Abstract
Free full text

WHO/IUIS Allergen Nomenclature: Providing a common language
Abstract
A systematic nomenclature for allergens originated in the early 1980s, when few protein allergens had been described. A group of scientists led by Dr. David G. Marsh developed a nomenclature based on the Linnaean taxonomy, and further established the World Health Organization/International Union of Immunological Societies (WHO/IUIS) Allergen Nomenclature Sub-Committee in 1986. Its stated aim was to standardize the names given to the antigens (allergens) that caused IgE-mediated allergies in humans. The Sub-Committee first published a revised list of allergen names in 1986, which continued to grow with rare publications until 1994. Between 1994 and 2007 the database was a text table online, then converted to a more readily updated website. The allergen list became the Allergen Nomenclature database (www.allergen.org), which currently includes approximately 880 proteins from a wide variety of sources. The Sub-Committee includes experts on clinical and molecular allergology. They review submissions of allergen candidates, using evidence-based criteria developed by the Sub-Committee. The review process assesses the biochemical analysis and the proof of allergenicity submitted, and aims to assign allergen names prior to publication. The Sub-Committee maintains and revises the database, and addresses continuous challenges as new “omics” technologies provide increasing data about potential new allergens. Most journals publishing information on new allergens require an official allergen name, which involves submission of confidential data to the WHO/IUIS Allergen Nomenclature Sub-Committee, sufficient to demonstrate binding of IgE from allergic subjects to the purified protein.
1. Introduction
Early reports of adverse health problems that are thought to represent allergies and asthma date back more than 3000 years from Egypt, Rome and China, but these are not well documented. Likely cases of allergy to various inhalation sources or insect bites or stings are often dismissed as the nature and cause of the reactions were unknown. Allergic diseases are complex and broad in terms of organs affected and severity, ranging from highly prevalent allergic rhinitis to life-threatening anaphylaxis (Cohen and Zelaya-Quesada, 2002). Progress in the allergy field started in 1869 when Dr. Charles H. Blackley demonstrated that pollen was the apparent cause of his hay fever (Taylor and Walker, 1973), and continued throughout the 1900s with the establishment of immunotherapy using whole allergen extracts as a routine practice by the 1970′s (summarized in Fig. 1). A need to develop a systematic nomenclature for allergens became apparent in the early 1980s, when only few allergens had been identified and allergen names were inconsistently used in publications. This article focuses on allergen nomenclature in the context of Immunoglobulin E (IgE)-mediated allergies, directed against apparently harmless substances, mostly proteins and glycoproteins from diverse biological origins. Allergen sources include pollen, mites, animal epithelia and saliva, fungi, insect venoms and a variety of plant and animal foods.
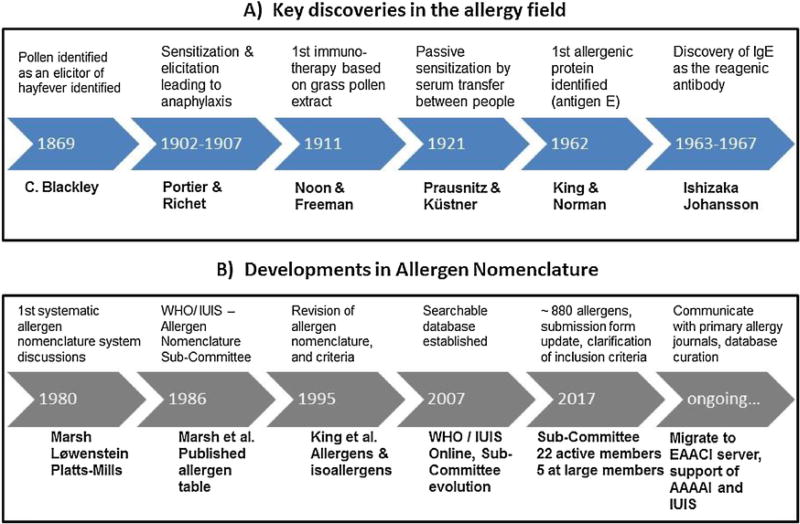
Historical perspective: A) Key discoveries in the allergy field. B) Developments in allergen nomenclature. Important milestones in development of clinical allergy and in nomenclature are described.
