Abstract
Free full text

Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumors
Tissue-resident memory CD8+ T cells (Trm) are positioned at common sites of pathogen exposure where they elicit rapid and robust protective immune responses1,2. However, the molecular signals controlling Trm differentiation and homeostasis are not fully understood. Here we show that mouse Trm precursor cells represent a unique CD8+ T cell subset that is distinct from the precursors of circulating memory populations at the levels of gene expression and chromatin accessibility. Exploiting computational and functional RNAi in vivo screens, we identified the transcription factor (TF) Runx3 as a key regulator of Trm differentiation and homeostasis. Runx3 was required to establish Trm populations in diverse tissue environments and supported expression of critical tissue-residency genes while suppressing genes associated with tissue egress and recirculation. Analysis of the accessibility of Runx3 target genes in Trm-precursor cells revealed a distinct regulatory role for Runx3 in controlling Trm differentiation despite relatively widespread and uniform expression among all CD8+ T cell subsets. Further, we show that human and murine tumor-infiltrating lymphocytes (TIL) share a core tissue-residency gene-expression signature with Trm. In a mouse model of adoptive T cell therapy for melanoma, Runx3-deficient CD8+ TIL failed to accumulate in tumors, resulting in greater rates of tumor growth and mortality. Conversely, overexpression of Runx3 enhanced TIL abundance, delayed tumor growth, and prolonged survival. In addition to establishing Runx3 as a central regulator of Trm differentiation, these results provide novel insight into the signals that promote T cell residency in tissues, which could be leveraged to enhance vaccine efficacy or adoptive cell therapy treatments that target cancer.
Long-lived memory T cells provide protection from reinfection and can serve as endogenous defenders against tumor growth3. Memory CD8+ T cell populations can be broadly segregated into circulating central and effector memory cells (Tcm and Tem) and tissue-resident memory cells (Trm) that primarily reside in non-lymphoid tissues without egress4. Circulating memory CD8+ T cells and Trm exhibit distinct gene-expression profiles5–7; however, the early transcriptional identity of differentiating Trm and the signals controlling their fate are not yet fully appreciated. Here, we utilized an established infection model where TCR transgenic CD8+ T cells responsive to lymphocytic choriomeningitis virus (LCMV) GP33–41 presented by MHC-class 2Db (P14) were transferred into recipient mice followed by infection with LCMV. In this acute infection model, P14 cells located in non-lymphoid tissues on day 7 of infection began to upregulate molecules characteristic of Trm8, including key tissue-retention molecules CD103 and CD69 (Extended Data Fig. 1a). Gene-expression analysis revealed that 90–96% of the genes upregulated in mature P14 Trm in the kidney parenchyma or intraepithelial lymphocyte (IEL) compartment of the small intestine were elevated in Trm-precursor cells relative to splenic effector cells on day 7 of infection (Fig. 1a). Furthermore, analysis of genes differentially expressed between splenic and non-lymphoid populations on day 7 of infection revealed two distinct gene-expression programs that segregated circulating (PBL, spleen, Tcm, and Tem) from non-lymphoid (kidney and IEL) P14 cells, independent of infection timepoint (Fig. 1b). Lymph node (LN) or splenic KLRG1loCD127hi memory-precursor (MP) cells preferentially give rise to circulating memory populations whereas shorter-lived KLRG1hiCD127lo terminal effector (TE) cells exhibit less memory potential3. Day 7 IEL P14 cells comprising the precursors of Trm, were transcriptionally distinct from splenic MP cells (Fig. 1c). This is notable as IEL Trm are predominantly KLRG1lo,9 and preferentially differentiate from lymphoid-derived KLRG1lo precursors seeding non-lymphoid tissues from days 4.5–7 of infection10 (Extended Data Fig. 1a–c), consistent with studies of skin Trm6. Thus, the Trm-precursor populations in non-lymphoid tissues are transcriptionally distinct from circulating effector cells as well as MP cells on day 7 of infection, and the majority of the Trm transcriptional program is already established at this time point, prior to contraction of the CD8+ T cell population.
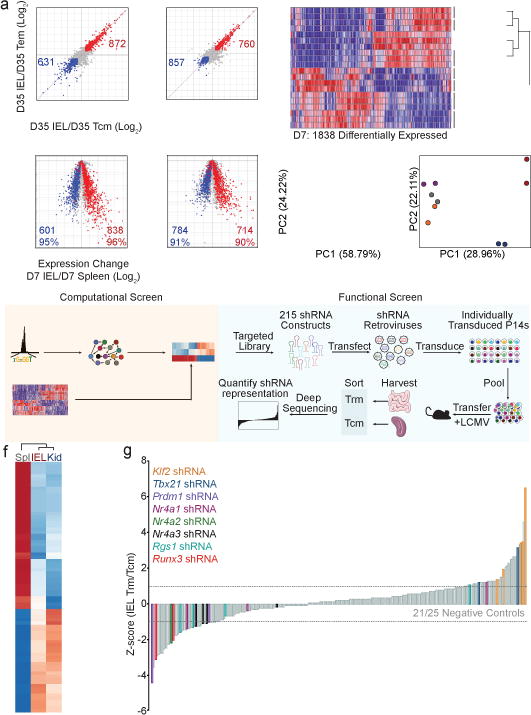
Computational and functional RNAi screens identify transcriptional regulators of Trm differentiation.
a, Comparison of gene-expression of IEL (left) and kidney Trm (right) relative to Tcm and Tem subsets on day 35 of LCMV infection; red, genes elevated in Trm relative to Tcm and Tem; blue, genes elevated in Tcm and Tem relative to Trm (top). Comparison of differentially-expressed genes in mature Trm (from top panel) in cells from the spleen, IEL, or kidney on day 7 of infection (bottom). b, Differentially-expressed genes between splenic, IEL, and kidney populations on day 7 of infection were compared among effector and memory CD8+ T cell subsets. Populations are ordered by hierarchical clustering with Pearson correlation. c, PCA of differentially-expressed genes among day 7 subsets and naive P14 cells. d, PCA of differential global chromatin accessibility of subsets on day 7 of infection identified by ATAC-seq analysis. e, Combinatorial screening approach. f, TFs with a PageRank score of ≥1.5-fold change in day 7 non-lymphoid cells compared to day 7 splenic cells are included in the heatmap; genes known to regulate Trm formation are in bold font. i, Relative enrichment of shRNAmirs in IEL Trm relative to splenic Tcm from the RNAi screen, reported as the average Z-score from 3 independent screens where each independent screen was performed by pooling DNA from sorted P14 cells from 15–18 mice. Each timepoint represents an individual experiment consisting of 2–3 biological replicates where n=2–10 mice were pooled for each replicate (a–d).
As chromatin accessibility is a key determinant of cell identity and fate, we profiled non-lymphoid and splenic effector populations using ATAC-seq on day 7 of infection. Uniquely accessible chromatin regions were identified in IEL P14 cells for genes characteristic of mature Trm (e.g. Cd69 and Nr4a1) whereas genes that promote T cell re-circulation (e.g. Klf2 and S1pr1) exhibited loss of accessible regions (Extended Data Fig. 2a). Principal component analysis (PCA) highlighted that, despite day 7 being an “effector” time point, the global chromatin landscape dramatically differs between effector CD8+ T cells located in the spleen, including MP cells, and those located in non-lymphoid tissues (Fig. 1d). The unique chromatin configuration of differentiating Trm is consistent with the striking transcriptional differences observed (Fig. 1a–c) and foreshadows the distinct fates of antigen-specific cells in the spleen relative to non-lymphoid tissues. Thus, precursors of Trm cells in non-lymphoid sites are a unique and distinct CD8+ T cell subset relative to effector cells in the lymphoid compartment, including the MP population.
Specification of CD8+ T cell fate during infection is dependent on the integrated activity of multiple TFs3, and notable regulators of Trm formation include Hobit6, Blimp16, Nr4a111, Eomes12, and T-bet12,13. To facilitate a broader understanding of the transcriptional network driving Trm differentiation, we utilized a combined screening approach, consisting of a computational strategy integrating ATAC-seq data, transcriptional profiling and personalized PageRank analysis to predict regulatory TFs, and a functional in vivo RNAi screen targeting putative Trm regulators identified through the computational approach (Fig. 1e). We recently demonstrated that analysis of accessible TF binding motifs and TF-target gene expression yielded insight into TFs with regulatory functions in the differentiation of circulating memory CD8+ T cells14. Leveraging this approach and the personalized PageRank analysis15, we predicted a number of TFs with established regulatory roles in controlling Trm differentiation (Blimp16, Nr4a111, Eomes12, T-bet12,13) and many with no previously described role in Trm (Fig. 1f, SI Table 1). We evaluated both barrier (IEL) and non-barrier (kidney) Trm sites to reveal TFs important to Trm differentiation independent of the tissue. Additionally, a key strength of this computational screen is that influential roles of differentially expressed TFs as well as TFs with homogenous expression can be anticipated (Extended Data Fig. 2b). To establish functional relevance for predicted regulators of Trm formation identified through PageRank analysis, we utilized an RNAi screening strategy16 to test hundreds of individual shRNAmir constructs in parallel for activity in promoting or repressing Trm differentiation in vivo (Fig. 1g, SI Table 2). Several TFs with established roles in regulating Trm were identified (Nr4a113, Blimp16, Klf217 and T-bet12,13) as well as TFs with previously unknown functions in controlling CD8+ Trm formation such as Nr4a3 and Runx3 (Fig. 1i).
