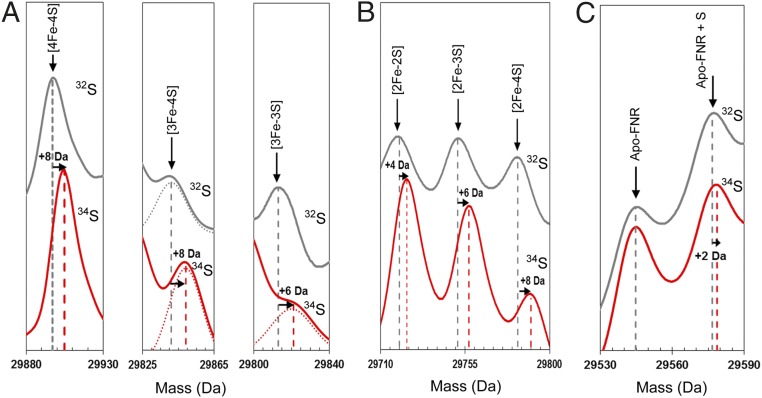
Mass shifts observed for the FNR [4Fe-4S] cluster, conversion intermediates, and cluster products on 34S substitution of cluster sulfides. (A) Deconvoluted mass spectra of natural abundance sulfur [4Fe-4S] S24F FNR and cluster conversion intermediates (black lines) and the equivalent 34S-substituted forms (red lines) as indicated. Dotted lines represent Gaussian fits of the MS data. (B) The same as in A except that spectra represent cluster conversion products as indicated. (C) The same as in A except that spectra show the apo-FNR peak and the first persulfide adduct. Predicted mass shifts for the assigned species are indicated.
To investigate the kinetic behavior of these species, [4Fe-4S] FNR was combined with excess dissolved O2 and continuously infused into the ESI source of the instrument. Fig. 2D shows the deconvoluted spectra obtained during the reaction time course over 120 min. Identical measurements of FNR that was not exposed to O2 before infusion showed that the changes observed were entirely caused by O2 exposure. At the earliest time points, a series of overlapping peaks identical in mass to those in Fig. 2B was observed. Peaks corresponding to the protein-bound cluster fragments (29,700–29,850 Da) increased in intensity, reaching a maximum ~15–30 min post-O2 exposure. During this period, the [4Fe-4S] peak remained the most abundant species before decaying away along with the protein-bound cluster fragment peaks at later time points. In contrast, peaks corresponding to sulfur adducts of apo-FNR continued to rise to a maximum ~30–50 min postexposure (Fig. 2D).
Abundances of the different cluster fragments and apoprotein species in Fig. 2D were plotted as a function of time. In MS experiments, abundances are reported relative to the most abundant species (arbitrarily set to 100%). For FNR experiments, the most abundant species over most of the time course was the [4Fe-4S] cluster, and therefore, these data fail to provide any information on the rate of [4Fe-4S] cluster decay. To enable global analysis to be performed, absorbance spectroscopy was, therefore, used to measure (at 406 nm) the decay of the cluster under conditions identical to those used for MS experiments. The absorbance data were converted to relative abundance (with the starting point set to 100%) for direct comparison with the MS data. Global analysis of multiple (n = 4) mass spectrometric kinetic datasets leads to the reaction scheme shown in Fig. 4A. This scheme was able to model the formation and/or decay of the peak intensities corresponding to the previously characterized [4Fe-4S], [3Fe-4S], [2Fe-2S], and apo-FNR species (Fig. 4B), indicating that the [3Fe-4S] cluster is the first intermediate formed (maximizing before 20 min) followed by [2Fe-2S] and apo-FNR. The temporal nature of [3Fe-3S], which maximized at ~20 min (Fig. 4C), is consistent with an intermediate in the [4Fe-4S] to [2Fe-2S] cluster conversion pathway. However, both the [2Fe-4S] and [2Fe-3S] species (persulfide-coordinating forms of [2Fe-2S]) also reach a maximum at ~20 min before the [2Fe-2S] cluster (~30 min) (Fig. 4 D and E, respectively). The data show that [2Fe-3S] is not formed from [2Fe-2S] or the reverse; rather, [3Fe-3S] clusters can give rise to both [2Fe-3S] and [2Fe-2S] clusters, and therefore, this part of the reaction represents a branch point in an otherwise largely linear reaction mechanism. Similarly, the [2Fe-4S] cluster must form from [3Fe-4S] (and not [3Fe-3S]), and therefore, the formation of this species represents a second branch point (Fig. 4A).
