Abstract
Free full text

Gene length and detection bias in single cell RNA sequencing protocols
Peer Review Summary
| Review date | Reviewer name(s) | Version reviewed | Review status |
|---|---|---|---|
| 2017 May 19 | Sam Buckberry and Timothy Stuart | Version 1 | Approved |
| 2017 May 17 | Wolfgang Huber | Version 1 | Approved |
| 2017 May 10 | Samuel W. Lukowski | Version 1 | Approved |
| 2017 May 10 | Charlotte Soneson | Version 1 | Approved |
Abstract
Background: Single cell RNA sequencing (scRNA-seq) has rapidly gained popularity for profiling transcriptomes of hundreds to thousands of single cells. This technology has led to the discovery of novel cell types and revealed insights into the development of complex tissues. However, many technical challenges need to be overcome during data generation. Due to minute amounts of starting material, samples undergo extensive amplification, increasing technical variability. A solution for mitigating amplification biases is to include unique molecular identifiers (UMIs), which tag individual molecules. Transcript abundances are then estimated from the number of unique UMIs aligning to a specific gene, with PCR duplicates resulting in copies of the UMI not included in expression estimates.
Methods: Here we investigate the effect of gene length bias in scRNA-Seq across a variety of datasets that differ in terms of capture technology, library preparation, cell types and species.
Results: We find that scRNA-seq datasets that have been sequenced using a full-length transcript protocol exhibit gene length bias akin to bulk RNA-seq data. Specifically, shorter genes tend to have lower counts and a higher rate of dropout. In contrast, protocols that include UMIs do not exhibit gene length bias, with a mostly uniform rate of dropout across genes of varying length. Across four different scRNA-Seq datasets profiling mouse embryonic stem cells (mESCs), we found the subset of genes that are only detected in the UMI datasets tended to be shorter, while the subset of genes detected only in the full-length datasets tended to be longer.
Conclusions: We find that the choice of scRNA-seq protocol influences the detection rate of genes, and that full-length datasets exhibit gene-length bias. In addition, despite clear differences between UMI and full-length transcript data, we illustrate that full-length and UMI data can be combined to reveal the underlying biology influencing expression of mESCs.
Introduction
Single cell RNA-Seq (scRNA-Seq) has rapidly gained popularity as the primary tool to profile gene expression of hundreds to thousands of single cells. This new technology enables researchers to examine transcription at the resolution of a single cell in a high-throughput manner, and has led to the discovery of novel cell types and revealed insights into the development of complex tissues as well as differentiation lineages. With the promise of novel discoveries, this new technology has been embraced by the scientific community.
Many technical challenges need to be overcome during data generation, and technology for performing scRNA-Seq is advancing at a rapid rate. The original Fluidigm C1 system has a 96-well plate, which limits how many single cells researchers can practically handle in an experiment. However, depth of sequencing is only limited by cost, with a sequencing depth of around 2 million reads per cell recommended ( Tung et al., 2016). Droplet based technology, such as InDrop ( Klein et al., 2015), Drop-Seq ( Macosko et al., 2015) and the more recent Chromium system from 10X Genomics ( Zheng et al., 2016), are cost effective methods to obtain relatively shallow sequencing of thousands to tens of thousands of single cells in one run. Lower sequencing depth limits the complexity of the expression profile attained per cell, as only the most highly expressed genes will be observed, however, it may be the case that researchers combine deeper sequencing of fewer single cells with shallow sequencing of tens of thousands of cells to answer their scientific questions of interest.
Not only are there different technologies for capturing single cells, there are also differences in library preparation protocols, which aim to amplify and process the minute amounts of RNA from each cell. Most RNA-Seq library preparation protocols include enrichment of mRNA by either polyA pulldown or ribosomal depletion, followed by fragmentation and PCR amplification before sequencing. The extensive PCR amplification that is required for scRNA-Seq increases technical variability in the data by introducing amplification biases ( Stegle et al., 2015). A solution for mitigating amplification biases is to include Unique Molecular Identifiers (UMIs), which are short (5–10bp) sequences ligated onto the 5’ end of the molecule prior to PCR amplification ( Islam et al., 2014). Transcript abundances are then estimated from the number of reads with unique UMIs aligning to a specific gene. PCR duplicates resulting in copies of the UMI are therefore not included in expression estimates. While some protocols, such as those used with Fluidigm C1 (e.g SMARTer), need to be modified to include UMIs ( Tung et al., 2016), some droplet based methods, for example the Chromium system ( Zheng et al., 2016), always include UMIs in the chemistry. It is worth noting that, while mechanisms such as alternative splicing can be studied using full-length transcript protocols, this type of analysis is not possible with data generated with protocols that include UMIs.
Gene length bias is well understood in bulk RNA-seq data. When cDNAs are fragmented, long genes result in more fragments for the same number of transcripts, resulting in higher counts and more power to detect differential expression ( Oshlack & Wakefield, 2009). As a result gene set testing is biased towards gene ontology categories containing longer genes ( Young et al., 2010). While there is much in common between scRNA-Seq and bulk RNA-Seq data, modifications to the protocols such as amplification and the inclusion of UMIs, may highlight different biases in the data.
Here we investigate the effect of gene length bias in scRNA-Seq across a variety of datasets that differ in terms of capture technology, library preparation, cell types and species. As hypothesised, we find that scRNA-seq datasets that have been sequenced using a full-length transcript protocol exhibit gene length bias akin to bulk RNA-seq data. Specifically, shorter genes tend to have lower counts and a higher rate of dropout. In contrast, protocols that include UMIs do not exhibit gene length bias. UMI protocols reveal that shorter genes are as highly expressed as longer genes, and dropout is mostly uniform across genes of varying length. These effects mean that different protocols have the ability to detect a different subset of genes, with shorter genes detected more readily using UMI protocols and longer genes detected by full-length protocols.
Methods
Processing of full-length datasets
We processed three datasets through our pipeline developed for full-length data:
Mouse embryonic stem cells ( Kolodziejczyk et al., 2015);
Human cerebral organoid cells ( Camp et al., 2015),
Mouse embryonic stem cells ( Buettner et al., 2015).