Runx3 is a well-established regulator of CD8+ T cell thymocyte development18, supports cytotoxic activity of mature CD8+ T cells19,20, and controls CD4+ T cell localization within the intestinal epithelium21. Although little is known regarding a role for Runx3 in CD8+ Trm, both computational and functional screens identified Runx3 as a putative regulator of Trm fate specification (Fig. 1f,g) despite relatively uniform Runx3 expression in circulating and resident CD8+ T cell subsets (Extended Data Fig. 2b, ,3a).3a). We validated a role for Runx3 through a 1:1 mixed transfer of P14 cells transduced with control (Cd19 shRNAmir) or Runx3 shRNAmir-encoding retroviruses into mice that were subsequently infected with LCMV (Fig. 2a). Runx3 shRNAmir suppressed Runx3 expression (Extended Data Fig. 3b) and impaired the formation of IEL Trm relative to circulating cells (Fig. 2a and Extended Data Fig. 3c,d), consistent with the RNAi screen. Further, Runx3-shRNAmir knockdown in the context of a localized enteric Listeria monocytogenes expressing GP33–41 (LM-GP33–41) infection similarly impaired Trm differentiation (Fig. 2b).
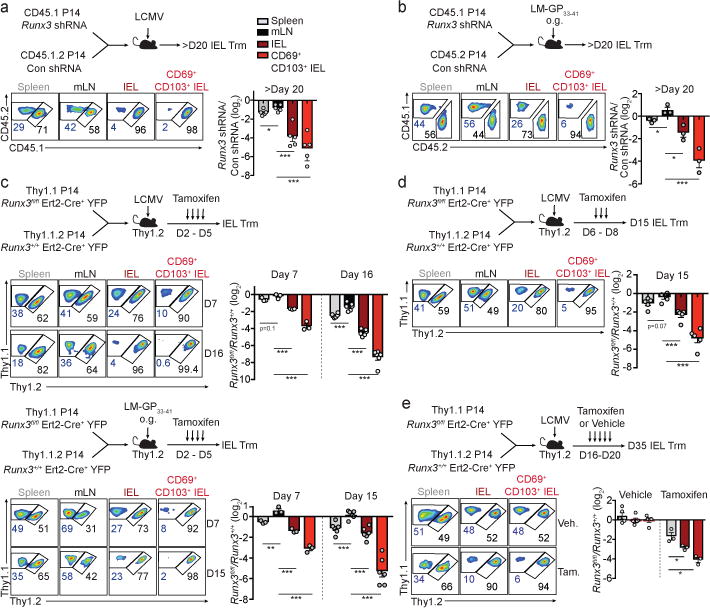
Runx3 is essential for the differentiation and long-term maintenance of CD8+ Trm cells.
a, Ratio of Runx3- or control-shRNAmir transduced P14 cells in indicated tissues on day 23 or 26 of LCMV infection. b, Ratio of transduced transferred cells in indicated tissues on day 32 of enteric LM-GP33–41 infection. c, Ratio of transferred Runx3fl/flErt2-Cre+YFP (Runx3fl/fl) and Runx3+/+Ert2-Cre+YFP (Runx3+/+) P14 cells in indicated tissues on days 6/7 and 15/16 of LCMV infection (top) or enteric LM-GP33–41 infection (bottom). d, Ratio Runx3fl/fl and Runx3+/+ P14 cells on day 15 of LCMV infection. e, Ratio of Runx3fl/fl and Runx3+/+ P14 cells on day 35 of infection. Graphs indicate mean ± s.e.m of n=5 (a), n=3 (b), n=3 (day 7) and n= 6 (day 15/16) (c), n=5 (d), and n=5 (vehicle) and n=3 (tamoxifen) (e). All data are from one representative experiment of 2 independent experiments except a is pooled from 2 independent experiments; *P<0.05, **P<0.01, ***P<0.005. Symbols represent an individual mouse (a–e).
Next, utilizing a tamoxifen-inducible deletion approach, Runx3fl/fl-Ert2-Cre+ P14 (Runx3fl/fl) or Runx3+/+-Ert2-Cre+ P14 (Runx3+/+) cells were mixed 1:1 and transferred into host mice followed by LCMV or enteric LM-GP33–41 infection (Fig. 2c). Runx3-deficiency resulted in a 2–6-fold loss of splenocytes and minimal loss of mLN cells by day 15/16 of infection. However, Runx3-deficiency resulted in a 50–150-fold loss of CD69+CD103+ Trm in both infection settings (Fig. 2c and Extended Data Fig. 3e). Moreover, delaying tamoxifen treatment to days 6–8 or 16–20 of infection further emphasized a distinct dependence of Trm differentiation on Runx3 (Fig. 2d) as well as a critical role for Runx3 in maintaining Trm homeostasis, respectively (Fig. 2e, Extended Data Fig. 3f). Furthermore, Runx3 was necessary for optimal Trm differentiation of H-2Db GP33–41 tetramer+ cells (Extended Data Fig. 4a–d). Taken together, these data demonstrate that Runx3 is critical for Trm differentiation and maintenance.
Runx3 deletion also resulted in a loss of Trm in non-barrier tissues (salivary gland and kidney, Extended Data Fig. 5a–b), and optimal Trm differentiation in the skin and lung parenchyma required Runx3 (Extended Data Fig. 5c–h). Thus, the loss of Trm in a range of non-lymphoid tissues indicated Runx3 drives Trm formation independently of the tissue site. Further, Runx3 was required for maximal granzyme B expression in Trm, although cytokine production was not affected (Extended Data Fig. 6a,b). Runx3-deficiency resulted in a greater frequency of Annexin V+ cells (Extended Data Fig. 6c,d), most prominently in CD69+CD103+ Trm; thus, the marked loss of Trm was at least in part due to a greater rate of apoptosis, as proliferation and trafficking were not impacted (Extended Data Fig. 6e,f).
We next assessed if ectopic expression of Runx3 could augment Trm differentiation. Overexpression of Runx3 accelerated IEL P14 CD69+CD103+ Trm differentiation on day 8 of infection, but did not impact migration to the small intestine (Fig. 3a). Evidence of enhanced Trm differentiation was further confirmed by the greater abundance of IEL Trm on day 12/13 of infection and enhanced CD103 expression, consistent with a reported role for Runx3 in regulating CD103 expression21,22 (Fig. 3b). Additionally, ectopic expression of Runx3 also boosted Trm differentiation in the lung parenchyma (Extended Data Fig. 7a–d).
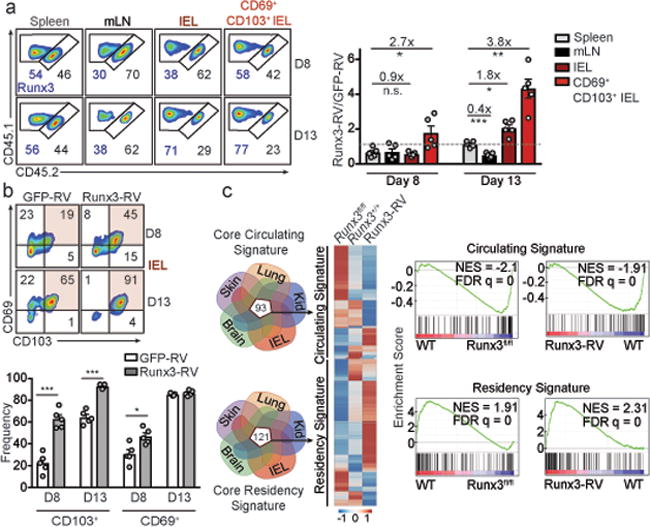
Runx3 programs CD8+ T cell tissue-residency.
a, Congenically distinct P14 cells were transduced with Runx3-RV (CD45.1+ cells) or GFP-RV (CD45.1.2+ cells), mixed at a 1:1 ratio, and transferred to recipient mice subsequently infected with LCMV. Ratio of transduced cells evaluated on days 8 and 12/13 of infection. b Frequency of CD69+ and CD103+ cells from a. c, Relative expression of the core “circulating” and “residency” genes between Runx3-RV, Runx3fl/fl, and Runx3+/+ CD8+ T cells (left) and gene set enrichment analysis (right). Graphs show mean ± s.e.m of n=5 mice (a,b) from one representative experiment of 2 independent experiments, *P<0.05, **P<0.01, ***P<0.005, n.s., not significant. Symbols represent an individual mouse (a,b).