Allergen extracts contain many other proteins and components in addition to allergenic proteins. The first identified allergenic protein, antigen E (AgE), was isolated from pollen of short ragweed (Ambrosia artemisiifolia) by King and Norman in 1962 (Marsh et al., 1981). Other ragweed allergens identified at an early stage included Ra3 and Ra5 (Marsh et al., 1981). Around the same time, proteins from ryegrass pollen (Rye I and Rye II, later called Lol p I and Lol p II) were identified as prominent allergens by Johnson and Marsh as reviewed in Freidhoff et al. (1986). In the course of this and subsequent discovery work originally aiming for a better understanding of HLA-associations with allergic immune responses, the potential of using specific allergenic molecules for more precise diagnosis and possibly for immunotherapy was gradually emerging (Yunginger and Gleich, 1972; Baer et al., 1980).
During the past decades, advances in protein biochemistry and molecular biology have accelerated the discovery and characterization of allergens, being generated by recombinant DNA technology for a variety of applications, including basic and clinical research, allergen product standardization, allergy diagnostics and development of novel therapeutic approaches. Investigation of individual patient sensitization profiles has recently become possible via application of their sera to solid phased purified allergens in single assays or on microarrays with over 100 purified allergens, to accurately identify IgE-binding proteins and sources that likely cause their symptoms. Clear IgE binding to 2S albumins of peanut or soybean or to oleosins in peanut are likely to indicate higher risks of severe reactions (Beyer et al., 2015; Ebisawa et al., 2013; Schwager et al., 2017). Measuring specific IgE patterns can also help guide clinicians to treat patients with allergen immunotherapy (Sastre et al., 2012). Clinicians seeing patients allergic to bee and wasp venom may also improve diagnostic and therapeutic success for patients with so-called double-sensitizations using valuable molecular markers to prove primary sensitizations to the culprit venom (Seyfarth et al., 2017). In the future, individual immunotherapeutic reagents and prescriptions may be available for improved therapy. These developments further underpin the need for a consistent and unambiguous nomenclature of allergens. In parallel, the Allergen Nomenclature Sub-Committee has adapted to these changes by updating the criteria for defining a new allergen and the information requested in the submission form. This article provides an update on these criteria and challenges facing the existing system. Publishing the criteria ensures consistency and transparency of the process. Researchers are strongly encouraged to address them, with support of appropriate data reported confidentially to the WHO/IUIS Sub-Committee, to demonstrate evidence of allergenicity in order to receive an official allergen name prior to publication.
2. The beginning of the systematic Allergen Nomenclature: three men in a boat 1980
The idea for the current allergen nomenclature system originated from a discussion among Drs. David Marsh (USA), Henning Løwenstein (Denmark) and Thomas Platts-Mills (UK) during a boat ride on Lake Constance (Bodensee), Konstanz, Germany, during the 13th Symposium of the Collegium Internationale Allergologicum in July 1980 (Marsh et al., 1986; Chapman, 2004). The revised nomenclature system was first described in the Bulletin of the World Health Organization by the committee of clinicians who joined the International Union of Immunological Society (IUIS) Sub-Committee for Allergen Nomenclature, with David Marsh as Chair (Marsh et al., 1986). Many of the Sub-Committee scientists listed in the 1986 publication have been active in the evolution of rules and decisions on proposed allergen nomenclature. Other members have chaired the Sub-Committee over time (Wayne Thomas, Heimo Breiteneder and now Richard E. Goodman), with Jørgen N. Larsen pioneering the development of a web site, which was refined by John Wise at the University of Nebraska. In 2017, there are 22 active members and five members at large (listed on the website). The website http://allergen.org/originally showed allergens and information as a simple table, while a searchable database was established in 2007 and entries are since then added as they are agreed upon by the Sub-Committee.
