Abstract
Free full text

Nucleic acid detection with CRISPR-Cas13a/C2c2
Associated Data
Abstract
Rapid, inexpensive, and sensitive nucleic acid detection may aid point-of-care pathogen detection, genotyping, and disease monitoring. The RNA-guided, RNA-targeting CRISPR effector Cas13a (previously known as C2c2) exhibits a “collateral effect” of promiscuous RNAse activity upon target recognition. We combine the collateral effect of Cas13a with isothermal amplification to establish a CRISPR-based diagnostic (CRISPR-Dx), providing rapid DNA or RNA detection with attomolar sensitivity and single-base mismatch specificity. We use this Cas13a-based molecular detection platform, termed SHERLOCK (Specific High Sensitivity Enzymatic Reporter UnLOCKing), to detect specific strains of Zika and Dengue virus, distinguish pathogenic bacteria, genotype human DNA, and identify cell-free tumor DNA mutations. Furthermore, SHERLOCK reaction reagents can be lyophilized for cold-chain independence and long-term storage, and readily reconstituted on paper for field applications.
The ability to rapidly detect nucleic acids with high sensitivity and single-base specificity on a portable platform may aid in disease diagnosis and monitoring, epidemiology, and general laboratory tasks. Although methods exist for detecting nucleic acids (1–6), they have trade-offs among sensitivity, specificity, simplicity, cost, and speed. Microbial Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated (CRISPR-Cas) adaptive immune systems contain programmable endonucleases that can be leveraged for CRISPR-based diagnostics (CRISPR-Dx). While some Cas enzymes target DNA (7, 8), single effector RNA-guided RNases, such as Cas13a (previously known as C2c2) (8), can be reprogrammed with CRISPR RNAs (crRNAs) to provide a platform for specific RNA sensing (9–12). Upon recognition of its RNA target, activated Cas13a engages in “collateral” cleavage of nearby non-targeted RNAs (10). This crRNA-programmed collateral cleavage activity allows Cas13a to detect the presence of a specific RNA in vivo by triggering programmed cell death (10) or in vitro by nonspecific degradation of labeled RNA (10, 12). Here we describe SHERLOCK (Specific High Sensitivity Enzymatic Reporter UnLOCKing), an in vitro nucleic acid detection platform with attomolar sensitivity based on nucleic acid amplification and Cas13a-mediated collateral cleavage of a reporter RNA (12), allowing for real-time detection of the target (Fig. 1A).

(A) Schematic of SHERLOCK.
(B) Schematic of ssRNA target detected via the Cas13a collateral detection. The target site is highlighted in blue.
(C) Cas13a detection of RNA with RPA amplification (SHERLOCK) can detect ssRNA target at concentrations down to ~2 aM, more sensitive than Cas13a alone. (n=4 technical replicates; bars represent mean ± s.e.m.)
(D) SHERLOCK is also capable of single-molecule DNA detection. (n=4 technical replicates; bars represent mean ± s.e.m.)
To achieve robust signal detection, we identified an ortholog of Cas13a from Leptotrichia wadei (LwCas13a), which displays greater RNA-guided RNase activity relative to Leptotrichia shahii Cas13a (LshCas13a) (10) (fig. S1). LwCas13a incubated with ssRNA target 1 (ssRNA 1), crRNA, and reporter (quenched fluorescent RNA) (Fig. 1B) (13) yielded a detection sensitivity of ~50 fM (Fig. 1C, S2). Although this sensitivity is an improvement on previous studies with LbCas13a (12), attomolar sensitivity is required for many diagnostic applications (14–16). We therefore explored combining Cas13a-based detection with different isothermal amplification steps (fig. S3, S4A) (17, 18). Of the methods explored, recombinase polymerase amplification (RPA) (18) afforded the greatest sensitivity and can be coupled with T7 transcription to convert amplified DNA to RNA for subsequent detection by LwCas13a. We refer to this combination of amplification by RPA, T7 RNA polymerase transcription of amplified DNA to RNA, and detection of target RNA by Cas13a collateral RNA cleavage-mediated release of reporter signal as SHERLOCK.
We first determined the sensitivity of SHERLOCK for detection of RNA (when coupled with reverse transcription) or DNA targets. We achieved single molecule sensitivity for both RNA and DNA, as verified by digital-droplet PCR (ddPCR) (Fig. 1C, D, S4B, C). Attomolar sensitivity was maintained when we combined all SHERLOCK components in a single reaction, demonstrating the viability of this platform as a point-of-care (POC) diagnostic (fig. S4D). SHERLOCK has similar levels of sensitivity as ddPCR and quantitative PCR (qPCR), two established sensitive nucleic acid detection approaches, whereas RPA alone was not sensitive enough to detect low levels of target (fig. S5A–D). Moreover, SHERLOCK shows less variation than ddPCR, qPCR, and RPA, as measured by the coefficient of variation across replicates (fig. S5E–F).