Given that manipulation of Runx3 impacted Trm formation in diverse tissue microenvironments, we constructed a core Trm transcriptional signature by computational integration of CD8+ Trm gene-expression datasets from the IEL, kidney, lung5, skin5 and brain7, to evaluate the hypothesis that Runx3 is a universal regulator of Trm specification (Fig. 3c, SI Table 3). Notably, we found the majority of the core tissue-residency signature genes were upregulated in Runx3-overexpressing cells and downregulated in Runx3-deficient cells. Conversely, the core signature of circulating memory cells was enriched in Runx3-deficient cells and depleted from Runx3-overexpressing cells (Fig 3c). Therefore, Runx3 promoted expression of tissue-residency signature genes and repressed genes characteristic of circulating cells, and this conclusion was further corroborated by ChIP-seq analysis23 indicating that Runx3 binding was enriched in both core tissue-residency and circulating genes relative to background sites (Extended Data Fig. 8a).
Through evaluation of accessible Runx3 binding motifs from ATAC-seq analysis, we generated a regulatory Runx3 binding network (Extended Data Fig. 8b) and found Runx3 putatively regulates a distinct network of genes in differentiating IEL-Trm precursor cells relative to splenic effector cells, including selective enrichment of genes linked to cell adhesion and regulation of TF activity. In connection, Runx3 has been shown to cooperate with the TF T-bet in multiple contexts19,24, yet T-bet is a potent suppressor of early Trm differentiation12,13. ChIP-seq data23 indicated Runx3 directly binds to multiple sites of the Tbx21 locus (encoding T-bet, Extended Data Fig. 8c), and Runx3-deficient CD8+ T cells exhibited elevated T-bet levels (Extended Data Fig. 8d). Knockdown of Tbx21 in Runx3-deficient cells enhanced Trm numbers in the IEL compartment and restored CD103 and CD69 expression (Extended Data Fig. 8e,f), but did not fully rescue Trm differentiation. These findings are consistent with Runx3 regulating multiple targets that influence Trm formation (Fig. 3c) including suppression of canonical tissue egress genes (Extended Data Fig. 8g,h).
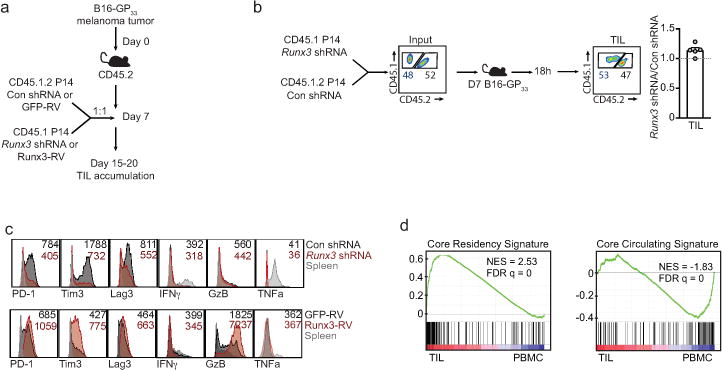
It has been noted that CD8+ tumor infiltrating lymphocytes (TIL) can exhibit characteristics of Trm, and a positive prognosis has been correlated with TIL that present qualities of Trm25,26. As Runx3 regulates core features of tissue-residency (Fig. 3c), we assessed the transcriptional similarities of Trm and TIL and evaluated a role for Runx3 in controlling TIL accumulation. TIL isolated from mouse melanoma27 or mammary tumors27 shared ~70% of the core tissue-residency gene-expression program relative to splenic CD8+ T cells (Fig. 4a), and this relationship was further highlighted through PCA (Fig. 4b). Utilizing an adoptive therapy model, Runx3-knockdown or Runx3-overexpressing P14 cells were mixed with control P14 cells at a 1:1 ratio and transferred into mice with established melanoma tumors expressing GP33–41 (Extended Data Fig 9a). Runx3-deficiency impaired TIL accumulation (Fig. 4c,d) without impacting migration to the tumor (Extended Data Fig. 9b). Conversely, Runx3-overexpression enhanced TIL abundance (Fig. 4c,d), expression of granzyme B (Extended Data Fig. 9c) and certain core tissue-residency genes while further suppressing core circulating genes (Fig. 4e). In clinical settings, TIL density strongly correlates with positive outcomes28, and we observed Runx3-deficient P14 cells were impaired in their ability to control tumor growth, resulting in greater mortality (Fig. 4f). Conversely, Runx3-overexpressing cells delayed tumor growth and prolonged survival (Fig. 4g). Notably, human CD8+ TIL also exhibited enrichment of the core tissue-residency signature relative to circulating CD8+ T cells25 (Extended Data Fig. 9d), and analysis of single-cell RNA-seq data from mouse29 and human melanoma TIL30 indicated that activated CD44+CD8+ T cells expressing Runx3 exhibited enrichment of the tissue-residency gene-expression signature relative to CD44+CD8+ TIL with low Runx3 expression levels (Fig. 4h). These data indicate that in both human and murine TIL, tissue-residency features are likely driven by Runx3. In connection, it was recently demonstrated that human lung cancer TIL enriched with certain qualities of Trm also strongly correlated with TIL abundance and a positive prognosis26. Taken together, manipulation of TFs promoting tissue-residency may yield more effective TIL and anti-viral memory T cells through supplementing CD8+ T cells with a gene-expression program that better supports features important to both Trm and TIL such as in situ survival, tissue retention, and repression of egress, ultimately fostering accumulation of protective T cells in tissues.
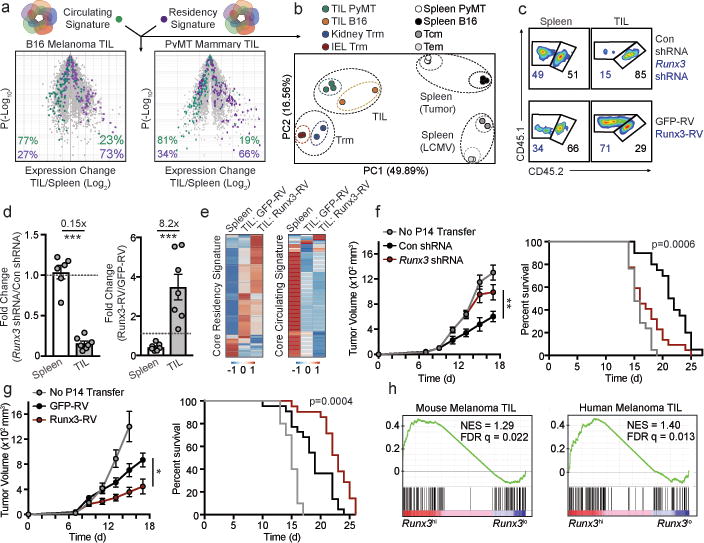
CD8+ TIL share transcriptional similarity with Trm and require Runx3 for tumor residency.
a, Comparison of the core tissue-residency signature and core circulating signature (from Fig. 3c) in B16 melanoma CD8+ TIL27 or PyMT mammary tumor CD8+ TIL27 relative to corresponding splenic cells. b, PCA of gene-expression of the core tissue-residency and circulating gene sets for TIL, Trm, or splenic subsets. c, d Congenically distinct P14 cells were transduced with Runx3 shRNAmir or Runx3-RV (CD45.1+ cells) and Con shRNAmir or GFP-RV (CD45.1.2+ cells), mixed at a 1:1 ratio and transferred into mice with established B16-GP33–41 melanoma tumors. Flow plots and graphs indicate ratio of transduced cells. e, Relative expression of the core tissue-residency and core circulating gene sets in GFP-RV splenocytes, GFP-RV TIL, and Runx3-RV TIL following the same approach as in c. f,g, Tumor growth and survival following adoptive transfer of the indicated cell population. h, Gene set enrichment analysis of the core tissue-residency signature in Runx3hi vs Runx3lo TIL from single-cell RNA-seq analyses of mouse29 and human30 melanoma TIL. Graphs indicate mean ± s.e.m of n=5 (Runx3 shRNAmir) or n=7 mice per group (Runx3-RV) from one representative experiment of 3 independent experiments (c,d) or data pooled from 3 independent experiments consisting of n=10–21 mice per group (f, g), *P<0.05, **P<0.01, ***P<0.005. Symbols represent an individual mouse (d).