Allergen names are assigned by the WHO/IUIS Allergen Nomenclature Sub-Committee through a defined submission process as described in Section 5. This process ensures that an appropriate, approved and non-redundant name is assigned to the allergen for any further publication. The benefit of this nomenclature system is that allergens are named in a properly documented, consistent and unambiguous manner, creating and maintaining clarity amongst the scientific, clinical and regulatory communities.
3. Evolution of allergen names
During the early 1980′s, the Sub-Committee established guidelines for naming allergens based on the taxonomic name of the source organism. Original allergen names comprised the first three letters of the genus and the first letter of the species epithet (both in italics) followed by a Roman numeral to indicate the allergen in the chronological order of isolation from the same source (Marsh et al., 1986). The publication described the requirements for data that should be provided as prerequisite for naming allergens including
- -
molecular weight (estimated mass by SDS-PAGE or gel-filtration),
- -
isoelectric point,
- -
amino acid composition or sequence, and
- -
immunochemical characterization, i.e. allergen recognition by patient IgE and/or animal antisera.
Epidemiology of allergens was called for in early publications of the Sub-Committee in terms of frequency of positive IgE binding from 20 to 30 human subjects. Over time, as biochemical, immunochemical and biophysical methods have improved, the expectations for description of allergens have grown and several nomenclature conventions have been changed including a requirement of fewer subjects.
In 1994, the nomenclature was revised, so that allergen names would not be in italics (reserved for genes), and the Roman numbers were replaced by Arabic ones (e.g. Amb a 1, Der p 1) (King et al., 1994). The 1994 update included a statement that sequence information would be required for new allergens (described in Radauer et al., 2014). The focus was on the amino acid (AA) sequence of the protein which is the relevant target for IgE. However, criteria for acceptance of sequences to the database have been adjusted over time as methods of determination have improved. A few new allergens were identified in a 1995 WHO/IUIS update (King et al., 1995).
In the 1990s, many allergens were being cloned and expressed as recombinant proteins and that led to the identification of multiple, highly homologous sequences. Then the WHO/IUIS Allergen Nomenclature Sub-Committee recognized the need to name homologous allergens in the same organism as either isoallergens or variants (isoforms), as described below.
4. Current allergen nomenclature conventions
As defined in the 1994 revision, allergen names consist of the first three letters from the genus, one letter from the species epithet, followed by an Arabic numeral. Occasionally, a letter is added to the genus (Sola for tomato, Solanum lycopersicum, as Sol was used for multiple ant species, Solenopsis sp.) or to the species (Hel as 1 for a snail Helix aspersa as Hel a was used for sunflower, Helianthus annuus) abbreviation to differentiate otherwise identical names of allergens from different species. Sometimes, the taxonomic name of the species is changed (e.g. Betula verrucosa became Betula pendula), but the Sub-Committee might decide to keep the same original allergen name if it has been widely used in the literature (e.g. Bet v 1 for the major birch pollen allergen).