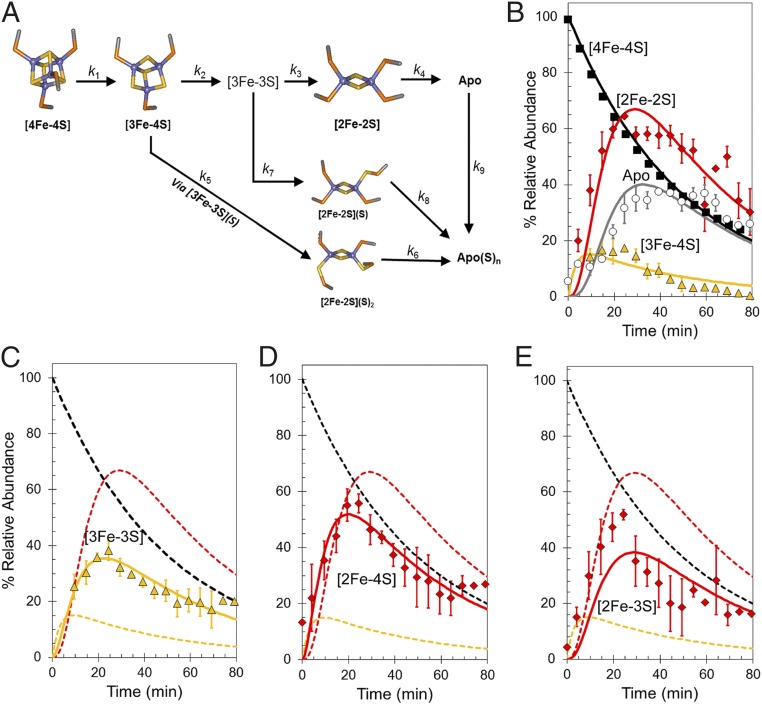
Mechanism of [4Fe-4S] cluster conversion. (A) Reaction scheme used to simulate the kinetic dependence native MS data. (B) Plots of relative abundances of [4Fe-4S] cluster (A406 nm; black squares), [3Fe-4S] (yellow triangles), [2Fe-2S] cluster (red diamonds), and apo (white circles) species as a function of time after exposure to excess O2. Global fitting to the experimental data using the reaction scheme depicted in A is shown as solid lines. (C–E) Plots of relative abundances of the [3Fe-3S] cluster (yellow triangles in C), the [2Fe-4S] cluster (red diamonds in D), and the [2Fe-3S] cluster (red diamonds in E). Global fits to the experimental data are shown as solid lines. Dashed lines in C–E show the responses of the [4Fe-4S], [3Fe-4S], and [2Fe-2S] clusters for easy comparison. Error bars show SEs for average MS datasets (n = 4). The global fitting model was initiated with 100% relative abundance of [4Fe-4S] clusters.
The time dependence of intensity in the apoprotein region indicates that apo-FNR was converted to sulfane-containing species, apo(S)n, with essentially full conversion at ~80 min (Fig. 4B). Indeed, the global fit was dependent on this being part of the mechanistic scheme (Fig. 4A). Similarly, the [2Fe-3S] and [2Fe-4S] clusters, which both contain persulfides, give rise to apo(S)n species after the disassembly of their ligated [2Fe-2S] cluster as shown in Fig. 4A. An important feature of the data (both from MS and absorbance spectroscopy) is that the rate at which the apoprotein is formed from [2Fe-2S] in ammonium acetate buffer is greater than that previously reported for other buffer systems (13, 14). The poor stability of [2Fe-2S] species under these conditions means that they decay away in the MS spectrum rather than accumulating.
Overall, the model describes the behavior of the main [4Fe-4S], [3Fe-4S], and [2Fe-2S] cluster species, previously identified by spectroscopy, and the appearance of the [3Fe-3S], [2Fe-4S], [2Fe-3S], and apo-FNR species, all observed here simultaneously by MS (Fig. 4 B–E). We note that the model indicates that direct conversion of the [4Fe-4S] cluster to the [3Fe-3S] species does not occur. Thus, loss of a single iron seems to be an obligatory first step in the cluster conversion process, with loss of sulfide following that step.
Table 2 shows the observed rate constants required to describe the reaction scheme depicted in Fig. 4A. The first reaction, corresponding to the conversion of the [4Fe-4S] cluster into the [3Fe-4S] cluster, has an observed rate constant (kobs1) of 0.02 min−1 (0.003 s−1), implying that it is rate limiting as previously observed (13, 14). Division of kobs1 by the O2 concentration (122 μM) used here provides an estimate of the apparent second-order rate constant, k = 25 M−1 s−1. This value is lower than the previously reported value of 80 M−1 s−1 for S24F FNR (28). It is known that the [4Fe-4S] to [2Fe-2S] cluster conversion is influenced by the nature of the buffer environment, in this case ammonium acetate.
Table 2.
Observed rate constants used to fit experimental data shown in Fig. 4 B–E to the cluster disassembly model shown in Fig. 4A
| Reaction step | Observed rate constant (min−1)* | Reaction |
| k1 | 0.020 ± 5 × 10−5 | [4Fe-4S] → [3Fe-4S] |
| k2 | 0.123 ± 0.002 | [3Fe-4S] → [3Fe-3S] |
| k3 | 0.072 ± 0.001 | [3Fe-3S] → [2Fe-2S] |
| k4 | 0.200 ± 0.003 | [2Fe-2S] → apo |
| k5 | 0.200 ± 0.003 | [3Fe-4S] → [2Fe-4S] |
| k6 | 0.145 ± 0.003 | [2Fe-4S] → apo(S0)n |
| k7 | 0.040 ± 0.001 | [3Fe-3S] → [2Fe-3S] |
| k8 | 0.210 ± 0.006 | [2Fe-3S] → apo(S0)n |
| k9 | 0.330 ± 0.008 | apo → apo(S0)n |
Kinetics of Persulfide Adduct Formation Determined by LC-MS.