Methods
Mice
Mice were maintained in specific-pathogen-free conditions in accordance with the Institutional Animal Care and Use Committees (IACUC) of the University of California, San Diego (UCSD) and The Scripps Research Institute, Jupiter, FL (TSRI-FL). All mice were of a C57BL6/J background and bred at UCSD and TSRI-FL or purchased from the Jackson Laboratory, including: WT or P14 mice with distinct expression of the congenic molecules CD45.1, CD45.2, Thy1.1, and Thy1.2 as well as control Thy1.1+Thy1.2+ Runx3+/+Ert2-Cre+YFP P14 mice and Runx3 inducible deletion Thy1.1+ Runx3fl/flErt2-Cre+YFP P14 mice. Runx3+/+dLck-Cre+YFP and Runx3fl/fldLck-Cre+YFP mice were used for studying polyclonal CD8+ T cell responses. The Rosa26 stop-flox eYFP reporter mice were used for all Runx3-deletion experiments. Cre-mediated deletion disrupts the Runx3 DNA-binding domain in exon 4, which exists in transcripts originating from both the distal and proximal promoter. Thus, both long and short Runx3 forms are inactivated in these alleles.
Naive T cell transfers, infection, and treatments
Naive P14 CD8+ T cells were transferred intravenously (i.v.) into congenically distinct sex matched recipient mice, or female P14 cells were transferred into male mice. For all microarray, RNA-seq, or ATAC-seq experiments, a total of 1×105 P14 cells were transferred. For co-transfer experiments, naive Thy1.1+Thy1.2+ Runx3+/+ Ert2-Cre YFP+ P14 cells and naive Thy1.1+ Runx3fl/fl Ert2-Cre YFP+ P14 cells were mixed 1:1 and a total of 3×104 P14 cells were transferred into Thy1.2+ recipient mice. Recipient mice were subsequently infected i.p. with 2×105 PFU of the Armstrong strain of lymphocytic choriomeningitis virus (LCMV) or 1010 CFU of Listeria monocytogenes expressing GP33–41 via oral gavage9 one day after cell transfer. For induced Runx3 deletion, recipient mice were treated with 1mg of tamoxifen diluted in sunflower oil i.p. on days 0–4, 2–5, or 6–8 of infection. For late deletion of Runx3 (days 16–20), recipient mice were treated with 2mg of tamoxifen via oral gavage.
For Trm precursor experiments, 1×105 P14 cells were transferred, recipient mice were infected with LCMV the next day, and KLRG1lo or KLRG1hi P14 cells from spleens and lymph nodes were sorted on day 5 of infection. Sorted cells (1×105) were transferred into recipient mice infected 4 days prior with LCMV. The number of CD62L+ Tcm, CD62L− Tem, or IEL Trm were evaluated on day 20–25 of infection using flow cytometry.
To distinguish vascular associated CD8+ T cells in non-lymphoid tissues, 3μg of CD8α (53–6.7) conjugated to APC eFlour780 was injected i.v. into mice four minutes prior to sacrifice and organ excision. CD8αneg cells were considered to be localized within non-lymphoid tissues.
Preparation of cell suspensions
Isolation of CD8+ T cells was performed similarly as described31. For isolation of CD8+ T cells from the small intestine intraepithelial lymphocyte (IEL) compartment, Peyer’s patches were removed and the intestine was cut longitudinally and subsequently cut laterally into 0.5–1cm2 pieces that were then incubated with 0.154mg/mL dithioerythritol (DTE) in 10% HBSS/HEPES bicarbonate for 30min at 37°C while stirring. Kidneys, salivary glands, and lungs were cut into pieces and digested for 30min with 100 U/mL type I collagenase (Worthington) in RPMI 1640, 5% FBS, 2mM MgCl2, 2mM CaCl2 at 37°C while shaking. Skin was processed similarly as described32 in which a 2cm2 area of the right flank was excised, pre-digested for 30min at 37°C and then enzymatically digested with 0.7 mg/mL collagenase D. After enzymatic incubations (skin, lungs, kidneys, salivary glands), tissues were further dissociated over a 70μm nylon cell strainer (Falcon). For isolation of lymphocytes, single-cell suspensions were then separated using a 44/67% Percoll density gradient. Spleens and lymph nodes were processed with the frosted ends of microscope slides. Red blood cells were lysed with ACK buffer (140 mM NH4Cl and 17 mM Tris-base, pH 7.4).
Antibodies, intracellular staining, flow cytometry, and cell sorting
The following antibodies were obtained from eBioscience: CD8α (53–6.7), CD8β (eBio H35–17.2), CD62L (MEL-14), CD127 (A7R34), KLRG1 (2F1), CD103 (2E7), CD69 (H1.2F3), CD45.1 (A20–1.7), CD45.2 (104), Thy1.1 (OX-7, HIS51), Thy1.2 (53–2.1), CCR9 (Ebio CW-1.2), CXCR3 (CXCR3–173), CD49d (R1–2), TNFα (MP6-XT22), GzB (GB11), PD-1 (J43), Tim3 (RMT3–23), Lag3 (eBioC9B7N), KI-67 (SolA15), and IFNγ (XMG1.2) or from BioLegend: CD62L (MEL-14), CD103 (2E7), CD69 (H1.2F3), CD45.1 (A20–1.7), Thy1.1 (OX-7), Thy1.2 (30-H12), and T-bet (4B10). For analysis of apoptosis, the Annexin V Apoptosis Detection Kit was used per manufacturer instructions (eBioscience); propidium iodide negative cells were analyzed for Annexin V staining. The H-2Db GP33–41 tetramer was obtained from the NIH Tetramer Core. For intracellular staining of cytokines or TFs while preserving ametrine or YFP reporter expression in transduced or Cre-YFP+ populations, cells were fixed and permeabilized through a 10min incubation with BD cytofix/cytoperm (BD Biosciences). Intracellular staining was subsequently performed using the Permeabilization Buffer of the Foxp3-Transcription Factor Staining Buffer Set (eBioscience). To assess cytokine production, CD8+ T cells were re-stimulated with the GP33–41 peptide in the presence of Protein Transport Inhibitor Cocktail (eBioscience). For flow cytometry analysis, all events were acquired on a BD LSRFortessa X-20 or a BD LSRFortessa. Cell sorting was performed on BD FACSAria or BD FACSAria Fusion instruments.
RNAi screening approach
We have described this screening approach in detail previously16. The targeted shRNAmir library was generated based on key genes identified from the computational screening approach as well as genes with known roles in regulating Trm from literature. The library was produced by cloning shERWOOD-designed shRNAmir sequences33, after PCR of synthetic 97mer oligos, into our pLMPd-Amt vector16. Purified DNA from sequence-verified clones was used to package retroviral particles in PLAT-E cells. For transfections, PLAT-E cells were seeded in the middle 60 wells of a 96-well flat bottom plate at a density of 4–6×104 cells/well one day prior to transfection. Next, each well was individually transfected with 0.2μg of DNA from each pLMPd-Amt clone and 0.2μg of pCL-Eco using TransIT-LT1 (Mirus). Retroviral supernatant was harvested 36, 48, and 60h after transfection, and RV sup from each well was used to individually transduce in vitro activated P14 cells in 96-well round bottom plates.
For CD8+ T cell activation in vitro, naive CD8+ T cells from spleen and lymph nodes were negatively enriched and 2×105 P14 cells were plated in the middle 60 wells of 96-well round bottom plates pre-coated with 100μg/mL goat anti-hamster IgG (H+L, Thermoscientific) and 1μg/mL anti-CD3 (145–2C11) and 1μg/mL anti-CD28 (37.51) (both from eBioscience). Culture media was removed 18h after activation, and replaced with retroviral supernatant supplemented with 50μM BME and 8μg/mL polybrene (Millipore) followed by spinfection (60min. centrifugation at 2000 rpm, 37°C). Two hours after the spinfection, the P14 cells were washed 3 times with cold PBS and 90% of each well of cells (individually transduced with distinct retroviral constructs) was harvested, pooled and 5×105 pooled P14 cells were transferred into recipient mice which were then infected 1h later with 1.5×105 PFU of LCMV clone 13 i.p. 1h later, resulting in an acute infection16. The remaining cells in vitro were cultured for an additional 24h and either pooled for “input” sequencing (6×105 P14 cells) or were used to test transduction efficiency of each construct using flow cytometry to detect the percentage of ametrine+ cells in each well.
Twelve days after infection, spleens and small intestines were harvested from 15–18 mice and splenocytes and IEL P14 cells were processed as described above. Prior to sorting, all IEL or splenic samples were pooled. CD62L+ P14 cells (Tcm) from the spleen as well as P14 cells from the IEL were sorted (2–6×105 cells total). Genomic DNA was then harvested from sorted cells using the FlexiGene kit (Qiagen). The integrated proviral passenger strand shRNAmir sequences in each cell subset were amplified from 20–100ng total genomic DNA per reaction, with 23–28 cycles of PCR using Ion Proton-compatible barcoded primers that anneal to the common 5′ mir30 and shRNAmir loop sequences. 2–3 replicate reactions were performed for each genomic DNA sample and the replicates were pooled after amplification. The pooled reactions were purified using AMPure XP beads, the amplicons in each sample were quantified using a Bioanalyzer, and then pooled in a 1:1 molar ratio for sequencing. In each replicate of the screen, a minimum of 2.5 million reads per sample were generated and retained, after filtering low-quality reads. Reads assigned to each barcode were aligned to a reference database of all shRNAmirs in the library using BLAST and a custom script to count the top alignment of each read and summarize the number of reads aligned to each shRNAmir.