The quality of the raw sequencing reads was examined using FastQC (v0.11.4). They were checked for contamination by aligning a sample of reads to multiple reference genomes using FastQ Screen (v0.6.4.). Reads were aligned to the appropriate reference using STAR (v2.5.2a) ( Dobin et al., 2013). For the mouse dataset, we used the mm10 version of the genome, using the chromFa.tar.gz file on http://hgdownload.soe.ucsc.edu/goldenPath/mm10/bigZips, and for the human datasets we used the hg38 version of the genome, using the hg38.chromFa.tar.gz file on http://hgdownload.cse.ucsc.edu/goldenPath/hg38/bigZips. Reads were summarised across genes using featureCounts (v1.5.0-p3) ( Liao et al., 2014), with GENCODE M9 annotation for mouse and GENCODE V22 annotation for human datasets. This pipeline was constructed in Bpipe (v0.9.9.3) ( Sadedin et al., 2012), and a report summarising the steps produced using MultiQC (v0.8) ( Ewels et al., 2016).
Gene filtering
Our gene filtering strategy was identical between datasets. Genes that had more than 90% zeroes across all cells, as well as ribosomal and mitochondrial genes, were filtered out. Genes that could not be annotated with an Entrez Gene ID were also removed in order to retain a set of well curated genes. Gene length information was taken as the sum of the exon lengths as outputted by the featureCounts software for the mm10 GENCODE VM4 annotation for all mouse datasets, and for all human datasets, we used the sum of exon lengths as outputted by featureCounts for the hg38 GENCODE V22 annotation. Genes that could not be annotated with gene length information were filtered out. We found that using these criteria helped reduce some of the variability in the datasets.
Processing of all datasets
Details of all datasets analysed in this study are listed in Supplementary Table 1.
Mouse embryonic stem cells, Kolodziejczyk et al., 2015, full-length. We downloaded the raw data from the ArrayExpress database under accession number E-MTAB-2600 and ran our full-length processing pipeline using the mm10 mouse genome to produce a counts matrix. We performed quality control on the cells and removed cells that had a dropout rate of greater than 80% and a library size of fewer than half a million. We calculated the proportion of sequencing reads taken up by the ERCC spike-ins and discarded three plates that had proportions of ERCC spike-ins that appeared excessive compared to the remaining plates. We performed gene filtering as described above. After cell and gene filtering, we were left with 530 cells and 12395 genes for further analysis.
Human primordial germ cells, Guo et al., 2015, full-length. We downloaded the processed data from Conquer. The data had been pseudo-aligned to the latest human reference genome, hg38, using the Salmon software tool, v0.6.0 ( Patro et al., 2017). The data is also available under the GEO accession number GSE63818. There did not appear to be any spike-in controls for this dataset, hence filtering was performed on the dropout rate and total sequencing depth for each cell. Cells with more than 85% dropout and fewer than half a million sequencing reads were filtered out. After cell and gene filtering, there were 226 cells and 15837 genes for further analysis.
Human cerebral organoid cells, Camp et al., 2015, full-length. We downloaded the data from SRA under accession SRP066834, and ran our full-length processing pipeline to produce a counts matrix, using the hg38 human genome. We removed cells that had greater than 90% dropout, library size smaller than half a million as well as cells that had more than 20% of the sequencing taken up by ERCC controls. After cell and gene filtering, we had 494 cells and 11325 genes for further analysis.
Mouse embryonic stem cells, Grün et al., 2014, UMI. We downloaded the processed data from GEO under accession number GSE54695. The data was aligned to the mm10 mouse genome using BWA and transcript number estimated from UMI counts by the authors. We removed cells that had > 80% dropout, library size smaller than 10000, as well as cells that had more than 5% of the sequencing taken up by ERCC controls. After cell and gene filtering, there were 127 cells and 9962 genes for further analysis.
Human induced pluripotent stem cells, Tung et al., 2016, UMI. We downloaded the processed molecule counts and sample information from the authors’ Github repository ( https://github.com/jdblischak/singleCellSeq). The data was aligned by the authors to the human genome hg19 using the Subjunc aligner ( Liao et al., 2013). The data is also available under GEO accession GSE77288. We removed cells that had > 70% dropout, fewer than 30000 sequencing reads per cell, as well as cells that had more than 3% of the sequencing taken up by ERCC spike-ins. After cell and gene filtering, we had 671 cells and 11971 genes for further analysis.
Human K562 cells (lymphoblastoma culture), Klein et al., 2015, UMI. The processed molecule count data was downloaded from GEO under accession GSM1599500. The data was aligned to the hg19 human genome using Bowtie v0.12.0 ( Langmead et al., 2009). Cells that had > 85% dropout, fewer than 10000 total sequencing reads, or an ERCC library size to total library size ratio > 0.01 were filtered out. After cell and gene filtering, we had 219 cells and 13418 genes for further analysis.
Mouse embryonic stem cells, Ziegenhain et al., 2016, UMI. We downloaded the molecule counts from GEO under accession GSE75790. The SCRB-Seq protocol, a 3’ digital gene expression RNA-Seq protocol, ( Soumillon et al., 2014), was used to generate the libraries. The data was processed by the authors through a dropseq pipeline, which included alignment to the mm10 mouse genome using STAR v2.4.0 ( Dobin et al., 2013). The cells all appeared good quality hence cell filtering wasn’t necessary. After gene filtering, we had 84 cells and 10519 genes for further analysis.
Mouse embryonic stem cells, Buettner et al., 2015, full-length. We downloaded the data from the European Nucleotide Archive, under accession PRJEB6989, and ran the data through our full-length pipeline, mapping to the mm10 mouse genome to produce a counts matrix. We filtered out cells with > 85% dropout and sequencing depth less than a million. After cell and gene filtering, we had 271 cells and 11700 genes for further analysis.
Combining mouse embryonic stem cell datasets
We combined the four different mouse embryonic stem cell datasets using the following approach. We performed gene and cell filtering on each dataset independently, and combined the datasets by taking the genes commonly detected across all four datasets (8678 genes, 1012 cells, each gene is detected in at least 10% of the cells for each dataset). This strategy ensured that the genes were detected in all four datasets, and hence larger datasets did not dominate gene filtering. It also ensured that the larger datasets did not dominate the principal components analysis.
Statistical analysis
All statistical analysis was performed in R-3.3.1, using the limma ( Ritchie et al., 2015), edgeR ( Robinson et al., 2010), scran ( Lun et al., 2016) and scater ( McCarthy et al., 2016) Bioconductor packages ( Gentleman et al., 2004). The UMI dataset was normalised using scran prior to differential expression analysis, as it clearly showed composition bias. Differential expression analysis in the mESCs was performed using edgeR, specifying a log-fold-change cut-off of 1 for the full-length dataset, and 0.5 for the UMI dataset. GO analysis was performed with hypergeometric tests using the goana function in the Bioconductor R package limma ( Ritchie et al., 2015). All scripts for analysing the datasets are available on the Oshlack lab Github page ( https://github.com/Oshlack/GeneLengthBias-scRNASeq).