LC-MS, in which protein samples are in denaturing solvent, can be used to follow the formation of sulfane adducts during Fe-S cluster reactions, because these adducts are covalent species that survive protein unfolding (29). LC-MS was applied to samples of FNR equivalent to those used for native MS experiments. The reaction was quenched at specific time points to halt the reaction and inhibit any subsequent sulfur exchange, so that the time evolution of sulfane adducts could be correlated with the native MS data and used to monitor cluster sulfide oxidation. In the absence of O2, the major peak at 29,547 Da corresponded to apo-FNR without sulfur adducts (Fig. 5). Shoulder peaks at +32, +64, and +96 Da are caused by the addition of one, two, and three sulfane sulfur atoms, respectively (29).

Tracking sulfide oxidation by LC-MS and native MS. (A) Deconvoluted LC-MS spectra before (black line) and after (red line) exposure to excess O2 reveal the formation of persulfide adducts. The broad feature at 29,900 Da in the anaerobic spectrum corresponds to poorly resolved cluster fragments. (B) Survey plot showing the formation of persulfide adducts during the reaction time course. (C) Abbreviated reaction scheme based on that in Fig. 3A used to simulate the kinetic behavior of FNR persulfide species. (D) Plots of relative abundances of the apo-S (blue squares), apo-SS (gray triangles), and apo-SSS (yellow circles) species detected in native MS experiments. Global fitting to the experimental data using the reaction scheme depicted in Fig. 4C is shown as solid lines. Dashed lines show the responses of the [4Fe-4S] (black), [2Fe-2S] (red), and apo (gray) species for comparison. Error bars show SEs for average MS datasets (n = 4). The global fitting model was initiated with 100% relative abundance of [4Fe-4S] clusters.
The addition of O2 resulted in a 2-Da shift in the apo-FNR to 29,545 Da, consistent with the presence of a single disulfide bond. In addition, there was an increased abundance of peaks corresponding to the sulfane adducts as previously observed (16) (Fig. 5). As can be seen from Fig. 5B, the single or double sulfane adducts of FNR become the major species with increasing time. The time dependence of their appearance shows that sulfide oxidation occurs simultaneously with cluster conversion. From the native MS experiments above, it was suggested that the [2Fe-3S] and [2Fe-4S] species give rise to the single and double sulfane forms of apo-FNR, respectively, after the disassembly of the persulfide-ligated [2Fe-2S] cluster. The temporal appearance of the single and double sulfane adducts, maximizing at ~30–40 min, supports this hypothesis (Fig. S3).

Tracking sulfide oxidation by LC-MS. Plots of relative abundances of apo-S (gray circles), apo-SS (red circles), and apo-SSS (blue circles) detected by LC-MS after exposure to excess O2. Global fits to the experimental data using the reaction scheme depicted in Fig. 5C are shown as solid lines. The predicted response of the apo species is shown in yellow (note the similarity to the apo species’ behavior in Fig. 4B). The global fitting model was initiated with 100% relative abundance of [4Fe-4S] clusters.
The apo(S)n species detected by LC-MS result from not only the presence of apo(S)n species in solution but also, denaturation of cluster-bound sulfane adducts present at the point of quenching, and these adducts cannot be distinguished in the LC-MS experiment. However, because the [2Fe-2S] forms are not stable in the ammonium acetate buffer of the native MS experiment, the kinetic profiles of formation of apo(S)n species in the LC-MS are similar to those in the native MS experiment, and the LC-MS data could be readily modeled by extending the initial mechanistic model (Fig. 5C) to reflect the proposed fate of the [2Fe-2S] clusters (Fig. S3). An important feature of the extended model is that apo(S)n = 1–4 species are in equilibrium with each other, implying that sulfide is readily mobilized from one species to another, presumably via disulfide exchange. The extended model, initiated with [4Fe-4S] FNR at 100% relative abundance, describes the appearance of the apo(S)n = 1–3 adducts detected via LC-MS and importantly, was also able to model the formation of apo(S)n species detected by native MS (Fig. 5D). The observed rate constants obtained from the experimental data fits (Fig. 5D and Fig. S3) are given in Table 3.
Table 3.
Observed rate constants used to fit experimental data shown in Fig. 5D and Fig. S3 to the apo(S)n model shown in Fig. 5C
| Reaction step | Observed rate constant (min−1)* | Reaction |
| k1–8 | Table 2 | Cluster disassembly model |
| k9 | 0.900 ± 0.02 | apo → apo(S) |
| k13 | 0.050 ± 0.004 | apo ← apo(S) |
| k10 | 1.000 ± 0.01 | apo(S) → apo(S)2 |
| k14 | 0.400 ± 0.006 | apo(S) ← apo(S)2 |
| k11 | 1.000 ± 0.009 | apo(S)2 → apo(S)3 |
| k15 | 0.900 ± 0.01 | apo(S)2 ← apo(S)3 |
| k12 | 0.980 ± 0.01 | apo(S)3 → apo(S)4 |
| k16 | 0.034 ± 6 × 10−4 | apo(S)3 ← apo(S)4 |
Cluster Reactivity of Dimeric [4Fe-4S] FNR.