Allergens from the same source are classified in biochemical groups (designated by Arabic numeral that contains two digits), usually according to the order in which they were identified (e.g. Der p 1 or Bet v 1). When possible, allergens from different species, related up to the taxonomic level of family or order, that belong to the same biochemical protein family will be assigned the same number across species. However, due to the historical naming convention, some allergens of the same biochemical protein family have been assigned different numbers (e.g. pectate lyases Amb a 1 and Art v 6, PR-10 proteins Bet v 1 and Ara h 8, or the profilins Amb a 8, Bet v 2, Phl p 12 and Art v 4). Within each group, allergens can be isoallergens or variants (isoforms) depending on their amino acid sequence identity. Isoallergens are homologous allergens that share the following common biochemical properties: similar molecular size, similar or identical biological function, if known, and an amino acid sequence identity of at least 67% (as a guideline not always followed if justified). Each isoallergen may have multiple forms of highly identical sequences (>90% identity, typically differing in only few amino acids), which are designated as variants (or isoforms). Isoallergens and their variants defined in this way are distinguished by numerical suffixes following a dot after the allergen number. The first two numerals 01–99 refer to a particular isoallergen (e.g. Amb a 1.01 and Amb a 1.02), and the two subsequent numerals 01–99 define each variant of a particular isoallergen (e.g. Amb a 1.0101 and Amb a 1.0102). Generally, allergen names should always be spelled out in full and partial abbreviation of allergen names (e.g. Ara h 1/2 or Ara h 1 and 2) is discouraged.
5. Review process by the Allergen Nomenclature Sub-Committee
The review process of a candidate allergen is shown in Fig. 2. The scientist submitting a protein as a candidate allergen should complete the submission form (www.allergen.org) and forward it to the Sub-Committee Chair by email. The scientist may suggest an allergen name (including isoallergen and variant number) based on current WHO/IUIS nomenclature.
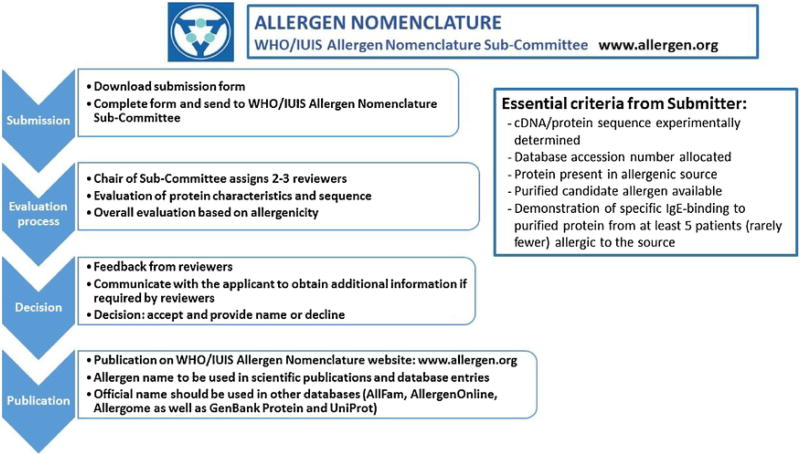
In many cases questions are forwarded to submitters in order to assure an appropriate science-based review. The goal is to provide a decision within two weeks, but sometimes decisions are delayed. Submitters should contact the Chair if they have questions.
Each submission is assigned for review to two or three members of the Sub-Committee, selected by the Chair of the Sub-Committee according to their area of expertise. Any possible conflict of interest with a potential reviewer can be indicated by the scientist submitting the form on the first page. The submission is held confidential within the Sub-Committee until a final decision is made. In case of uncertainty regarding particular criteria, descriptions or methods used, additional information may be requested. Final decisions are made by the Sub-Committee, based on the current criteria for allergen nomenclature, and communicated to the investigators who should then use the assigned name in their publication and in sequence databases. The process normally requires two weeks from submission to review, but may require longer depending on a need for additional data.
6. Essential criteria for acceptance of a protein as an allergen in the database
Acceptance of a protein as an allergen in the database essentially relies on the demonstration of its presence in the source that causes allergies, characterization of the protein by standard methods of biochemistry and molecular biology (Section 6.1), and, most importantly, the proof of specific recognition by relevant human serum IgE (Section 6.2). The main criteria for acceptance of a candidate allergen, summarized in the decision chart of Fig. 3, are:
Description of the allergenic source, including taxonomic name.
Information on the purification and characterization of the candidate allergenic protein including amino acid sequence.
Description of the allergic human serum donors used to test IgE binding to the candidate protein.
Demonstration of specific IgE binding with five sera of relevant patients.