Results
Gene length bias is apparent in scRNA-Seq in non-UMI based protocols
Initially, we analysed three different datasets generated using full-length transcript protocols: mouse embryonic stem cells ( Kolodziejczyk et al., 2015), human primordial germ cells ( Guo et al., 2015) and human brain whole organoids ( Camp et al., 2015). For a full list of the datasets analysed see Supplementary Table 1. Quality control of the single cells was performed and problematic cells filtered out (see methods), leaving 530 mouse embryonic stem cells, 226 human primordial germ cells and 494 human brain organoid cells. For each gene, the average log-counts, normalised for sequencing depth, and the proportion of zeroes across the cells (i.e. the dropout rate per gene) were calculated. Gene-wise abundances were estimated for all datasets by dividing the gene-level counts by gene length to obtain reads per kilobase per million (RPKM). In order to assess gene length bias, genes were assigned to 10 bins based on gene length, such that each bin had roughly 1000 genes. The results are summarised in the boxplots in Figure 1.
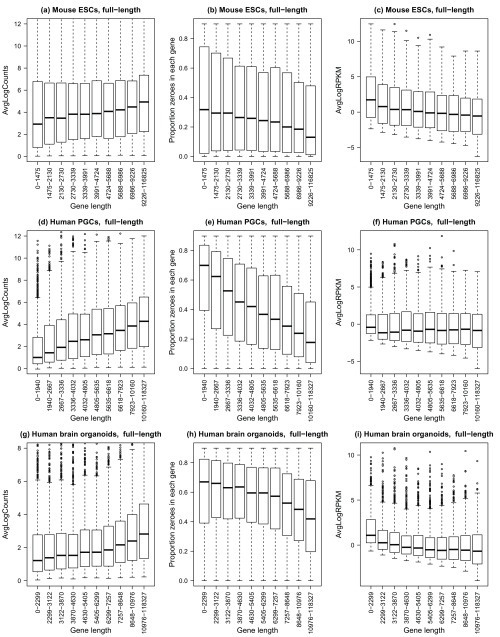
Three different datasets were analysed: ( a– c) mouse embryonic stem cells, n=530 ( Kolodziejczyk et al., 2015), ( d– f) human primordial germ cells, n=226 ( Guo et al., 2015), ( g– i) human brain whole organoids, n=494 ( Camp et al., 2015). For all plots ( a– i), the x-axis shows 10 gene length bins all containing roughly equal numbers of genes. The left panel shows gene-wise average log counts, the middle panel shows proportion of zeroes in each gene (dropout rate per gene), and the right panel shows average log counts corrected for gene length (RPKM).
For all three full-length protocol datasets, shorter genes have lower count level expression proportions compared to longer genes, with a clear trend of increasing log-counts as gene length increases ( Figure 1a, d, g). This was accompanied by a decreasing trend in the dropout rate per gene as gene length increased, highlighting the fact that shorter genes are more difficult to detect using full length protocols ( Figure 1b, e, h). These trends are stronger for the human PGCs and human brain organoid datasets, while not as severe for the mouse ESCs. Calculating transcript abundance by dividing gene-level counts by gene length mostly removed the gene length bias for the human PGCs and brain organoid datasets ( Figure 1f, i), however for the mouse ESCs calculating RPKMs appeared to induce a trend with gene length such that shorter genes appeared more highly expressed relative to the longer genes ( Figure 1c).
UMI-based protocols do not suffer from gene length bias
We hypothesised that because UMI protocols tag each transcript molecule separately we would not see a similar gene length bias in these protocols. In order to assess gene length bias in scRNA-Seq datasets with included UMIs, we analysed three different datasets: mouse embryonic stem cells generated using a CEL-Seq protocol ( Grün et al., 2014; Hashimshony et al., 2012), human induced pluripotent stem cells generated using a modified SMARTer protocol with the Fluidigm C1 system ( Tung et al., 2016) and human leukemia cell line K562 cells using the CEL-Seq protocol with InDrop ( Klein et al., 2015). After quality control and filtering of problematic cells, 127 single cells remained for the mouse embryonic stem cells, 671 for human induced pluripotent stem cells and 219 human K562 cells.
We found that for the human iPSCs and human K562 datasets, the average log-counts were fairly uniform across the 10 gene length bins, and for the mouse ESCs, the shorter genes appear to be more highly expressed than the longer genes ( Figure 2a, d, g). Comparing medians, the dropout rate per gene is slightly lower for shorter genes in the mouse ESCs, while for the human iPSCs and K562 cells, the dropout is fairly uniform across the gene length bins, although slightly more variable for the shortest genes ( Figure 2b, e, h). However, calculating RPKMs by dividing by gene length induces a clear trend with gene length where shorter genes appear to be more highly expressed relative to longer genes, with the median log RPKM decreasing with increasing gene length ( Figure 2c, f, i).
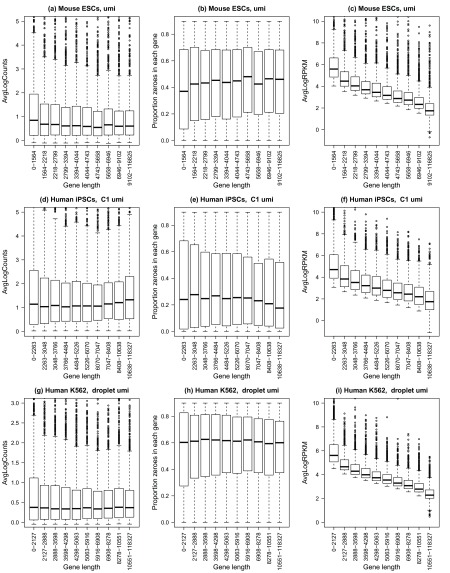
Three different datasets were analysed: ( a– c) mouse embryonic stem cells n=127 ( Grün et al., 2014), ( d– f) human induced pluripotent stem cells n=671 ( Tung et al., 2016), and ( g– i) human leukemia cell line K562 cells, n=219 ( Klein et al., 2015). For all plots ( a– i), the x-axis shows 10 gene length bins all containing roughly equal numbers of genes. The left panel shows gene-wise average log counts, the middle panel shows proportion of zeroes in each gene (dropout rate per gene), and the right panel shows average log expression corrected for gene length (RPKM).