We next examined whether SHERLOCK would be effective in infectious disease applications that require high sensitivity. We produced lentiviruses harboring genome fragments of either Zika virus (ZIKV) or the related flavivirus Dengue (DENV) (19) (Fig. 2A). SHERLOCK detected viral particles down to 2 aM and could discriminate between ZIKV and DENV (Fig. 2B). To explore the potential use of SHERLOCK in the field with paper-spotting and lyophilization (1), we first demonstrated that Cas13a-crRNA complexes lyophilized and subsequently rehydrated (20) could detect 20 fM of non-amplified ssRNA 1 (fig. S6A) and that target detection was also possible on glass fiber paper (fig. S6B). The other components of SHERLOCK are also amenable to freeze-drying: RPA is provided as a lyophilized reagent at ambient temperature, and we previously demonstrated that T7 polymerase tolerates freeze-drying (2). In combination, freeze-drying and paper-spotting the Cas13a detection reaction resulted in comparable levels of sensitive detection of ssRNA 1 as aqueous reactions (fig. S6C–E). Although paper-spotting and lyophilization slightly reduced the absolute signal of the readout, SHERLOCK (Fig. 2C) could readily detect mock ZIKV virus at concentrations as low as 20 aM (Fig. 2D).
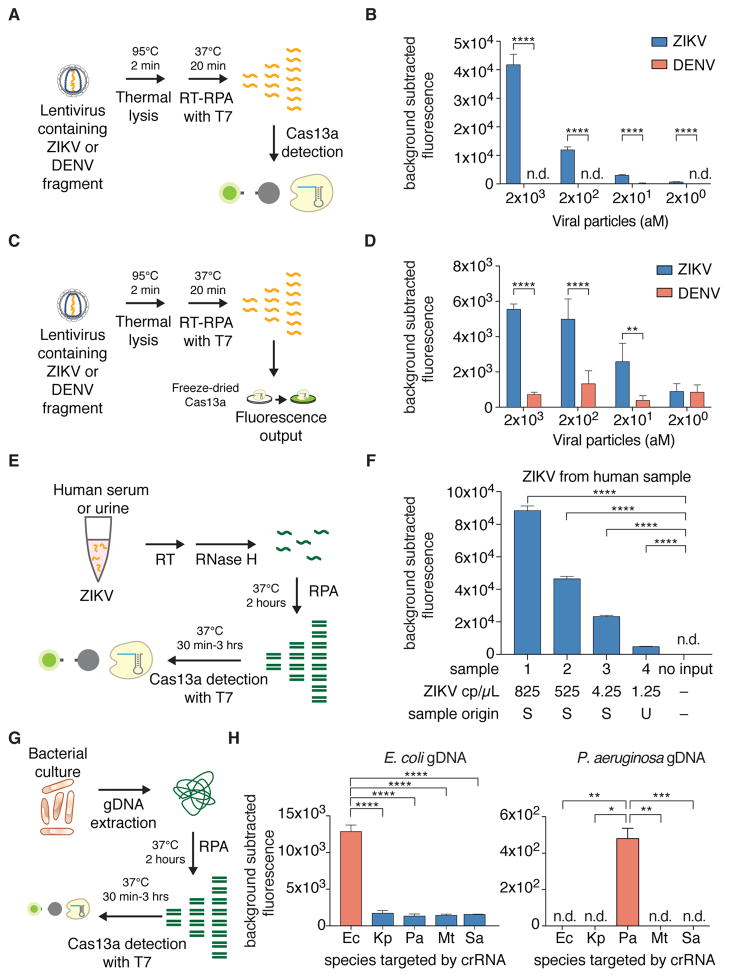
(A) Schematic of ZIKV RNA detection by SHERLOCK.
(B) SHERLOCK is capable of highly sensitive detection of the ZIKV lentiviral particles. (n=4 technical replicates, two-tailed Student t-test; ****, p < 0.0001; bars represent mean ± s.e.m.; n.d., not detected)
(C) Schematic of ZIKV RNA detection with freeze-dried Cas13a on paper
(D) Paper-based SHERLOCK is capable of highly sensitive detection of ZIKV lentiviral particles. (n=4 technical replicates, two-tailed Student t-test; **, p < 0.01; ****, p < 0.0001; bars represent mean ± s.e.m.)