Under conditions where the ionization of dimeric FNR was optimized, addition of O2 resulted in three low-intensity peaks that increased in intensity with time, reaching a maximum at ~15 min, before decaying away along with all cluster-bound forms (Fig. 2B). These peaks correspond to dimeric FNR containing a [3Fe-4S] and [4Fe-4S] cluster (59,740 Da), two [3Fe-4S] clusters (59,685 Da), and a [3Fe-3S] and [3Fe-4S] cluster (59,653 Da) (Fig. S4). The low intensity of these peaks is consistent with cluster conversion initiating the dimer to monomer transition, such that only the [4Fe-4S] cluster can maintain a significant population of dimeric FNR. The equilibrium between monomer and dimer is apparently shifted toward monomer forms when [3Fe-4S] or [3Fe-3S] clusters are bound. With increasing time, a large, broad, and poorly resolved feature appeared. The average mass of this feature was ~59,348 Da, and it is most likely caused by disulfide-linked dimeric FNR, with an average of eight additional sulfur atoms.
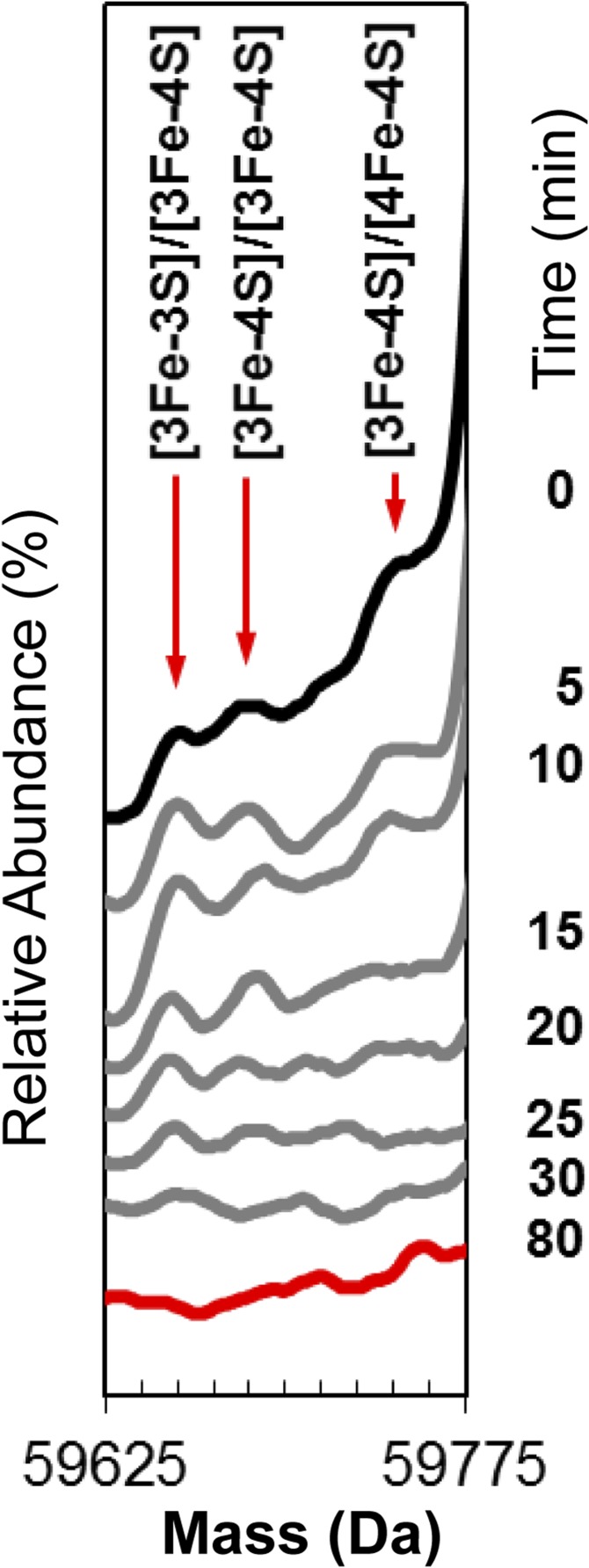
Dimeric FNR cluster conversion species. Stack plot of deconvoluted native mass spectra recorded as a function of time (as indicated) after exposure of dimeric [4Fe-4S] FNR to O2. The starting spectrum is in black (0 min), the final spectrum at 80 min is in red, and spectra at intervening time points are in gray. Minor peaks corresponding the formation of [3Fe-4S] and [3Fe-3S] clusters on dimeric FNR were observed to form and decay but at very low intensity.
Discussion
Here, we have used mass spectrometry to investigate the O2-mediated conversion of the [4Fe-4S] cluster of FNR to a [2Fe-2S] form, which causes dissociation of the FNR dimer into monomers and loss of high-affinity DNA binding (7–14). The S24F variant of FNR was used for this study, because previous studies showed that it reacts via the same two-step mechanism (1 and 2) as WT FNR but at a slower rate (28), enabling real-time mass spectrometry measurements. Thus, it is reasonable to conclude that the intermediates uncovered in this work also occur in WT FNR. The data presented here provide remarkable mechanistic detail of the conversion process.