Comparing gene length bias between different mouse embryonic stem cell datasets
To ensure the gene length bias is not due to the specific biology of the different cell types, we analysed four different mouse embryonic stem cell datasets generated using both UMI and full-length transcript protocols ( Buettner et al., 2015; Grün et al., 2014; Kolodziejczyk et al., 2015; Ziegenhain et al., 2016). When we combined all four datasets together (see methods) and performed principal components analysis, we noted that the cells clustered by dataset, with the UMI datasets on the left and full-length datasets on the right of the plot ( Figure 3a). Interestingly, in principal components two and three, we saw some biological structure in the datasets emerging, with cells grown in different media clustering together ( Figure 3b). In particular, three different datasets (two full-length, one UMI), grown in standard media with 2i inhibitors all cluster together on the left of the plot. This shows great promise for obtaining biologically interesting results from combining multiple datasets generated in separate labs using different technology.
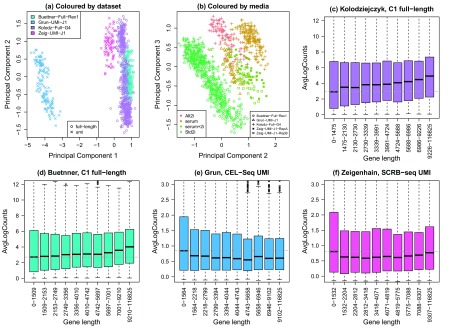
Four different mouse embryonic stem cell datasets were combined, two full-length transcript ( Buettner et al., 2015; Kolodziejczyk et al., 2015) and two UMI datasets ( Grün et al., 2014; Ziegenhain et al., 2016). ( a) Principal component analysis plot (coloured by dataset) shows the major source of variation between the cells is the dataset, with the UMI datasets on the left and the full-length datasets on the right. ( b) Examining principal components two and three reveals that the next major source of variation in the data is the media in which cells are grown. In particular three datasets (two full-length and one UMI) which have cells grown in standard media with 2i inhibitors all cluster together on the left. J1, Rex1 and G4 refer to the mESC cell line. The Ziegenhain dataset has single cells profiled in two batches. ( c– d) Gene length bias is present in full-length mESC datasets; dotted grey line is the median log-count in the first gene length bin. ( e– f) Gene length bias is absent in UMI mESC datasets; dotted grey line is the median log-count in the first gene length bin.
In terms of the gene length bias across the multiple datasets, it is clear that data generated from full length protocols exhibit gene length bias, with shorter genes having lower average log-counts compared to longer genes ( Figure 3c, d). This is not as pronounced compared to other full-length datasets ( Figure 1d, g), however compared to the UMI mESC datasets it is quite noticeable. For the UMI datasets, the gene length bias is mostly uniform across the gene length bins, however the shortest genes in the first bin appear to have slightly higher average log-counts and are more variable compared to the longer genes ( Figure 3e, f).
Detection differences in UMI and full-length mESC datasets
In order to investigate whether choice of protocol impacts which genes are detected, we compared genes detected in both UMI mESC datasets to genes detected in both full-length mESC datasets. Across all datasets, 13434 genes were detected in at least one of the four datasets. Across both UMI datasets, 8866 genes were detected with counts in at least 10% of the cells for each dataset. For the full-length datasets, 11328 genes were detected using the same criteria. The full-length datasets had much greater sequencing depth (median ~ 3million reads, Supplementary Table 1) and more cells compared to the UMI datasets (median ~33,000 reads, Supplementary Table 1), hence it is unsurprising that more genes are detected across both full-length datasets. However, there were 188 genes detected in the UMI datasets that were not detected in the full-length datasets ( Figure 4a). The genes unique to the UMI datasets tended to be shorter compared to the gene lengths of the 2644 genes uniquely detected in the full-length datasets ( Figure 4b, p-value=0.000297, Wilcoxon Rank Sum Test). The genes uniquely detected in either the full-length or UMI datasets tended to be lowly expressed, hence more difficult to detect in general ( Supplementary Figure 1).

( a) A Venn diagram comparing the number of genes detected in two UMI mESC datasets, with the number detected in the two full-length datasets. We find that while the majority of genes are detected in all datasets (n=8689), there are genes that are uniquely detected when using either a full-length or UMI protocol. ( b) Density plots of gene length for the subsets of genes corresponding to the Venn diagram in ( a). The uniquely detected genes for the UMI datasets (blue line) tend to be shorter than the uniquely detected genes in the full-length datasets (red line), p=0.000297. ( c) A Venn diagram showing the number of enriched GO categories in the 188 genes unique to UMIs and the 2649 genes unique to the full-length protocols. This reveals that these genes interrogate different biology, with only 3 GO categories in common. ( d) Density plots of average gene length for each GO category corresponding to the significantly enriched GO categories in ( c). We assigned each GO category an average length by calculating the median of the lengths of all genes annotated to each GO category. While there is not a significant shift in location in the density plots we noted a much greater spread of median length in the enriched GO categories for the uniquely detected UMI genes, largely driven by the presence of GO categories that tend to have very short genes.
Comparing differential expression between two media (2i inhibitors versus serum) in one UMI dataset ( Grün et al., 2014), revealed that 31% (59/188) of the uniquely detected genes were defined as significantly differentially expressed (total differentially expressed = 1641/9962, 16%). For a similar comparison in a full length dataset (2i inhibitors versus serum, Kolodziejczyk et al., 2015), 20% (531/2644) of the uniquely detected genes in full length datasets were significantly differentially expressed (total differentially expressed = 1653/12395, 13%). This highlights that protocol choice may impact ability to detect differential expression of some genes.
Examining which GO terms are over-represented for the 188 genes unique to the UMI dataset revealed that categories such as neural crest cell migration, negative regulation of megakaryocyte differentiation and stem cell development were among the 26 statistically significantly enriched categories ( Supplementary Table 2). There were 4/26 GO categories with extremely short average gene length (<1000, median gene length across all GO categories = 4039), with the top two GO categories, “nucleosome” and “DNA packaging complex”, having median gene length in GO categories = 614, 706. However there were also statistically significant categories comprised of longer than average genes (13/26 categories with median length > 4039), indicating that pathways enriched for the unique UMI genes were not heavily biased towards categories only containing short genes.