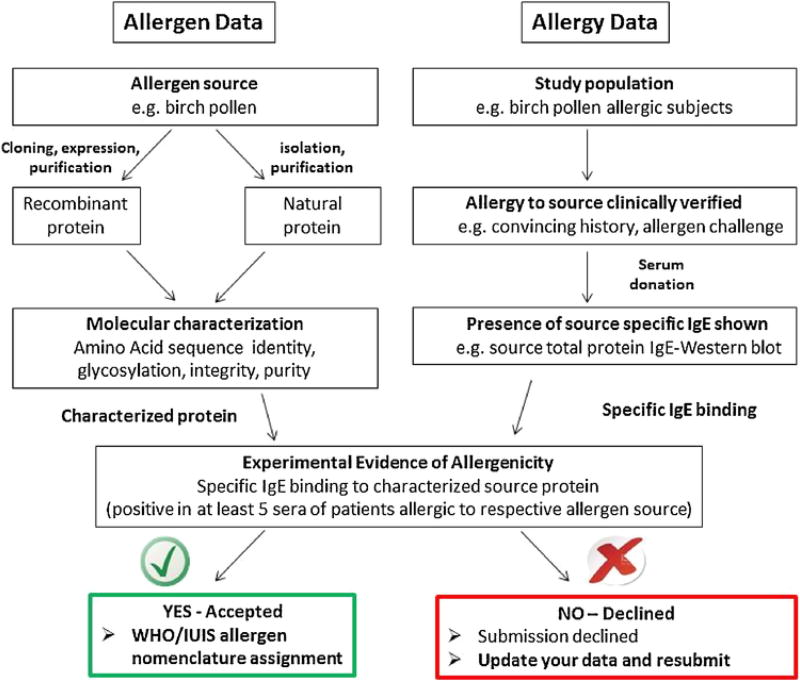
The primary questions of the submission form are designed to allow the reviewers to understand what was done, how complete the characterization of the allergen candidate was, indicating the protein sequence and physical characteristics, the selection of serum donors, tests for IgE binding as well as biological tests (SPT or BAT). The scientists submitting the form should provide enough information to demonstrate that their protein is most likely to be an allergen.
Over time, the WHO/IUIS Sub-Committee has refined the criteria for inclusion of proteins in the allergen database, as methods of testing by investigators and knowledge of allergen structures have improved (Chapman et al., 2007, Breiteneder and Chapman, 2014). The database currently includes 880 allergens, about 100 of which have had their three-dimensional structure determined (Pomés et al., 2015). Demonstrated biochemical function and tertiary structure are not necessary to obtain a name from the Sub-Committee, but may help in the overall understanding of allergenicity and potential cross-reactivity.
6.1. Analysis of candidate allergens by biochemistry and molecular biology
The WHO/IUIS allergen database includes only proteins and glycoprotein allergens. Ideally, the full amino acid and nucleotide sequence of the allergen should be provided on the submission form. If the full sequence is not available (i.e. for purified native allergens that have not been cloned), a partial amino acid sequence must be determined (i.e. by mass spectrometry).
The apparent MW of the protein should be estimated based on migration compared to standards in SDS-PAGE, by size exclusion chromatography, by mass spectrometry, or by calculation based on the amino acid sequence. Mass spectrometry determines the absolute molecular mass and is more definitive than SDS-PAGE, but its use is not a mandatory requirement.
The amino acid sequence of the protein should be deposited in a general protein database (UniProt or GenBank Protein) and the accession number specified in the application form. The experimentally determined protein sequence, as well as the determined DNA sequence, should be entered in the submission form. If the amino acid sequence is incomplete (due to gaps for example), each amino acid gap should be entered as “xxx”. In cases where only a partial sequence was determined by N-terminal or LC–MS/MS sequencing, which is identical to a published protein sequence from the same species, the accession number of the published sequence should be added with a clear note in the submission form. If the sequence has not been published before, the authors have the option to request that a non-public sequence remains confidential until publication.