ESI-MS experiments, including cluster-specific 34S isotopic substitution, and global analysis are consistent with previous spectroscopic studies showing that the first step of the reaction is the loss of Fe2+ from the [4Fe-4S]2+ cluster to form a [3Fe-4S]1+ cluster intermediate (13, 14, 30). This process is assumed to occur via oxidation of the cluster to an oxidized state, [4Fe-4S]3+, that immediately ejects an Fe2+ ion, forming the [3Fe-4S]1+ species (13, 30). After it is formed, the [3Fe-4S]1+ cluster is only transiently stable, ejecting an Fe3+ ion together with two sulfide ions (S2−) to generate the [2Fe-2S]2+ cluster form of FNR. Little is known about the rearrangement of the [3Fe-4S]1+ cluster to the [2Fe-2S]2+ form. The ESI-MS data reported here provide key insights into this. The detection of a [3Fe-3S] cluster that kinetic modeling shows is a cluster conversion intermediate results from the loss of one sulfide ion from the [3Fe-4S]1+ cluster, implying that the product is a [3Fe-3S]3+ species. An inorganic model [3Fe-3S]3+ cluster has recently been described for the first time, in which all iron and sulfide ions are in the same molecular plane forming a hexagonal arrangement (31). Previous studies of the [3Fe-4S] cluster of FdI ferredoxin from Pyrococcus furiosus by nondenaturing ESI Fourier transform ion cyclotron resonance MS revealed the presence of a [3Fe-3S]3+ cluster form along with other cluster breakdown species (26). In that case, the instability of the cluster was caused by ionization (the initial [3Fe-4S] cluster was stable in negative ion mode), but it provides clear independent evidence for the existence of a protein-bound [3Fe-3S] cluster. In the case of FNR, the charge state of the [3Fe-3S] cluster is not established. Assumption of a 3+ overall charge resulted in a reproducible difference between observed and predicted masses (Table 1). However, if 1+ charge is assumed, a very close match between observed and predicted masses was obtained. The same is true for the dimeric FNR species with [3Fe-3S] clusters bound. An overall charge of +1 on the cluster may arise from oxidation of the first sulfide released from the [3Fe-4S]1+ intermediate (that is, by release of sulfane).
A key feature of our kinetic model of the [3Fe-4S] to [2Fe-2S] conversion process is that the [3Fe-4S]1+ intermediate rapidly loses either a sulfide ion or a sulfane atom to form the [3Fe-3S]1+ or [3Fe-3S]3+ cluster, which undergoes additional reaction relatively slowly. Hence, conversion of the [3Fe-3S] intermediate is the rate-limiting step of this process. The observed rate constant (k3 in Fig. 4 and Table 2) of 0.072 min−1 (1.2 × 10−3 s−1) is comparable with that previously reported for the [3Fe-4S] to [2Fe-2S] conversion for both the S24F variant (5 × 10−4 s−1) and WT FNR (1.7 × 10−3 s−1) (28). This similarity suggests that the intermediate cluster species previously detected by EPR may actually be a [3Fe-3S] cluster rather than a [3Fe-4S] species as previously concluded, or it could be a mixture of the two. Analysis of the magnetic properties of the model [3Fe-3S]3+ cluster species revealed a paramagnetic S = 1/2 ground state (31), which gives rise to an X-band EPR spectrum that is similar to those of [3Fe-4S]1+ cluster proteins (31, 32) as well as that recorded for the FNR cluster conversion intermediate in both WT and S24F FNR (14, 28). This similarity could indicate that [3Fe-3S]3+ is formed on FNR, but we note that [3Fe-3S]1+ would also be paramagnetic, although the EPR properties of such a cluster are unknown.
Assuming that the hexagonal planar arrangement of the three Fe3+ and sulfide ions in the model [3Fe-3S] cluster is also a feature of the FNR [3Fe-3S] species (Fig. S5), this arrangement suggests how the cuboid [4Fe-4S] cluster may rearrange to form the planar [2Fe-2S] rhomb, the process that drives the key structural rearrangement of FNR leading to monomerization and loss of DNA binding. Support for this proposal comes from the dimeric FNR MS data, which strikingly shows that, although the [4Fe-4S]/[4Fe-4S] FNR dimer was readily detected, only very minor amounts of [3Fe-4S]/[4Fe-4S], [3Fe-4S]/[3Fe-4S], and [3Fe-3S]/[3Fe-4S] species were observed. We note that these species could alternatively correspond to different combinations of clusters (e.g., the [3Fe-4S]/[3Fe-4S] species could also be a [2Fe-4S]/[4Fe-4S] species), but the fact that they are minor compared with the [4Fe-4S]/[4Fe-4S] form remains. This ambiguity emphasizes the importance of the monomer region for definitive identification of cluster forms. If the [3Fe-3S] cluster formed is planar, then the accompanying structural rearrangement, involving conversion from a tetrahedral to a planar arrangement of coordinating Cys residues, would be expected to disrupt the dimer as observed in the MS data. Furthermore, the relatively rapid conversion of [3Fe-4S] to [3Fe-3S] would account for the failure of [3Fe-4S] species to accumulate in the MS spectra of the FNR dimer region. The temporal behavior of the low-intensity FNR dimer cluster conversion species matches well that observed for the monomeric FNR [3Fe-4S] and [3Fe-3S] intermediates, consistent with dissociation of dimeric FNR species into monomer forms.