For the full-length datasets, the GO categories that were significantly enriched (n=111) were different to those pathways enriched for the unique UMI genes, with only 3 GO categories overlapping ( Figure 4c, Supplementary Table 3). GO categories such as those involved in plasma membrane, cell signalling, and ion and cation channel activity, were over-represented for the 2649 unique genes. While there were no significantly enriched GO categories that had extremely small average gene length (<1000), 14% (16/111) had median gene length < 2632 (the 5 th percentile of median gene length across the GO categories). There was one statistically significant GO category with extremely large average gene length (> 10,000). Although there was no significant shift in median gene length of GO categories between the UMI and full-length GO categories, we noted that the variation in median GO length for the uniquely detected UMI genes was 3.5 times greater than for the uniquely detected full-length genes, largely driven by prevalence of very small sets ( Figure 4d, p-value = 5.6x10 -6, F-test).
Discussion
While single cell RNA-sequencing technology is advancing at a rapid rate and novel discoveries are being made, the datasets being generated have many technical biases. Here, we have investigated the role that gene length plays in protocols that include UMIs as well as full-length transcript protocols. Unsurprisingly, we find that for full-length protocols, genes that tend to be shorter have lower counts and a higher rate of dropout, while UMI based protocols have a more even distribution of dropout across genes of varying length. In addition, a UMI protocol is more likely to detect lowly expressed genes that are shorter compared to a full-length protocol, where lowly expressed genes that are longer are easier to detect ( Supplementary figure 1). Of course, UMI protocols are unable to provide information on transcript structure such as which isoforms are expressed in a sample, and only provide overall gene level expression measures. Since UMI counts are already molecule counts, expression levels should be expressed as normalised counts (e.g. counts per million) rather than dividing by gene length to obtain RPKMs, as this latter measure will artificially inflate the expression estimates of shorter genes relative to longer genes.
While datasets generated using a UMI based protocol tend to have much lower sequencing depths, and hence lower counts, we found that in mESCs we were still able to detect uniquely expressed genes in the UMI datasets that were not detected in full-length datasets. However, a larger set of genes were detected in the mESC full-length datasets. Performing GO analysis on genes uniquely detected by each protocol revealed that they interrogate different biology, and hence the choice of protocol may affect which pathways can be studied. In particular, the genes unique to either the UMI or the full-length datasets appeared to be biologically relevant, as a subset were found to be significantly differentially expressed when comparing cells grown in two different media.
We combined four different datasets generated from mESCs that had strikingly different sequencing depths and protocols. Despite these differences, we found that we were able to recover biologically relevant structure. In particular, three different datasets (two full-length, one UMI), grown in standard media with 2i inhibitors, all cluster together when examining higher principal components. Although promising, the greatest source of variation between the cells was the dataset they belonged to, highlighting the known issues with large batch effects in scRNA-seq ( Tung et al., 2016; Hicks et al., 2015). Hence, analysis methods including data cleaning and normalisation are crucial when combining datasets in order to extract biologically meaningful relationships.
Data and software availability
Latest source code for scripts used to analyse the datasets:
https://github.com/Oshlack/GeneLengthBias-scRNASeq
Information on the repositories and accession numbers of all datasets used in this study:
Mouse embryonic stem cells, Kolodziejczyk et al., 2015, full-length: ArrayExpress database under accession number E-MTAB-2600.
Human primordial germ cells, Guo et al., 2015, full-length: GEO under accession number GSE63818
Human cerebral organoid cells, Camp et al., 2015, full-length: SRA under accession number SRP066834
Mouse embryonic stem cells, Grün et al., 2014, UMI: GEO under accession number GSE54695
Human induced pluripotent stem cells, Tung et al., 2016, UMI: author’s GitHub repository, https://github.com/jdblischak/singleCellSeq.
Human K562 cells (lymphoblastoma culture), Klein et al., 2015, UMI: GEO under accession number GSM1599500
Mouse embryonic stem cells, Ziegenhain et al., 2016, UMI: GEO under accession number GSE75790
Mouse embryonic stem cells, Buettner et al., 2015, full-length: European Nucleotide Archive under accession PRJEB6989
Funding Statement
Luke Zappia is supported through an Australian Government Research Training Program Scholarship. Alicia Oshlack is supported through a National Health and Medical Research Council Career Development Fellowship APP1126157.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary material
Supplementary Figure 1: Average log counts for detected genes in UMI and full-length transcript protocols. The average log counts tend to be much lower for UMI datasets compared to the full-length datasets. The genes uniquely detected for each protocol tend to be lowly expressed, hence more difficult to detect.
.
Supplementary Table 1: Details of the datasets analysed in the paper.
.
Supplementary Table 2: Enrichment of GO categories for the 188 genes uniquely detected in the UMI mESC datasets.
.
Supplementary Table 3: Enrichment of GO categories for the 2649 genes uniquely detected in the full-length mESC datasets.
.