For analysis of shRNAmir representation in Tcm relative to IEL Trm, the total number of reads in each of the samples was normalized, and the number of reads for each shRNAmir was scaled proportionally. Subsequently, the normalized number of reads in the IEL Trm cells for a given shRNAmir was divided by the normalized number of reads for the same shRNAmir in the Tcm sample and then log2 transformed. The mean and standard deviation of the ratios of each of the 25 negative control shRNAmir constructs (targeting Cd19, Cd4, Cd14, Ms4a1, Cd22, Hes1, Klf12, Mafb, Plagl1, Pou2af1, and Smarca1) were used to calculate the Z-score for each shRNAmir construct. The screen was repeated three times and the Z-score of each construct from each individual screen was averaged and plotted (Fig. 1i, SI Table 2). Certain constructs were added after the first screen or were not detectable in one of the screens, but all constructs were successfully screened 2–3 times except for 13 constructs, which are marked by an asterisk in SI Table 2. Eighty-four percent (21/25) of all negative control shRNAmir constructs had an average Z-score between −0.9 and 0.9.
CD8+ T cell transduction, cell transfer, and infection for individual analysis of retroviral constructs
Activation, transfections, and transductions were carried out as described for the RNAi screening approach except in some experiments 2×106 P14 cells were activated per well in 6-well plates. Congenically distinct P14 cells transduced with the Runx3.2 shRNAmir or Cd19.1 shRNAmir (control) retroviruses were mixed 1:1 within 24h of transduction and a total of 1–5×105 P14 cells were transferred i.v. into recipient mice. One hour after adoptive transfer, recipient mice were infected i.p. or intratracheally (i.t.) with 2×105 PFU LCMV armstrong or intradermally (i.d.) with 2×104 PFU clone 13. In similar experiments, P14 cells were transduced with MigR1-based retroviruses34 that were empty (GFP-RV) or that contained Runx3 cDNA (Runx3-RV), mixed 1:1 and transferred to recipient mice for subsequent infections. For T-bet rescue experiments, Thy1.2+ Runx3+/+ Ert2-Cre YFP+ P14 cells were transduced with Cd19.1 shRNAmir and Thy1.1+ Runx3fl/fl Ert2-Cre YFP+ P14 cells were transduced with Tbx21.3 shRNAmir, mixed 1:1 and transferred into recipient mice, which were infected 1h later with LCMV armstrong i.p. and treated with 1mg tamoxifen i.p. for five consecutive days starting with the day of infection.
Adoptive therapy tumor model
For adoptive therapy experiments, 5×105 B16-GP33 cells, treated for mycoplasma contamination and authenticated in in vitro killing assays, were transplanted subcutaneously into the right flank of wild-type mice. After tumors became palpable, 7–8 days post-transplant, in vitro expanded P14 cells were transferred i.v. For comparison of TIL accumulation in a mixed transfer setting, naive P14 cells were activated, transduced, and expanded with 100U/mL of IL-2 for 2–3 days; cells transduced with control constructs (Cd19.1 shRNAmir or GFP-RV) or experimental constructs (Runx3.2 shRNAmir or Runx3-RV) were mixed 1:1 and 0.5–1×106 P14 cells were transferred i.v. For efficacy studies, transduced cells were expanded for 5–6 days; transduced cells were then sorted (or not sorted with a Runx3-RV and GFP-RV transduction efficiency >83%), and 1–2.5×106 cells were transferred i.v. into mice with established B16-GP33 tumors. Tumors were monitored daily and mice with ulcerated tumors or tumors exceeding 1500 mm3 were euthanized, in accordance with UCSD IACUC .
qPCR, Microarray, RNA-seq, and ATAC-seq analysis
For validation of the Runx3-RV overexpression construct and Runx3.2 shRNAmir construct, enriched CD8+ T cells were activated, transduced, and expanded for 4–6 days in 100U/mL IL-2. Cells were sorted on ametrine (Runx3 shRNAmir or Con shRNAmir) or GFP (Runx3-RV or GFP-RV) directly into TRIzol (Life Technologies) and RNA was extracted per manufacturer’s specifications. Next, cDNA was synthesized using Superscript II (Life Technologies) and qPCR was performed using the Stratagene Brilliant II Syber Green master mix (Agilent Technologies). Runx3 expression levels were normalized to the housekeeping gene Hprt. We have previously validated the Tbx21.3 shRNAmir16. The following primers were used for qPCR: Runx3 forward, 5′-CAGGTTCAACGACCTTCGATT-3′, and Runx3 reverse, 5′-GTGGTAGGTAGCCACTTGGG-3′; Hprt forward, 5′-GGCCAGACTTTGTTGGATTT-3′, and Hprt reverse, 5′-CAACTTGCGCTCATCTTAGG-3′.
On day 7 of infection, tissues from 2–3 mice were pooled and 2–3×104 P14 cells from the IEL, kidney, spleen, or blood were sorted into TRIzol. On day 35 of infection, tissues from 5–10 mice were pooled and 1–2×104 CD62L+ Tcm, CD62L− Tem, kidney Trm, and IEL Trm P14 cells were sorted into TRIzol. As described previously, RNA was amplified and labeled with biotin and hybridized to Affymetrix Mouse Gene ST 1.0 micrroarrays (Affymetrix)35. Analyses were performed using GenePattern Multiplot Studio. Differentially expressed genes in IEL Trm compared to Tcm and Tem as well as kidney Trm compared to Tcm and Tem were identified with a fold change (FC) >1.5 and an expression value (EV) >120 (Fig. 1a). Genes with >1.5 FC and >120 EV between day 7 spleen, day 7 IEL, and day 7 kidney samples were identified (1838 probes) and evaluated in day 7 and day 35 subsets, which were ordered with Pearson correlation using the HierarchicalClustering module of GenePattern (Fig. 1b); data was row centered, row normalized, and visualized with the HierarchicalClusteringViewer module within GenePattern.
The core Trm and circulating signatures were generated by integrating differential expression (>1.5 FC) data comparing Trm from the following tissues to circulating splenic memory cells (or splenic Tcm if both Tcm and Tem datasets were available): D35 IEL (LCMV), D35 kidney parenchyma (LCMV), D30 skin CD103+CD8+ (herpes simplex virus)5, D30 lung CD103+CD8+ (influenza virus)5, and D20 CD103+ brain (vesicular stomatitis virus)7; overlapping genes upregulated in all Trm populations comprised the core tissue-residency signature (121 genes) and genes downregulated in all populations comprised the circulating signature (93 genes). The mouse TIL microarray datasets were generated previously27.
For RNA-seq analysis of D7 IEL, D7 MP, and D7 TE, the populations were sorted on day 7 of LCMV Armstrong infection as well as naive P14 cells; spleens or IEL samples from 2–3 mice were pooled and 5×103 cells were sorted. For RNA-seq analysis of TIL, congenically distinct P14 cells were transduced with Runx3-RV or GFP-RV, mixed 1:1 and 1×106 were transferred to mice with day 7 established melanoma B16-GP33 tumors. Eight days later, 1×103 transduced TIL or splenocytes were sorted from 4 mice for each replicate. For library preparation, isolation of polyA+ RNA was performed as detailed online (www.immgen.com/Protocols/11cells.pdf). For RNA-seq analyses of Runx3-manipulated cells, CD8+ T cells from naive Runx3+/+ YFP+ (WT) and Runx3fl/fl YFP+ (Runx3fl/fl) mice were enriched by negative isolation and transduced (as detailed above) with a Cre cDNA expressing retrovirus (Cre-RV). Runx3-overexpressing cells were generated similarly by transducing Runx3+/+ YFP+ CD8 T cells with a Runx3-cDNA expressing retrovirus (Runx3-RV). Forty-eight hours after TCR activation, the CD8+ T cells were resuspended and re-cultured in fresh media supplemented with 100U/mL rhIL-2; twenty-four hours later, YFP+ (“WT” or “Runx3fl/fl”) or GFP+ (Runx3-RV) were FACS-purified and then recultured in 100U/mL IL-2. The cells were expanded until day 6 by reculturing at 5×105 cells/mL every 24h in fresh 100U/mL IL-2 media. On day 6 post-activation, cells were harvested and total RNA was extracted in TRIzol. Purified RNA was depleted of ribosomal RNA and strand-specific paired-end libraries were prepared and sequenced using an Illumina Nextseq 500. Samples were generated from two biological replicates, and approximately 20 million paired-reads were generated per sample. Reads were mapped using Tophat36 and aligned reads in transcripts were counted with HTseq37. Gene-set-enrichment analysis (GSEA) was performed by using the GSEA module in GenePattern, and the normalized enrichment scores and false-discovery rate q values were determined by using the permutation test.