(E) Schematic of SHERLOCK detection of ZIKV RNA isolated from human clinical samples.
(F) SHERLOCK is capable of highly sensitive detection of human ZIKV-positive serum (S) or urine (U) samples. Approximate concentrations of ZIKV RNA shown were determined by qPCR. (n=4 technical replicates, two-tailed Student t-test; ****, p < 0.0001; bars represent mean ± s.e.m.; n.d., not detected)
(G) Schematic of using SHERLOCK to distinguish bacterial strains using a universal 16S rRNA gene V3 RPA primer set.
(H) SHERLOCK achieves sensitive and specific detection of E. coli or P. aeruginosa gDNA. (n=4 technical replicates, two-tailed Student t-test; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; bars represent mean ± s.e.m.). Ec, Escherichia coli; Kp, Klebsiella pneumoniae; Pa, Pseudomonas aeruginosa; Mt, Mycobacterium tuberculosis; Sa, Staphylococcus aureus.
SHERLOCK is also able to detect ZIKV in clinical isolates (serum, urine, or saliva) where titers can be as low as 2 × 103 copies/mL (3.2 aM) (21). ZIKV RNA extracted from patient serum or urine samples and reverse transcribed into cDNA (Fig. 2E) could be detected at concentrations down to 1.25 × 103 copies/mL (2.1 aM), as verified by qPCR (Fig. 2F). Furthermore, the signal from patient samples was predictive of ZIKV RNA copy number and could be used to predict viral load (Fig. S6F). To simulate sample detection without nucleic acid purification, we measured detection of ssRNA 1 spiked into human serum, and found that Cas13a could detect RNA in reactions containing as much as 2% serum (fig. S6G).
Another important epidemiological application for CRISPR-dx is the identification of bacterial pathogens and detection of specific bacterial genes. We targeted the 16S rRNA gene V3 region, where conserved flanking regions allow universal RPA primers to be used across bacterial species and the variable internal region allows for differentiation of species. In a panel of five possible targeting crRNAs for different pathogenic strains and gDNA isolated from E. coli and Pseudomonas aeruginosa (Fig. 2G), SHERLOCK correctly genotyped strains and showed low cross-reactivity (Fig. 2H). Additionally, we were able to use SHERLOCK to distinguish between clinical isolates of Klebsiella pneumoniae with two different resistance genes: Klebsiella pneumoniae carbapenemase (KPC) and New Delhi metallo-beta-lactamase 1 (NDM-1) (22) (fig. S7).
To increase the specificity of SHERLOCK, we introduced synthetic mismatches in the crRNA:target duplex that enable LwCas13a to discriminate between targets that differ by a single-base mismatch (fig. S8A, B). We designed multiple crRNAs with synthetic mismatches in the spacer sequences to detect either the African or American strains of ZIKV (Fig. 3A, B) and strain 1 or 3 of DENV (Fig. 3C, D). Synthetic mismatch crRNAs detected their corresponding strains with significantly higher signal (two-tailed Student t-test; p < 0.01) than the off-target strain, allowing for robust strain discrimination based off single mismatches (Fig. 3B, D, S8C). Further characterization revealed that Cas13a detection achieves maximal specificity while maintaining on-target sensitivity when a mutation is in position 3 of the spacer and the synthetic mismatch is in position 5 (fig. S9 and S10).

(A) Schematic of ZIKV strain target regions and the crRNA sequences used for detection. SNPs in the target are highlighted red or blue and synthetic mismatches in the guide sequence are colored red.
(B) Highly specific detection of strain SNPs allows for the differentiation of ZIKV African versus American RNA targets using Cas13a. (n=2 technical replicates, two-tailed Student t-test; **, p < 0.01; ***, p < 0.001; bars represent mean ± s.e.m.)
(C) Schematic of DENV strain target regions and the crRNA sequences used for detection. SNPs in the target are highlighted red or blue and synthetic mismatches in the guide sequence are colored red.
(D) Highly specific detection of strain SNPs allows for the differentiation of DENV strain 1 versus strain 3 RNA targets using Cas13a. (n=2 technical replicates, two-tailed Student t-test; *, p < 0.05; **, p < 0.01; ***, p < 0.001; bars represent mean ± s.e.m.)
The ability to detect single-base differences opens the opportunity of using SHERLOCK for rapid human genotyping. We chose five loci spanning a range of health-related single-nucleotide polymorphisms (SNPs) (Table S1) and benchmarked SHERLOCK detection using 23andMe genotyping data as the gold standard at these SNPs (23) (Fig. 4A). We collected saliva from four human subjects with diverse genotypes across the loci of interest, and extracted genomic DNA either through column purification or direct heating for five minutes (20). SHERLOCK distinguished alleles with high significance and with enough specificity to infer both homozygous and heterozygous genotypes (Fig. 4B, S11, S12).