References
- Buettner F, Natarajan KN, Casale FP, et al. : Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015;33(2):155–60. 10.1038/nbt.3102 [Abstract] [CrossRef] [Google Scholar]
- Camp JG, Badsha F, Florio M, et al. : Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci U S A. 2015;112(51):15672–7. 10.1073/pnas.1520760112 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, et al. : STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. 10.1093/bioinformatics/bts635 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Ewels P, Magnusson M, Lundin S, et al. : MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–3048. 10.1093/bioinformatics/btw354 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, et al. : Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. 10.1186/gb-2004-5-10-r80 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Grün D, Kester L, van Oudenaarden A: Validation of noise models for single-cell transcriptomics. Nat Methods. 2014;11(6):637–640. 10.1038/nmeth.2930 [Abstract] [CrossRef] [Google Scholar]
- Guo F, Yan L, Guo H, et al. : The Transcriptome and DNA Methylome Landscapes of Human Primordial Germ Cells. Cell. 2015;161(6):1437–1452. 10.1016/j.cell.2015.05.015 [Abstract] [CrossRef] [Google Scholar]
- Hashimshony T, Wagner F, Sher N, et al. : CEL-Seq: Single-Cell RNA-Seq by Multiplexed Linear Amplification. Cell Rep. 2012;2(3):666–673. 10.1016/j.celrep.2012.08.003 [Abstract] [CrossRef] [Google Scholar]
- Hicks SC, Teng M, Irizarry RA: On the widespread and critical impact of systematic bias and batch effects in single-cell RNA-Seq data. bioRxiv. 2015. 10.1101/025528 [CrossRef] [Google Scholar]
- Islam S, Zeisel A, Joost S, et al. : Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods. 2014;11(2):163–166. 10.1038/nmeth.2772 [Abstract] [CrossRef] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, et al. : Droplet Barcoding for Single-Cell Transcriptomics Applied to Embryonic Stem Cells. Cell. 2015;161(5):1187–1201. 10.1016/j.cell.2015.04.044 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kolodziejczyk AA, Kim JK, Tsang JC, et al. : Single Cell RNA-Sequencing of Pluripotent States Unlocks Modular Transcriptional Variation. Cell Stem Cell. 2015;17(4):471–485. 10.1016/j.stem.2015.09.011 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Langmead B, Trapnell C, Pop M, et al. : Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. 10.1186/gb-2009-10-3-r25 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Liao Y, Smyth GK, Shi W: featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–30. 10.1093/bioinformatics/btt656 [Abstract] [CrossRef] [Google Scholar]
- Liao Y, Smyth GK, Shi W: The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41(10):e108. 10.1093/nar/gkt214 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lun AT, Bach K, Marioni JC: Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016;17(1):75. 10.1186/s13059-016-0947-7 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Macosko EZ, Basu A, Satija R, et al. : Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161(5):1202–1214. 10.1016/j.cell.2015.05.002 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- McCarthy DJ, Campbell KR, Lun AT, et al. : scater: pre-processing, quality control, normalisation and visualisation of single-cell RNA-seq data in R. bioRxiv. 2016. 10.1101/069633 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Oshlack A, Wakefield MJ: Transcript length bias in RNA-seq data confounds systems biology. Biol Direct. 2009;4:14. 10.1186/1745-6150-4-14 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Patro R, Duggal G, Love MI, et al. : Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14(4):417–419. 10.1038/nmeth.4197 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, et al. : limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. 10.1093/nar/gkv007 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK: edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. 10.1093/bioinformatics/btp616 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Sadedin SP, Pope B, Oshlack A: Bpipe: a tool for running and managing bioinformatics pipelines. Bioinformatics. 2012;28(11):1525–1526. 10.1093/bioinformatics/bts167 [Abstract] [CrossRef] [Google Scholar]
- Soumillon M, Cacchiarelli D, Semrau S, et al. : Characterization of directed differentiation by high-throughput single-cell RNA-Seq. bioRxiv. 2014. 10.1101/003236 [CrossRef] [Google Scholar]
- Stegle O, Teichmann SA, Marioni JC: Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015;16(3):133–145. 10.1038/nrg3833 [Abstract] [CrossRef] [Google Scholar]
- Tung PY, Blischak JD, Hsiao C, et al. : Batch effects and the effective design of single-cell gene expression studies. bioRxiv. 2016;62919 10.1101/062919 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, et al. : Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11(2):R14. 10.1186/gb-2010-11-2-r14 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Zheng GX, Terry JM, Belgrader P, et al. : Massively parallel digital transcriptional profiling of single cells. bioRxiv. 2016. 10.1101/065912 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Ziegenhain C, Vieth B, Parekh S, et al. : Comparative analysis of single-cell RNA sequencing methods. bioRxiv. 2016. 10.1101/035758 [Abstract] [CrossRef] [Google Scholar]
Phipson, Zappia and Oshlack present evidence against the existence of systematic gene length bias in single cell RNA sequencing experiments that use unique molecular identifiers (UMI). In contrast, methods that measure read counts across full length transcripts appear similar to bulk RNA sequencing methods in that they are biased against short transcripts. Although these results are somewhat unexpected by those working in the field, the thorough analysis presented by Phipson et al. will be a valuable reference to those wishing to design single cell RNA-seq experiments. The article is written in a clear and accessible manner, and it is also nice to see all the analysis code has been made available. However, there are a number of minor issues with the paper in it's current form that we think should be addressed.
Of particular note, the paper appears to conflate UMI methods with 3' counting methods. We see this as incorrect as i) long-read sequencing technology may allow profiling of full-length transcripts while incorporating UMIs, and ii) 3' counting methods can be used without UMIs. The effect of 3' counting on gene length bias could be separated from the effect of using UMIs by ignoring UMIs in a 3' counting experiment and testing to see if substantial gene length bias exists. Our guess is that it would not, due to the simple fact that the effective gene length is approximately equal for all genes when you measure only the last ~300 bp. Therefore, for the examination of gene length bias, it seems to us that the emphasis should be on 3' counting and not UMIs. Of course, not using UMIs would introduce substantial PCR amplification bias, but this is a separate issue to that being addressed by the paper.
Minor comments:
Introduction:
"...technology enables researchers to examine transcription at the resolution of a single cell..." -- The technology measures mRNA abundance, not transcription itself. (paragraph 1)
"...alternative splicing... analysis is not possible with data generated with protocols that include UMIs." -- it is possible that long-read technologies (eg. Pacbio or Oxford Nanopore) could be coupled with UMI tagged cDNA generated using drop-seq methods before cDNA fragmentation to capture full-length transcripts. (paragraph 3)
It may be beneficial to include supplemental table 1 in the main text.
Processing of all datasets:
Why are different cutoffs used for filtering out cells between experiments? eg. 80% dropout for Kolodziejczyk, 85% dropout for Guo, 90% for Camp, 70% for Tung, 85% Klein. Similar with the library size cutoff and percent ERCC cutoff.
In the Klein methods section, ERCC percentage is reported as >0.01 total library size rather than the percentage. For readability it may be better to have consistent style throughout the manuscript (eg. percent total for everything).
For Ziegenhain methods, it's stated that all cells appeared high quality and so weren't filtered. What constitutes high quality, and how was this assessed? As the count matrix was used in this case, were the cells pre-filtered by the original authors?
Statistical analysis:
"UMI dataset was normalized using scran ...as it clearly showed composition bias." What method was used for normalization, and what exactly is meant by 'compositional bias' and how was this assessed? We believe the scran package depends on scater for implementation of it's normalization methods.
Why use different fold change parameters for UMI and full-length methods? Also, a log (is this log2?) fold change of 1 is 0 fold change. Furthermore, how were the log transformed values calculated for datasets with many zeros?
Figure 1:
More informative axis labels, eg. "Average normalized read counts (log2 scale)" rather than "AvgLogCounts" would increase readability.
Please note that the Tung et al. paper is now published in Scientific Reports and the Ziegenhain paper is published in Molecular Cell.