Loss of sulfide during conversion of [3Fe-4S] to [3Fe-3S] might be compensated for through reengagement of the Cys residue that detaches when the initial Fe2+ is lost (Fig. S5). We note that a dimeric form of AfFNR, containing a partly degraded cluster (possibly a 3Fe cluster), displays a disorganized cluster binding loop (residues 20–29) after the loss of iron(s) (6). Thus, the data strongly indicate that, as soon as an Fe2+ ion is lost from one [4Fe-4S]2+ cluster (on one of the subunits of the dimer), the stability of the dimer is reduced and that the monomer–dimer equilibrium becomes heavily favored in the direction of the monomer.
Recently, it was shown that cluster sulfide is oxidized to sulfane, with incorporation into Cys residues as persulfides that coordinate the [2Fe-2S] form of FNR (16). Cys persulfides, as a stored form of sulfur, allow the original cluster to be repaired on supply of electrons and Fe2+ as shown in vitro (16). The formation of persulfides raises questions about when sulfide oxidation occurs in relation to cluster conversion: simultaneously with the conversion process or subsequent to it in an additional reaction. The MS data resolve this question. Analysis of the kinetic data shows that sulfide oxidation occurs at the same time as formation of the [2Fe-2S] cluster, leading to [2Fe-2S] and singly and doubly persulfide-coordinated species forming simultaneously. The [3Fe-3S] intermediate identified here undergoes loss of Fe3+ and loss/oxidation of sulfide to [2Fe-2S](S) and [2Fe-2S], representing a branch point of the mechanism. Clearly, [2Fe-2S](S)2 cannot be formed directly from [3Fe-3S] but instead, must form from [3Fe-4S], and therefore, it represents a second branch point. By analogy with the above, this reaction is likely to occur via a [3Fe-3S](S) intermediate, where the sulfide is oxidized to sulfane and released from the cluster but retained on a Cys as a persulfide. This species cannot be distinguished from the [3Fe-4S] intermediate in the MS spectrum. This ambiguity may be the reason why the global fit to the [3Fe-4S] species kinetic profile is not as good as for the other cluster intermediates; the data actually represent a combination of [3Fe-4S] and [3Fe-3S](S) species. A fit of the data based on a revised model in which this additional step is included (Fig. S6) is consistent with this proposal. The simultaneous nature of cluster conversion and sulfide oxidation was confirmed by LC-MS experiments carried out under identical conditions to the native MS experiments.

Formation of [3Fe-4S] and [3Fe-3S](S) species. (A) Reaction scheme from Fig. 3A amended to include an additional step, in which the ejected sulfide from the [3Fe-4S] cluster intermediate is oxidized and retained on the protein as a persulfide. (B) Plot of relative abundance of the species at mass of 29,843 Da corresponding to [3Fe-4S] and [3Fe-3S](S) species (yellow triangles) detected by native MS as a function of time after exposure to excess O2. Global fitting to the experimental data using the reaction scheme depicted in A is shown as a solid line. Dashed lines show the responses of the [4Fe-4S] (black), [2Fe-2S] (red), and apo (gray) species for comparison. Error bars show SE for average MS dataset (n = 4). The global fitting model was initiated with 100% relative abundance of [4Fe-4S] clusters.
The formations of a Cys persulfide by oxidative coupling of a sulfide from the trisulfide face of the [3Fe-4S] cluster or the putative hexagonal ring of a [3Fe-3S] cluster with a nearby free cysteine are both plausible paths. Presumably, electrons from sulfide oxidation reduce O2 to either H2O2 or water, accounting for the observed stoichiometry for the cluster conversion reaction of ~1.5 O2 consumed per cluster (30). Cluster iron oxidation yields only one electron. The other electrons must be derived from sulfide oxidation, resulting, overall, in a mixture of superoxide (where no sulfide oxidation occurs), hydrogen peroxide, and water (14, 16).
In conclusion, the ESI-MS results, with the benefit of previously published spectroscopic and kinetic analyses, provide insight into the mechanism by which O2 sensing occurs in FNR. A global kinetic analysis of the starting, intermediate, and product species yields unsuspected aspects of the mechanistic model. Not only has a previously unrecognized [3Fe-3S] cluster intermediate species been identified, but the nature of sulfide oxidation to generate persulfide species has also been resolved. This work further shows the feasibility of time-resolved ESI-MS for the study of iron-sulfur clusters and their reactions within a protein framework, with potential broad application to studies of other systems involving interactions/reactions of protein cofactors with small molecules.
Materials and Methods
Protein Purification.