ATAC-seq was performed as described in detail previously24. Sorted cells (2.5×104) were resuspended in 25μL of lysis buffer and spun down 600g for 30min at 4°C. The nuclear pellet was resuspended in 25μL of Tn5 transposase reaction mixture (Nextera DNA Sample Prep Kit, Illumina) and incubated for 30min at 37°C. Transposase-associated DNA was subsequently purified (Zymo DNA clean-up kit). For library amplification, DNA was amplified using indexing primer from Nextera kit and NEBNext High-Fidelity 2X PCR master mix. Then, the amplified DNA was size-selected to fragments less than 800 bp using SPRI beads. The library was sequenced using Hiseq 2500 for single-end 50-bp sequencing to yield at least 10 million reads. We used bowtie to map raw reads to the Mus musculus genome (mm10) with following parameters: “–best -m 1”. We called peaks for each individual replicate as well as the pooled data from the two replicates using MACS2 with a relaxed threshold (P-value 0.01).
For the single cell RNA-seq analysis of human30 and mouse melanoma TIL29, the preprocessed single cell TIL gene expression data was downloaded from GEO database GSE72056 or GSE86042, respectively. Activated CD8+ TIL (CD8a expression >5 and CD44 expression>2) was used and classified into Runx3hi TIL, which express relatively high levels of Runx3 (Runx3 expression>3) and Runx3lo TIL with no Runx3 expression (Runx3 expression=0). For the human TIL, melanoma #75 was used. GSEA was performed to evaluate enrichment of the core tissue-residency gene expression signature in Runx3hi TIL relative to Runx3lo TIL.
Computational Screen: TF regulatory networks and personalized PageRank analysis
TF regulatory networks and PageRank analysis was performed similarly as described24 except that gene expression and ATAC-seq data from D7 IEL, D7 kidney and D7 spleen samples were used. To construct the TF regulatory network, TF-binding motifs were first scanned on ATAC-seq peaks using an algorithm described previously14 and a P-value cutoff of 1×10−5. Then, we connected a TF to a gene if the TF had any predicted binding motif in the ATAC-seq peak of the nearest gene. We assembled all the interactions between TFs and genes into a regulatory network. To identify important TF regulators for Trm differentiation, we performed personalized PageRank analysis in the TF regulatory network constructed above using the pipeline described previously14. The importance of a TF is based on the quantity and quality of its regulated gene targets. A TF would receive a higher PageRank score if it regulates more important genes where the importance is evaluated by differential expression from microarray or RNA-seq analyses. Extended Data Fig. 2b and SI Table 1 indicate the PageRank score and expression value of all TFs expressed (>120 EV) in the spleen, kidney or IEL cells.
Statistical analysis
Student’s t-test (two-tailed) was used for comparisons between two groups. Log-rank (Mantel-Cox) test was used to compare survival curves. All microarray, RNAseq, and ATACseq samples were performed independently in 2–3 replicates. All statistical tests were performed with GraphPad Prism software and P<0.05 was considered statistically significant.
Data Availability
RNA-seq, microarray, and ATAC-seq data are available in the GEO database: accession codes (will be provided). Source Data are provided in the online version of the manuscript. Additional information and materials will be made available upon request.
Extended Data
Extended Data Figure 1
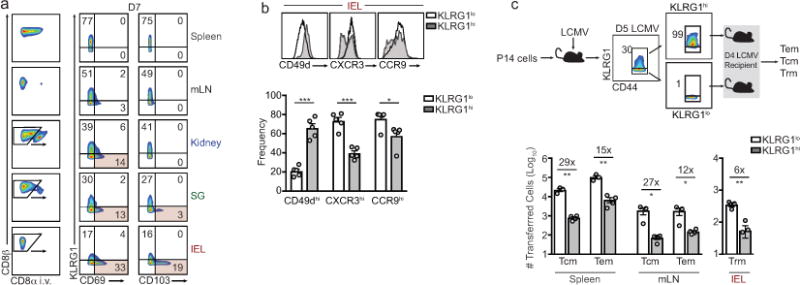
a, Representative flow cytometric gating strategy for distinguishing P14 cells located in non-lymphoid tissues following CD8α i.v. administration in LCMV infected mice (left). Right, in vitro activated P14 cells were transferred to recipient mice and infected with LCMV and the frequency of CD69+ and CD103+ P14 cells among KLRG1hi and KLRG1lo on day 7 of infection is indicated. b, Frequency of CCR9, CXCR3, and CD49d on KLRG1lo and KLRG1hi cells in the IEL compartment on day 7 of infection. c, Schematic of experimental design (top). KLRG1lo and KLRG1hi P14 cells were sorted from spleens and LNs on day 5 of LCMV infection and transferred into recipient mice infected 4 days prior with LCMV. Tcm, Tem, and Trm P14 cells were enumerated on days 20 or 25 of infection using flow cytometry (bottom). Graphs indicate mean ± s.e.m of n=5 mice (a,b) or n=3–4 mice (c) from one representative of 2 independent experiments, *P<0.05, **P<0.01, ***P<0.005. Symbols represent an individual mouse (c).
Extended Data Figure 2
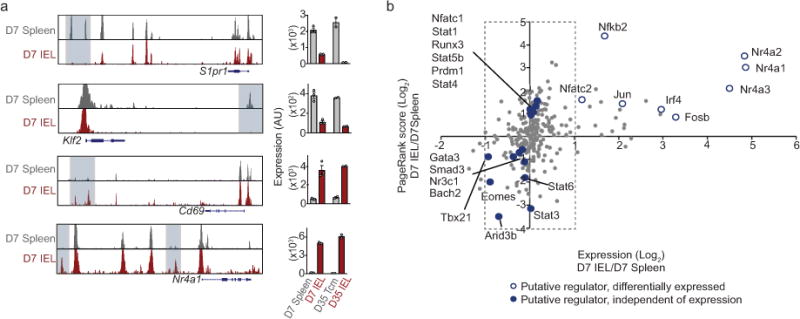
a, ATAC-seq analysis of the indicated loci on day 7 of infection (left) and corresponding gene expression (right). b, Personalized PageRank score and gene-expression of TFs with select TFs highlighted.
Extended Data Figure 3
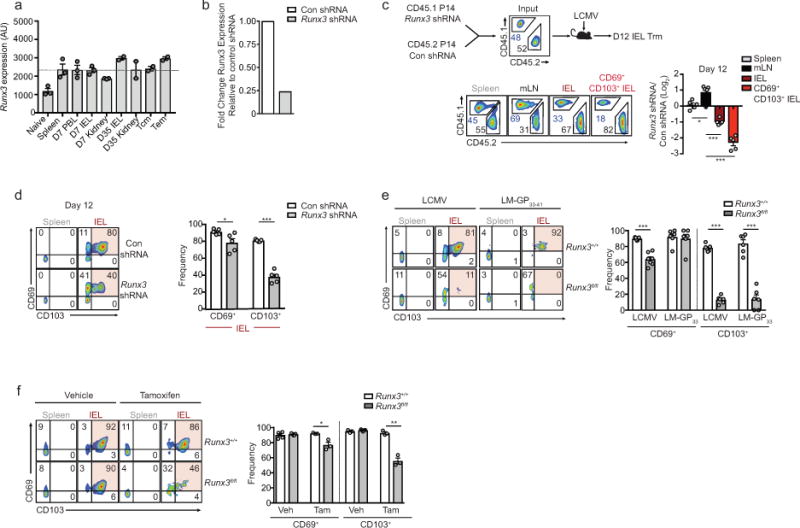
a, Runx3 mRNA levels from indicated cells determined by microarray analyses. b, Relative Runx3 mRNA expression of in vitro cultured cells transduced with Con shRNAmir or Runx3 shRNAmir-encoding retroviruses measured by qPCR. c, Congenically distinct P14 cells were transduced with Runx3 shRNAmir or Con shRNAmir encoding retroviruses, mixed at a 1:1 ratio, and transferred to recipient mice that were subsequently infected with LCMV. Representative flow cytometry plots (bottom, left) and quantification of the ratio of Runx3 shRNAmir or control shRNAmir transduced P14 cells in indicated tissues on day 12 of infection (bottom, right). d, Representative flow cytometry plots (left) and quantification of the frequency of CD69+ and CD103+ cells of Con shRNAmir or Runx3 shRNAmir cells (right) from experimental schematic in c. e, Representative flow cytometry plots and quantification of the frequency of CD69+ and CD103+ cells from Fig. 2 c,d. f, Representative flow cytometry plots and quantification of the frequency of CD69+ and CD103+ cells from Fig. 2f. Graphs indicate mean ± s.e.m and representative of two independent experiments (b) with n=5 (c,d), n=5 (LM-GP33–41) or n=6 (LCMV) (e), and n=5 (vehicle) or n=3 (tamoxifen) (f), *P<0.05, **P<0.01 ***P<0.005. Symbols represent an individual mouse (c–f).