(A) Circos plot showing location of human SNPs detected with SHERLOCK.
(B) SHERLOCK can correctly genotype four different individuals at four different SNP sites in the human genome. The genotypes for each individual and identities of allele-sensing crRNAs are annotated below each plot. (n=4 technical replicates, two-tailed Student t-test; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; bars represent mean ± s.e.m.)
(C) Schematic of cell-free DNA detection of cancer mutations using SHERLOCK.
(D) Sequences of two genomic loci assayed for cancer mutations in cell-free DNA. Shown are the target genomic sequence with the SNP highlighted in blue and the mutant/wild-type sensing crRNA sequences with synthetic mismatches colored in red.
(E,F) Cas13a can detect the mutant minor allele in mock cell-free DNA samples for the EGFR L858R (E) or the BRAF V600E (F) minor allele. (n=4 technical replicates, two-tailed Student t-test; *, p < 0.05; **, p < 0.01; ****, p < 0.0001; bars represent mean ± s.e.m.)
Finally, we sought to determine if SHERLOCK could detect low frequency cancer mutations in cell free (cf) DNA fragments, which is challenging because of the high levels of wild-type DNA in patient blood (24–26). We first found that SHERLOCK could detect ssDNA 1 at attomolar concentrations diluted in a background of genomic DNA (fig. S13A). Next, we found that SHERLOCK was also able to detect single nucleotide polymorphism (SNP)-containing alleles (fig. S13B, C) at levels as low as 0.1% of background DNA, which is in the clinically relevant range. We then demonstrated that SHERLOCK could detect two different cancer mutations, EGFR L858R and BRAF V600E, in mock cfDNA samples with allelic fractions as low as 0.1% (Fig. 4C–F) (20).
The SHERLOCK platform lends itself to further applications including (i) general RNA/DNA quantitation in lieu of specific qPCR assays, such as TaqMan, (ii) rapid, multiplexed RNA expression detection, and (iii) other sensitive detection applications, such as detection of nucleic acid contamination. Additionally, Cas13a could potentially detect transcripts within biological settings and track allele-specific expression of transcripts or disease-associated mutations in live cells. We have shown that SHERLOCK is a versatile, robust method to detect RNA and DNA, suitable for rapid diagnoses including infectious disease applications and sensitive genotyping. A SHERLOCK paper test can be redesigned and synthesized in a matter of days for as low as $0.61/test (Table S2) with confidence, as almost every crRNA tested resulted in high sensitivity and specificity. These qualities highlight the power of CRISPR-Dx and open new avenues for rapid, robust and sensitive detection of biological molecules.
Acknowledgments
We thank F. Chen, V. Rusu, R. Gupta, D. Daniels, C. Garvie, I. Finkelstein, V. Adalsteinsson, A. Das, E.S. Lander, R. Macrae, and R. Belliveau for discussions and support. Human genotyping data was collected with informed consent by the subjects and in consent with the guidelines of the approved MIT IRB protocol IRB-4062. O.A.A. is supported by a Paul and Daisy Soros Fellowship and National Defense Science and Engineering Fellowship. J.S.G. is supported by a D.O.E. Computational Science Graduate Fellowship. R.P.B, J.L, and D.T.H. are supported by the NIH through NIAID (R01AI117043). A.D. is supported by an NSF Graduate Research Fellowship and a Air Force Office of Scientific Research grant (FA9550-14-1-0060). Zika work was partially funded by Marc and Lynne Benioff to P.C.S. and antibiotic resistance work was partially funded by Josh and Anita Bekenstein to D.T.H. A.R. is supported by the Howard Hughes Medical Institute. F.Z. is a New York Stem Cell Foundation-Robertson Investigator. F.Z. is supported by the NIH through NIMH (5DP1-MH100706 and 1R01-MH110049), NSF, Howard Hughes Medical Institute, the New York Stem Cell, Simons, Paul G. Allen Family, and Vallee Foundations; and James and Patricia Poitras, Robert Metcalfe, and David Cheng. A.R. is a member of the Scientific Advisory Board for ThermoFisher Scientific. J.S.G., O.O.A, R.P.B., A.R., E.V.K., D.T.H., P.C.S., J.J.C and F.Z. have filed patent applications relating to the work in this manuscript, including J.S.G., O.O.A, E.V.K. and F.Z on International application no. PCT/US2016/038258 filed June 18, 2015 (CRISPR-C2c2 systems and uses thereof); J.S.G., O.O.A, and F.Z on US provisional patent application no. 62/351,662 filed June 17, 2016 (CRISPR-C2c2 systems and diagnostic uses thereof); J.S.G., O.O.A, J.J.C and F.Z on US provisional patent application no. 62/432,553 filed December 9, 2016 (SHERLOCK diagnostic); J.S.G., O.O.A, P.C.S., J.J.C and F.Z on US provisional patent application no. 62/471,917 filed March 15, 2017 (viral application of SHERLOCK); J.S.G., O.O.A, A.R., J.J.C and F.Z on US provisional patent application no. 62/471,931 filed March 15, 2017 (mutation detection with SHERLOCK); J.S.G., O.O.A, R.P.B., D.T.H., J.J.C and F.Z on US provisional patent application no. 62/471,936 filed March 15, 2017 (bacterial applications of SHERLOCK); and J.S.G., O.O.A, J.J.C and F.Z on US provisional patent application no. 62/471,940 filed March 15, 2017 (devices), each relating to CRISPR-C2c2 systems, specific uses and improved uses thereof for diagnostic application filed by Broad, Harvard, MGH, MIT and NIH. Cas13a/C2c2 expression plasmids are available from Addgene under UBMTA.