Figure 4:
The comparison of the number of genes detected by UMI vs full-length methods is somewhat confounded by the differing sequencing depth between the methods. This is stated in the text, but a better comparison could be made by sub-sampling reads from the experiments to equivalent numbers of reads per cell.
We have read this submission. We believe that we have an appropriate level of expertise to confirm that it is of an acceptable scientific standard.
The paper presents a very useful investigation of the dependence of detection efficiency in single cell RNA sequencing on (a) gene length and (b) certain choices in the experimental protocol, namely, shotgun sequencing of full transcripts versus transcript end sequencing such as in methods that employ UMIs. Overall the article is well written, clear, and likely to be useful for practitioners of experimental design and data analysis in the field.
I have a few points that a revised version might address:
The term ‘dropout’ is used in many places, but not properly defined, neither mathematically nor biophysically. At some point in the middle of the manuscript the authors seem to imply that they use ‘dropout’ as a synonym to ‘occurrence of a zero count’ in the data. What is the rationale behind giving the name ‘dropout’ to such an event? What is dropping, and out of what? I understand that some colleagues use this term to point to high probabilities of seeing a zero count for (low abundant?) genes due to the sparse sampling, but I wonder whether (or in which datasets, protocols) this is really something that is more ominous than what is trivially implied by Poisson or Gamma-Poisson statistics, and if so, whether only 0s are ominous or also 1s, 2s, …? Given that this is a paper by statisticians on detection biases it would be great to see a more careful treatment of this aspect of the data.
Why are the parameter choices in Section “Processing of all datasets” (for fraction of dropouts and number of reads) so different between the different datasets? There seems to be a potential for the introduction of biases or artefacts in the computed statistics (of Figs. 1 and 2) through choices made here, and it would be good to demonstrate that such biases, if any, are inconsequential.
In Figs. 1 and 2, how are the ‘average log counts’ computed for data that contain a lot of zeros? The logarithm is not defined for 0. And whatever is the answer to this question, how did the authors make sure that it introduces no biases/artefacts that affect the shown trends? In particular, in conjunction with the filtering steps mentioned above in Point 2?
In Figs. 1 and 2, how is the set of genes selected that enter the calculation of ‘Proportion of zeros in each gene’? Again, how can we be sure that the choices made in the filtering do not affect the conclusions made here?
It is recommendable that the scripts are provided in a github repository. I wonder whether the authors would be willing to go the full length and upload the scripts to a repository that also does regular “live” testing of the scripts for functionality (e.g. installation, dependencies, versions, data availability), such as Bioconductor or CRAN.
On p.5., the authors report differing trends for human PGCs and human brain organoids, compared to mouse ESCs. Do they imply that this is a biological observation, and if so, what does it mean? Or could there be confounding with experimental circumstances? (In which case the effect would perhaps better be reported in association with that than with the names of biological conditions).
In the Discussion and on p.5, results from applying RPKM to UMI-based data are reported. Perhaps the point could be strengthened that already for very basic theoretical reasons this is a nonsensical thing to do. Finding this also empirically is nice, but perhaps it can be said that this confirms basic reasoning rather than being ‘news’.
Minor:
On p.1, a wording is used that implies that datasets are being sequenced. But nucleotides are sequenced, and datasets are produced.
I think the term “pseudo-aligned / pseudo-alignment” is ugly, and “mapped / mapping” is better and more widely used in the field.
On the bottom right of p.4, the term “log-fold change cut-off of 1” is unclear. Which base? Also, do you perhaps mean absolute logarithmic fold change?
The boxplots in Figs. 1 and 2 are a bit dull. Use of geom_hex with aes(x=rank(genelength)) in ggplot2 could present an alternative.
In the caption of Fig.1, ambiguity in the term ‘log counts corrected by gene length’ could be avoided by more explicit mathematical terminology (e.g. corrected = divided?)
Discussion: the conclusion that the choice of protocol may affect which pathways can be studied is a bit wild, and probably also not helpful if not translated into concrete advice to readers for how to address it when doing their experimental designs.
I have read this submission. I believe that I have an appropriate level of expertise to confirm that it is of an acceptable scientific standard.
General comments
This is a well-written and concise study that reveals some very interesting results. Firstly, pertaining to the enrichment of biological processes in data processed using different protocols, the fact that the genes detected in full-length transcript and 3' transcript-end protocols show markedly different specificity for enriched pathways is intriguing. With 3' being highly specific and biologically relevant, compared to the generic pathways identified for full-length data, it raises the possibility that a much higher resolution of biological function and greater classification accuracy might be attained if full-length transcript data was re-analyzed as 3' transcript-end. Secondly, the importance of using correct normalization methods for UMI data is shown here to be of critical importance for accurate analysis.
Also, many thanks to the authors for making their analysis code available on GitHub.
Summary
In this manuscript, the authors asked whether single-cell RNA-seq data would be biased by choice of protocol. Specifically, they looked at the difference between data generated using full-length transcript protocols compared to those generated using 3' transcript end-only protocols that incorporate unique molecular identifiers. To do this, they used publically available scRNA-seq datasets from mouse and human.
Their conclusion is that full-length transcript methods exhibit gene length bias, such that short genes have less mapped reads than longer genes, which translates to lower transcript counts and a higher dropout rate. Conversely, UMI-based methods do not suffer from either of these effects. They also demonstrated that a combination of both methods can enhance the biological interpretation of the scRNA-seq data.
Comments for the authors:
For each of the datasets that were pre-processed (not raw data), it is possible that the different reference genomes (hg19, complete GRCh38, transcriptome-only GRCh38) and the use of different software packages could create artifacts that affect data analysis, particularly if the mapping software was an old version. I note that, with respect to pre-processed data, five different aligners were used. It is clear that all alignment packages have their strengths/weaknesses, especially if they haven't been updated regularly. Could the differences between (i) these packages, and (ii) the different references, contribute in any way to the results obtained in this study?
Related to question 1, would isoform/ splice junction-aware aligner yield different results compared to those that aren't designed for that type of mapping? Would you expect a difference in the full-length data sets that were processed with transcriptome-only reference (Guo 2015 1) compared to the complete hg38 genome reference (Camp 2015 2)?
Each pre-processed dataset was filtered using slightly different parameters. How did the authors establish the dropout percentage threshold for removing cells (none, 70, 80, 85)? How were the library size and sequencing read thresholds determined for each sample? It's not clear to me why these should all be different. Is it to maximize the cell numbers on a per-sample basis? Have the authors tried using the same threshold for all pre-processed samples as for the in-house filtering (90%), or applying the other thresholds to raw data?