Extended Data Figure 4
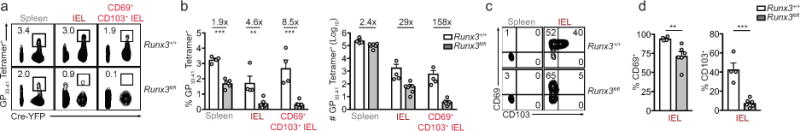
a, Representative flow cytometry plot of H-2Db GP33–41 tetramer staining of lymphocytes from Runx3fl/fl dLck-Cre+YFP and Runx3+/+dLck-Cre+YFP mice on day 12 of LCMV infection (gated on total lymphocytes). b, Quantification of the proportion (left) and absolute number (right) of tetramer+ cells. c,d, Representative flow cytometry plots and quantification of the frequency of CD69+ and CD103+ cells. Graphs indicate mean ± s.e.m with n=4 (Runx3+/+) or n=5 (Runx3fl/fl) mice pooled from two independent experiments, **P<0.01, ***P<0.005. Symbols represent an individual mouse (b,d).
Extended Data Figure 5
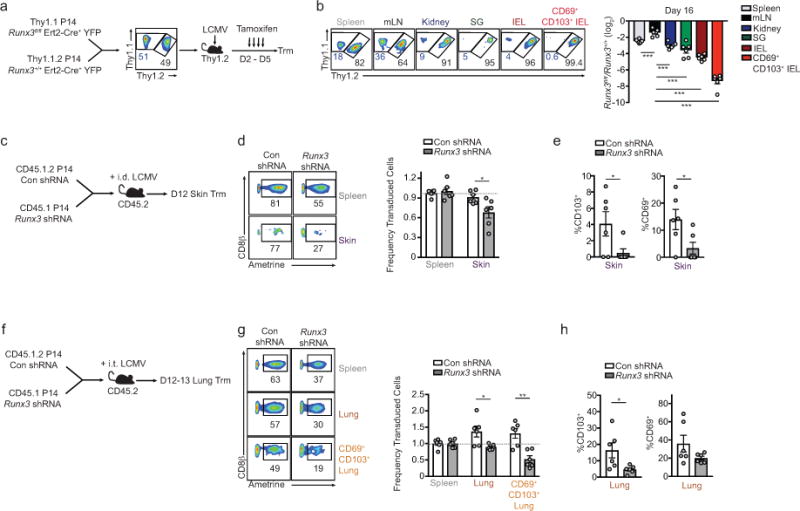
a, Schematic of experimental design. b, Representative flow cytometry plots (left) and quantification (right) of the ratio Runx3fl/fl and Runx3+/+ P14 cells (gated on YFP-Cre+ cells) in lymphoid and non-lymphoid compartments on days 15/16 of LCMV infection (same data as in Fig. 2d but including SG and kidney populations). c, Schematic for experimental design. d, Representative flow cytometry plots (left) and quantification (right) of the ratio of transduced cells in the skin relative to the spleen for Con shRNAmir or Runx3 shRNAmir P14 cells on day 12 of an intradermal (i.d.) LCMV infection. e, Frequency of CD69+ and CD103+ cells. f, Schematic for experimental design. g, Representative flow cytometry plots (left) and quantification (right) of the ratio of transduced cells in the lung parenchyma relative to the spleen for Con shRNAmir or Runx3 shRNAmir P14 cells on day 12 of an intratracheal (i.t.) LCMV infection. h, Frequency of CD69+ and CD103+ cells. Graphs indicate mean ± s.e.m and representative of two independent experiments with n=6 (b), or data pooled from two individual experiments with n=6 per group (c–h), *P<0.05, **P<0.01, ***P<0.005. Symbols represent an individual mouse (b,d,e,g,h).
Extended Data Figure 6
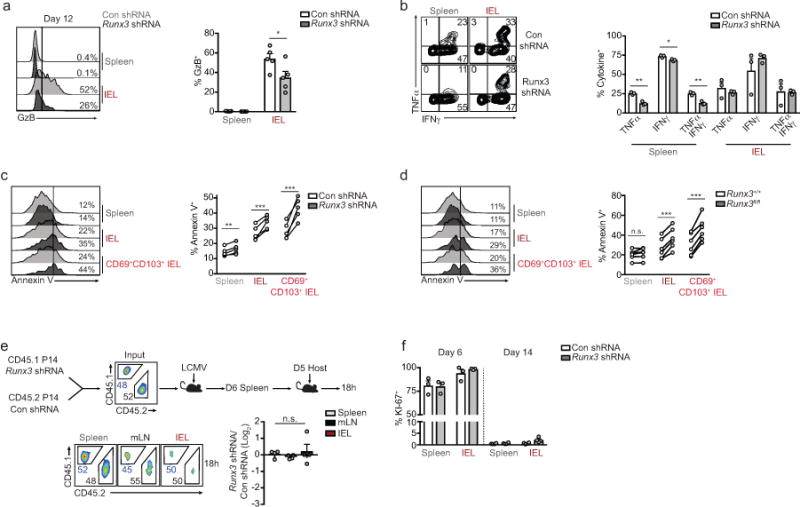
a, Representative flow cytometry histogram of granzyme B (GzB) staining (left) and quantification of frequency of GzB+ cells on day 12 or 14 of infection. b, Representative flow cytometry plots (left) and quantification (right) of the frequency of IFNγ- and TNFα-producing Con shRNAmir or Runx3 shRNAmir P14 cells on day 6 of LCMV infection, restimulated with GP33–41 peptide. c,d, Representative histograms and quantification of Annexin V+ cells from shRNAmir mixed transfers on day 14 of LCMV infection (c) or from day 8 Runx3fl/fl and Runx3+/+ mixed P14 transfers where tamoxifen was administered on days 2–5 of LCMV infection (d). e, Congenically distinct P14 cells were transduced with Con shRNAmir or Runx3 shRNAmir encoding retroviruses, mixed at a 1:1 ratio, and transferred to recipient mice that were subsequently infected with LCMV. On day 6 of infection, splenocytes were harvested and retransferred to day 5 infected host mice and 18h later spleen, mLN and small intestine were harvested to assess trafficking. Representative flow cytometry plots (bottom, left) and quantification of the ratio of Con shRNAmir and Runx3 shRNAmir transduced P14 cells (bottom, right) in indicated tissues 18h after transfer. f, Frequency of KI-67+ Con shRNAmir or Runx3 shRNAmir transduced P14 cells in a mixed transfer setting on days 6 and 12 or 14 of LCMV infection. Graphs indicate mean ± s.e.m and representative of two independent experiments with n=5 (a), n=3 (b), n=5 (c), n=6 (d), n=4 (e), and n=3 on day 6 or n=4 on day 14 (f) except d is pooled from two independent experiments, *P<0.05, **P<0.01, ***P<0.005, n.s., not significant. Symbols represent an individual mouse (a–f).
Extended Data Figure 7
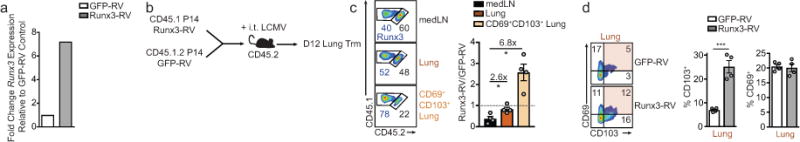
a, Runx3 mRNA expression of in vitro cultured cells transduced with GFP-RV or Runx3-RV. b, Schematic for experimental design of intratracheal (i.t.) LCMV infection. c, Representative flow cytometry plots (left) and quantification (right) of the ratio of GFP-RV or Runx3-RV cells in the mediastinal LN (medLN), lung parenchyma, or CD69+CD103+ lung parenchyma population on day 12 or 13. d, Representative flow cytometry plots (left) and quantification (right) of the frequency of CD69+ and CD103+ P14 cells in the lung parenchyma. Graphs indicate mean ± s.e.m and data representative of one of two independent experiments (a) and n=4 per group (c,d), *P<0.05, ***P<0.005. Symbols represent an individual mouse (c,d).