A plasmid for the expression of C-terminal (His)6-tagged E. coli S24F FNR, in which the mutant fnr gene was ligated into pGS21a plasmid (Genscript) at NdeI and HindIII sites, was purchased (Genscript) (Fig. S7). Protein was overproduced in E.coli (BL21 λDE3 Star) cultures as previously described (28), except that 0.4 mM isopropyl β-d-thiogalactoside was used to induce protein expression. For minimal media growth preparations, cultures were grown in M9 minimal media as previously described (33). Cells were harvested by centrifugation, lysed on ice by sonication in buffer A (25 mM Hepes, 2.5 mM CaCl2, 100 mM NaCl, 100 mM NaNO3, pH 7.5) with 500 mM KCl, and centrifuged at 40,000 × g for 45 min at 4 °C. Inside an anaerobic cabinet (O2 < 2 ppm; Belle Technology), the cleared cell lysate was loaded onto a HiTrap (GE Healthcare) Ni2+-chelating column (2 × 5 mL) with 14% (vol/vol) buffer B (buffer A with 500 mM KCl, 500 mM imidazole, pH 7.5) and washed until A280 nm ≤ 0.1. Bound proteins were eluted using a linear gradient (10 mL) from 14 to 100% buffer B. Fractions (1 mL) containing FNR were pooled and immediately desalted into buffer C (buffer A with 2 mM DTT, pH 7.5) via a HiTrap desalting column (4 × 5 mL). As isolated, samples were ≤50% replete with cluster. To increase cluster loading, [4Fe-4S] clusters were inserted via an NifS catalyzed in vitro reconstitution reaction, from which the protein was reisolated using a HiTrap Heparin column as previously described (33). Bound protein was eluted using buffer A with 500 mM KCl, pH 7.5. Residual apo-S24F (~30 kDa) was separated from holo-S24F (~60 kDa) via gel filtration with a calibrated Sephacryl S100HR column previously equilibrated with buffer D (50 mM Tris, 300 mM KCl, 2 mM DTT, pH 8.0). Dimeric holo-S24F–containing fractions were pooled and frozen as described previously (14). 34S-substituted [4Fe-4S] S24F FNR was prepared using 34S-labeled cysteine in a reconstitution reaction as previously described (16).
Mass spectrometry.
For native ESI-MS, an aliquot of S24F FNR was exchanged into buffer E (250 mM ammonium acetate, pH 6.7) using Zeba spin (Thermo Scientific) or midi-PD10 (GE Healthcare) desalting columns; the volume of the eluent was increased to 2 mL, and the concentration of [4Fe-4S] S24F FNR was determined via absorbance at 406 nm (see below). An aliquot of the sample was combined with an aliquot (1.6 mL) of buffer E containing dissolved atmospheric O2 (168 μM O2; final concentration) in an anaerobic cuvette to give ~14 μM [4Fe-4S] S24F FNR. Buffer E lacking dissolved O2 was used for control experiments. The sample was immediately loaded into a 1-mL gas-tight syringe (Hamilton) and infused directly using a syringe pump (0.3 mL/h) into the ESI source of a Bruker micrOTOF-QIII mass spectrometer (Bruker Daltonics) operating in the positive ion mode. The ESI-TOF was calibrated using ESI-L Low Concentration Tuning Mix (Agilent Technologies). Before the introduction of sample, the gas-tight syringe (Hamilton) and associated PEEK tubing (Upchurch Scientific) were flushed with 5 mL anaerobic buffer E. The oxygen permeability of PEEK tubing is 14 mL/250 cm2 (1 atm/25 °C) over 24 h (Upchurch Scientific). MS data were acquired over the m/z range of 700–3,500 continuously for 120 min, with acquisition controlled using Bruker oTOF Control software and parameters as follows: dry gas flow of 4 L/min, nebulizer gas pressure of 0.8 Bar, dry gas at 180 °C, capillary voltage of 4,500 V, offset of 500 V, ion energy of 5 eV, collision radio frequency of 200 Vpp, and collision cell energy of 10 eV. Optimization of experimental conditions for the transmission of dimeric species (2,500–5,000 m/z) was achieved by increasing the equivalent of the cone voltage (in-source collision-induced dissociation of 75 eV), reducing the collision cell energy (4 eV), and increasing the collision RF (1,500 Vpp) (34).
For LC-MS, an aliquot of S24F FNR was exchanged into buffer E (120 μL; ~252 μM [4Fe-4S]) using a Zeba spin column, and the volume of the eluent increased to 500 μL. For O2 reactivity measurements, the sample was combined with an aliquot (1.6 mL) of oxygenated buffer E (~14 μM [4Fe-4S], 168 μM O2; final concentration) in an anaerobic cuvette and allowed to react. Samples (50 μL) were removed at varying time points and diluted to ~1.5 μM (final concentration) with an aliquot (406 μL) of quenching solution [an aqueous mixture of 0.8 mM EDTA, 0.7% (vol/vol) formic acid, 2% (vol/vol) acetonitrile]. For experiments probing the reversibility of adduct formation before and after reaction with O2, samples were treated with 20 mM DTT. Samples were sealed, removed from the anaerobic cabinet, and injected (1 µL) onto a ProSwift reversed phase RP-1S column (4.6 × 50 mm; Thermo Scientific) at 25 °C using an UltiMate 3000 HPLC system (Dionex).