Gene ontology analysis is widely used and can provide insight into biological functions. I wonder whether the authors also considered using more specific databases such as Reactome or KEGG that can highlight enriched pathways that are not detected by GO analysis. These may show less disparity than GO terms.
Minor point: Throughout the text and in Figure 3 and 4, UMI is capitalised, but in Fig 2 it is shown in lower case in the plot titles.
I have read this submission. I believe that I have an appropriate level of expertise to confirm that it is of an acceptable scientific standard.
References
This is a nice evaluation of the extent of gene length and detection bias in single-cell RNA-seq data sets generated with different types of protocols. Overall, it is clearly written and the results are well presented and agree with expectations. All analysis code is available in a GitHub repository. An additional step towards full reproducibility would be to also make the processed data objects and additional scripts, not present in the GitHub repository, accessible.
Otherwise, my main comment concerns the representation of gene abundances, specifically the calculation of RPKMs by dividing the library size-normalized gene counts with the "exon-union" length of the gene. Without information about which isoform is contributing to the expression of a given gene, this length may be far from the true number of base pairs "contributing" to the observed reads. An alternative approach would be to aggregate isoform-level TPM estimates (from methods like Salmon 1, RSEM 2 or kallisto 3) to the gene level, and I am wondering whether that would affect the conclusions. Similarly, it could be interesting to investigate whether suggested alternatives to actual or expected read counts, such as "scaled TPMs" 4 or census counts 5, would mitigate the observed gene length bias.
In a couple of places, I think that the manuscript would benefit from some clarifications:
In the last lines of the "Gene filtering" paragraph, it is mentioned that genes that could not be annotated with gene length information were filtered out. How many genes are affected by this, and in what way can they be assigned reads (i.e., correspond to well-defined genomic regions) but not a length?
From the "Processing of all datasets" paragraph, it is not completely clear whether cells are filtered out only if they have both more than (e.g.) 85% dropout and fewer than (e.g.) 500,000 reads, or if one of these criteria alone is enough. It is also not fully clear from the text whether cell filtering or gene filtering was performed first (e.g., the "Gene filtering" paragraph mentions "all cells", but in the following paragraph and in the code it seems that the cell filtering was performed first).
On what values was the principal component analysis applied? Could you expand a bit more on how the data set merging strategy ensures that the larger datasets do not dominate the PCA (they still make up a larger part of the final dataset)?
In the "Statistical analysis" paragraph, how was the UMI data set normalized with scran? Was there an actual normalization step, or a calculation of normalization factors used later in the analysis?
For the four mouse mESC data sets, it might be useful to provide a table listing the conditions (=colors in Figure 3b) that were included in each of them, since it is a bit difficult to discern all color/symbol combinations in Figure 3b.
The numbers in Figure 4a and b do not match (the numbers in Figure 4b match those given in the text, while those in Figure 4a match the figure legend).
Are the two densities in Figure 4d generated with the same kernel width? If not, the differences may be visually exaggerated.
For the preprocessing of the Guo et al. data set, the pseudo-alignment with Salmon was done to the reference transcriptome rather than the genome.
Finally, for the gene set analysis, in addition to the observation that there are some gene sets with short median gene lengths that are among the most enriched in the "UMI-specific" genes, it might be interesting to see whether these gene sets were in fact top-ranked because of the short genes contained in them, or if it was the longer genes in these gene sets that were the significant ones.
I have read this submission. I believe that I have an appropriate level of expertise to confirm that it is of an acceptable scientific standard.
References
Articles from F1000Research are provided here courtesy of F1000 Research Ltd
Citations & impact
Impact metrics
Article citations
Deep learning-based models for preimplantation mouse and human embryos based on single-cell RNA sequencing.
Nat Methods, 14 Nov 2024
Cited by: 0 articles | PMID: 39543284
Quantification and statistical modeling of droplet-based single-nucleus RNA-sequencing data.
Biostatistics, 25(3):801-817, 01 Jul 2024
Cited by: 5 articles | PMID: 37257175 | PMCID: PMC11247185
Data normalization for addressing the challenges in the analysis of single-cell transcriptomic datasets.
BMC Genomics, 25(1):444, 06 May 2024
Cited by: 0 articles | PMID: 38711017 | PMCID: PMC11073985
Review Free full text in Europe PMC
Comparative Analysis of Single-Cell RNA Sequencing Methods with and without Sample Multiplexing.
Int J Mol Sci, 25(7):3828, 29 Mar 2024
Cited by: 1 article | PMID: 38612639 | PMCID: PMC11011421
Comparative analysis of 10X Chromium vs. BD Rhapsody whole transcriptome single-cell sequencing technologies in complex human tissues.
Heliyon, 10(7):e28358, 19 Mar 2024
Cited by: 3 articles | PMID: 38689972 | PMCID: PMC11059509
Go to all (38) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
BioProject
- (2 citations) BioProject - PRJEB6989
Functional Genomics Experiments
- (2 citations) ArrayExpress - E-MTAB-2600
GEO - Gene Expression Omnibus (5)
- (2 citations) GEO - GSE54695
- (2 citations) GEO - GSE75790
- (2 citations) GEO - GSE63818
- (2 citations) GEO - GSM1599500
- (1 citation) GEO - GSE77288
Nucleotide Sequences
- (2 citations) ENA - SRP066834
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Detecting differential alternative splicing events in scRNA-seq with or without Unique Molecular Identifiers.
PLoS Comput Biol, 16(6):e1007925, 05 Jun 2020
Cited by: 10 articles | PMID: 32502143 | PMCID: PMC7299405
Full-Length Single-Cell RNA-Sequencing with FLASH-seq.
Methods Mol Biol, 2584:123-164, 01 Jan 2023
Cited by: 3 articles | PMID: 36495447
Quantile normalization of single-cell RNA-seq read counts without unique molecular identifiers.
Genome Biol, 21(1):160, 03 Jul 2020
Cited by: 15 articles | PMID: 32620142 | PMCID: PMC7333325
Computational solutions for spatial transcriptomics.
Comput Struct Biotechnol J, 20:4870-4884, 01 Sep 2022
Cited by: 32 articles | PMID: 36147664 | PMCID: PMC9464853
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Australian Federal Government
National Health and Medical Research Council (1)
Grant ID: APP1126157