Footnotes
Materials and Methods
References
Full text links
Read article at publisher's site: https://doi.org/10.1126/science.aam9321
Read article for free, from open access legal sources, via Unpaywall:
https://science.sciencemag.org/content/sci/356/6336/438.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
End-point diagnostics of Giardia duodenalis assemblages A and B by combining RPA with CRISPR/Cas12a from human fecal samples.
Parasit Vectors, 17(1):463, 12 Nov 2024
Cited by: 0 articles | PMID: 39533301 | PMCID: PMC11558988
Prophylactic and therapeutic vaccine development: advancements and challenges.
Mol Biomed, 5(1):57, 11 Nov 2024
Cited by: 0 articles | PMID: 39527305 | PMCID: PMC11554974
Review Free full text in Europe PMC
Insight into the natural regulatory mechanisms and clinical applications of the CRISPR-Cas system.
Heliyon, 10(20):e39538, 18 Oct 2024
Cited by: 0 articles | PMID: 39502233 | PMCID: PMC11535992
Review Free full text in Europe PMC
Detection Methods for Pine Wilt Disease: A Comprehensive Review.
Plants (Basel), 13(20):2876, 14 Oct 2024
Cited by: 0 articles | PMID: 39458823 | PMCID: PMC11511408
Review Free full text in Europe PMC
Recent Advances in the CRISPR/Cas-Based Nucleic Acid Biosensor for Food Analysis: A Review.
Foods, 13(20):3222, 10 Oct 2024
Cited by: 0 articles | PMID: 39456285 | PMCID: PMC11507162
Review Free full text in Europe PMC
Go to all (1,156) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Detecting pathogens with Zinc-Finger, TALE and CRISPR- based programmable nucleic acid binding proteins.
J Microbiol Methods, 152:98-104, 02 Aug 2018
Cited by: 21 articles | PMID: 30076867
Review
Field-deployable viral diagnostics using CRISPR-Cas13.
Science, 360(6387):444-448, 01 Apr 2018
Cited by: 520 articles | PMID: 29700266 | PMCID: PMC6197056
A CRISPR Way to Diagnose Infectious Diseases.
N Engl J Med, 377(17):1685-1687, 01 Oct 2017
Cited by: 14 articles | PMID: 29069564
Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6.
Science, 360(6387):439-444, 15 Feb 2018
Cited by: 845 articles | PMID: 29449508 | PMCID: PMC5961727
Funding
Funders who supported this work.
Howard Hughes Medical Institute (1)
Grant ID: award301050
NCI NIH HHS (1)
Grant ID: F30 CA210382
NIAID NIH HHS (3)
Grant ID: K08 AI119157
Grant ID: T32 AI007061
Grant ID: R01 AI117043
NIH Office of the Director (4)
Grant ID: 1R01-MH110049
Grant ID: 5DP1-MH100706
Grant ID: award301047
Grant ID: award301048
NIMH NIH HHS (2)
Grant ID: DP1 MH100706
Grant ID: R01 MH110049
NSF Office of the Director (1)
Grant ID: award301053
New York Stem Cell Foundation (1)
Grant ID: award301049
Paul G. Allen Family Foundation (1)
Grant ID: award301054
Simons Foundation (1)
Grant ID: award301051
Vallee Foundation (1)
Grant ID: award301052