Gradient elution was performed at a flow rate of 0.2 mL/min using a linear gradient (15 min) from 2 to 100% (vol/vol) acetonitrile and 0.1% (vol/vol) formic acid. The eluent was continuously infused into a Bruker micrOTOF-QIII mass spectrometer running Hystar (Bruker Daltonics) using positive mode ESI. The mass spectrometer was calibrated with ESI-L tuning mix (Agilent Technologies). MS acquisition parameters were as follows: dry gas flow of 8 L/min, nebulizer gas pressure of 1.8 Bar, dry gas at 240 °C, capillary voltage of 4,500 V, offset of 500 V, and collision RF of 650 Vpp.
Processing and analysis of MS experimental data were carried out using Compass DataAnalysis, version 4.1 (Bruker Daltonik). Neutral mass spectra were generated using the ESI Compass, version 1.3 Maximum Entropy deconvolution algorithm over a mass range of 29,000–31,000 Da for the monomer and 59,000–60,500 Da for the dimer. For kinetic modeling, to clearly resolve overlapping peaks, multiple Gaussian functions were fitted to the experimental data using a least squares regression function in Origin 8 (Microcal) (34). Exact masses are reported from peak centroids representing the isotope average neutral mass. For apoproteins, these masses are derived from m/z spectra, for which peaks correspond to [M + nH]n+/n. For cluster-containing proteins, where the cluster contributes charge, peaks correspond to [M + FeSx+ + (n − x)H]n+/n, where M is the molecular mass of the protein, FeS is the mass of the iron-sulfur cluster of x+ charge, H is the mass of the proton, and n is the total charge. In the expression, the x+ charge of the cluster offsets the number of protons required to achieve the observed charge state (n+) (26). Predicted masses are given as the isotope average of the neutral protein or protein complex in which cofactor binding is expected to be charge compensated (35). For time-resolved MS intensity data, kinetic schemes were modeled using Dynafit 4 (BioKin Ltd) (36).
Other Analytical Techniques.
Protein concentrations were determined using the method of Bradford (Bio-Rad), with BSA as the standard and a previously determined correction factor of 0.83 (30). The iron and sulfide content of the protein was determined as described previously (15, 37), and the [4Fe-4S]2+ cluster concentrations were determined using an ε406 nm of 16.22 (±0.14) mM−1 cm−1 (13, 28). An Oxygraph+ (Hansatech Instruments) was used to determine the dissolved oxygen content of buffer E (244 ± 3 μM). Absorption and CD measurements were made using a Jasco V550 UV-visible spectrophotometer and a Jasco J-810 spectropolarimeter, respectively. Kinetic data at A406 nm were recorded via a fiber optic link as previously described (14).
Acknowledgments
We thank the University of East Anglia for funding the purchase of the electrospray ionization mass spectrometer instrument. This work was supported by Biotechnology and Biological Sciences Research Council Grant BB/L007673/1 (to J.C.C., A.J.T., and N.E.L.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.R.D. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/10.1073/pnas.1620987114/-/DCSupplemental.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.1620987114
Read article for free, from open access legal sources, via Unpaywall:
https://www.pnas.org/content/pnas/114/16/E3215.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.1620987114
Article citations
Binding of a single nitric oxide molecule is sufficient to disrupt DNA binding of the nitrosative stress regulator NsrR.
Chem Sci, 15 Oct 2024
Cited by: 0 articles | PMID: 39464610 | PMCID: PMC11500311
CyaY and TusA regulate ISC- and SUF-mediated l-cysteine desulfurase activity.
RSC Chem Biol, 27 Sep 2024
Cited by: 0 articles | PMID: 39372677 | PMCID: PMC11446229
Binding of IscU and TusA to different but competing sites of IscS influences the activity of IscS and directs sulfur to the respective biomolecular synthesis pathway.
Microbiol Spectr, e0094924, 09 Jul 2024
Cited by: 1 article | PMID: 38980029 | PMCID: PMC11302665
Proteomic strategies to interrogate the Fe-S proteome.
Biochim Biophys Acta Mol Cell Res, 1871(7):119791, 25 Jun 2024
Cited by: 0 articles | PMID: 38925478
Review
The methylerythritol phosphate pathway as an oxidative stress sense and response system.
Nat Commun, 15(1):5303, 21 Jun 2024
Cited by: 0 articles | PMID: 38906898 | PMCID: PMC11192765
Review Free full text in Europe PMC
Go to all (32) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity.
Proc Natl Acad Sci U S A, 94(12):6087-6092, 01 Jun 1997
Cited by: 180 articles | PMID: 9177174 | PMCID: PMC21006
Reversible cycling between cysteine persulfide-ligated [2Fe-2S] and cysteine-ligated [4Fe-4S] clusters in the FNR regulatory protein.
Proc Natl Acad Sci U S A, 109(39):15734-15739, 10 Sep 2012
Cited by: 60 articles | PMID: 23019358 | PMCID: PMC3465412
Influence of the environment on the [4Fe-4S]2+ to [2Fe-2S]2+ cluster switch in the transcriptional regulator FNR.
J Am Chem Soc, 130(5):1749-1758, 11 Jan 2008
Cited by: 34 articles | PMID: 18186637
Funding
Funders who supported this work.
Biotechnology and Biological Sciences Research Council (2)
Grant ID: BB/L007673/1
Grant ID: BB/L007673/1