Extended Data Figure 8
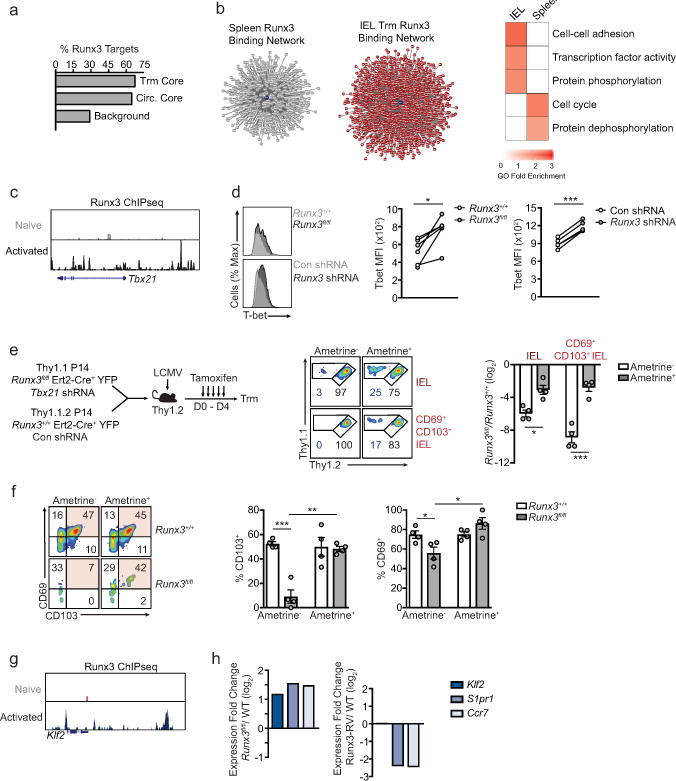
a, Percentage of genes of the core tissue-residency signature, core circulating signature, or background sites that exhibit direct Runx3 binding by ChIP-seq analysis23. b, Predicted Runx3 binding network, generated from ATAC-seq analysis, in IEL P14 cells and splenic P14 cells on day 7 of infection (left). Red indicates genes putatively regulated by Runx3 in IEL cells; grey indicates genes putatively regulated by Runx3 in splenic cells. Gene Ontology (GO) enrichment analysis (right) of gene sets in the predicted Runx3 binding network in each tissue. c, Runx3 ChIP-seq of the Tbx21 locus in naive and activated CD8+ T cells from Lotem et al.23. d, Representative flow cytometry histograms (left) and MFI quantification (right) of T-bet expression in splenic P14 cells on day 8 of infection. e, Schematic for experimental design (left) in which Runx3+/+lErt2-Cre+YFP were transduced with Con shRNAmir and Runx3+/+Ert2-Cre+YFP P14 cells were transduced with Tbx21 shRNAmir, mixed 1:1 and transferred into recipient mice subsequently infected with LCMV. Recipient mice were treated with tamoxifen on days 0–4 of infection. Representative flow cytometry plots (middle panel) and quantification of the ratio of untransduced (ametrine−) Runx3+/+ and Runx3fl/fl P14 cells and the ratio of transduced (ametrine+) Runx3+/+/Con shRNAmir and Runx3fl/fl/Tbx21 shRNAmir (right) were evaluated on day 12 of LCMV infection. f, Representative flow cytometry plots (left) and quantification (right) of the frequency of CD69+ and CD103+ cells. g, Runx3 ChIPseq of the Klf2 locus in naive and activated CD8+ T cells23. h, Fold change in gene-expression of Klf2, S1pr1, and Ccr7 in Runx3fl/fl and Runx3-RV cells relative to Runx3+/+ WT cells, from RNA-seq analysis consisting of 2 replicates per sample. Graphs indicate mean ± s.e.m and data representative of one of two independent experiments with n=6 (Runx3fl/fl) or n=4 (Runx3 shRNA) (d) and n=4 per group(e,f). *P<0.05, **P<0.01 ***P<0.005. Symbols represent an individual mouse (d–f).
Extended Data Figure 9
a, Schematic of adoptive therapy experimental design. b, Congenically distinct P14 cells were transduced with Runx3 shRNAmir or Con shRNAmir encoding retroviruses, mixed at a 1:1 ratio, and transferred into mice with established B16-GP33 melanoma tumors. Eighteen hours after transfer, tumors were harvested to assess the ratio of Runx3 shRNAmir or Con shRNAmir P14 cells. c, Representative flow cytometry histograms of Con shRNAmir, Runx3 shRNAmir, GFP-RV, or Runx3-RV TIL in mixed transfer settings. Control P14 splenocytes were included in histograms for reference. d, Gene set enrichment analysis of the core tissue-residency and core circulating gene signatures in human lung CD8+ TIL relative to corresponding CD8+ PBMCs25. Graphs indicate mean ± s.e.m and combined of two independent experiments with n=5 mice per group (b) or representative of two independent experiments with n=3–6 per group (b). Symbols represent an individual mouse (b).
Acknowledgments
We thank all the members of the Goldrath and Pipkin laboratories for their contributions. We also thank the Flow Cytometry Core at the La Jolla Institute for Allergy and Immunology. This study was funded in part by the US National Institutes of Health U19AI109976 (S.C., M.E.P., A.W.G), California Institute for Regenerative Medicine RB5-07012 (W.W.).
Footnotes
Author Contributions: J.J.M. designed and performed experiments, analyzed the data and wrote the manuscript; C.T. assisted with the RNAi screen, transfections, transductions, tissue processing, and tumor models; B.Y. performed the computational analyses and ATAC-seq experiment; K.Z. assisted with computational analyses; K.O. assisted with tissue processing, analysis, and qPCR; A.T.P assisted with tissue processing, inducible deletion experiments, and analysis; D.W and A.J.G. helped with the inducible deletion experiments, RNA-seq analysis, and tumor models; S.C provided reagents and advice; W.W. supervised the computational analysis and contributed advice; M.E.P. and A.W.G. supervised the project, designed experiments, and wrote the manuscript.
Author Information: The authors declare no competing financial interests.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature24993
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc5747964?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Epstein-Barr virus infection induces tissue-resident memory T cells in mucosal lymphoid tissues.
JCI Insight, 9(20):e173489, 22 Oct 2024
Cited by: 0 articles | PMID: 39264727 | PMCID: PMC11530129
Transcriptional rewiring in CD8<sup>+</sup> T cells: implications for CAR-T cell therapy against solid tumours.
Front Immunol, 15:1412731, 27 Sep 2024
Cited by: 0 articles | PMID: 39399500 | PMCID: PMC11466849
Review Free full text in Europe PMC
A Prime-Boost Vaccination Approach Induces Lung Resident Memory CD8+ T Cells Derived from Central Memory T Cells That Prevent Tumor Lung Metastasis.
Cancer Res, 84(19):3173-3188, 01 Oct 2024
Cited by: 0 articles | PMID: 39350665 | PMCID: PMC11443216
Injury-induced myosin-specific tissue-resident memory T cells drive immune checkpoint inhibitor myocarditis.
Proc Natl Acad Sci U S A, 121(42):e2323052121, 08 Oct 2024
Cited by: 0 articles | PMID: 39378095
Phenotypic and spatial heterogeneity of CD8+ tumour infiltrating lymphocytes.
Mol Cancer, 23(1):193, 09 Sep 2024
Cited by: 1 article | PMID: 39251981 | PMCID: PMC11382426
Review Free full text in Europe PMC
Go to all (358) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus (2)
- (1 citation) GEO - GSE86042
- (1 citation) GEO - GSE72056
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Armed and Ready: Transcriptional Regulation of Tissue-Resident Memory CD8 T Cells.
Front Immunol, 9:1770, 30 Jul 2018
Cited by: 68 articles | PMID: 30131803 | PMCID: PMC6090154
Review Free full text in Europe PMC
The Emerging Role of CD8+ Tissue Resident Memory T (TRM) Cells in Antitumor Immunity: A Unique Functional Contribution of the CD103 Integrin.
Front Immunol, 9:1904, 15 Aug 2018
Cited by: 85 articles | PMID: 30158938 | PMCID: PMC6104123
Review Free full text in Europe PMC
Transcriptional programming of tissue-resident memory CD8+ T cells.
Curr Opin Immunol, 51:162-169, 02 Apr 2018
Cited by: 73 articles | PMID: 29621697 | PMCID: PMC5943164
Review Free full text in Europe PMC
Runx3 drives a CD8+ T cell tissue residency program that is absent in CD4+ T cells.
Nat Immunol, 23(8):1236-1245, 26 Jul 2022
Cited by: 41 articles | PMID: 35882933
Funding
Funders who supported this work.
NCRR NIH HHS (1)
Grant ID: S10 RR027366
NHGRI NIH HHS (1)
Grant ID: R01 HG009626
NIAID NIH HHS (3)
Grant ID: U19 AI109976
Grant ID: R37 AI067545
Grant ID: R01 AI095634
NIH HHS (1)
Grant ID: S10 OD016262