To enable evaluation of potentially confounding data, scientists submitting a name are encouraged to provide additional information such as glycosylation state and purity of the protein isolated from the natural source or recombinant allergen. For glycosylated proteins, although not required, possible involvement of the glycan in IgE binding could be assessed following deglycosylation or by performing an inhibition assay using a high glycan source such as phytohemagglutinin (PHA) from Phaseolus vulgaris. Alternatively, IgE reactivity with a non-glycosylated recombinant allergen protein provides evidence that carbohydrate binding is not essential for allergen recognition if the recombinant protein has been demonstrated to fold like the native allergen. If the protein identity has been verified by monoclonal antibodies, the identity of the hybridoma and the assay method used should be described to allow the Sub-Committee to evaluate the quality and relevance of the data presented. If the cDNA sequence was determined by polymerase chain reaction (PCR) the sequence and position of primers should be described. If the sequence has already been published, the reference should be provided. If the sequence and description was already presented at a scientific conference, the abstract and poster or relevant presentation slides should be provided.
Finally, IgE-mediated allergies are also induced by small molecules if bound to carrier proteins (e.g. human serum albumin). These are low-MW organic compounds such as various drugs (e.g. antibiotics, muscle relaxants, opioids) and various organic compounds that can act as IgE-binding haptens and cause clinical reactions (Deak et al., 2016). There are also certain carbohydrate structures, such as specific plant- or insect-derived asparagine-linked glycans, commonly called cross-reactive carbohydrate determinants (CCD) that can bind IgE, but rarely or never elicit allergic reactions (Van der Veen et al., 1997; Foetisch et al., 2003; Mari et al., 2008). The carbohydrate structure galactose alpha-1,3 galactose (alpha-gal), present on many non-primate mammalian proteins, has recently been found to cause delayed IgE-mediated reactions following consumption of red meat in those who have been sensitized by salivary proteins from tick bites (Commins et al., 2009; Commins and Platts-Mills, 2013). Similar relationships between tick bites and meat allergy have been found in Europe (by the tick Ixodes ricinus; Hamsten et al., 2013a; Hamsten et al. Allergy 2013b), as well as Asia. These glycoproteins are unusual targets of IgE binding. Since the antibody recognition is directed to the glycan structure rather than the protein structure, they are not included in the WHO/IUIS database as allergens unless IgE binding has been demonstrated against the deglycosylated (unglycosylated) protein.
6.2. Proof of allergenicity of the newly identified proteins
Allergens are usually only accepted in the database if the purified protein is demonstrated to specifically bind IgE antibodies from at least five patients allergic to the respective allergen source. Specificity is demonstrated by lack of IgE binding in atopic and non-atopic control donors. Exceptions to the patients’ number may be made in particularly justified cases (such as in the field of occupational allergy or allergens in novel foods or new products), where it might be difficult to find five specifically allergic patients to prove allergenicity. In these cases, additional proof of allergenicity might then be requested, such as IgE immunoblots, dot blots, ELISA, basophil activation tests or mediator release assays (i.e. RBL) with the pure allergen. The IgE binding should be tested in a described and well controlled assay with the purified (natural or recombinant) allergen. Data from in vivo provocation of patients is not required, but adds value in demonstrating mast cell degranulation. Such tests should only be performed following informed consent of subjects, ethical review panel approval and appropriately controlled, sterile antigens.
Proof of allergy to the allergen source is also preferred. In general, the route of exposure might contribute to identify symptoms. For example, the natural route of pollen, animal dander, fungal spores and airborne occupational allergen exposure is inhalation. Thus, rhino conjunctivitis or asthma is the expected clinical outcome. Food allergens should be linked to objective symptoms of oral allergy syndrome, angioedema of the larynx, symptoms from the gastro-intestinal tract or systemic symptoms such as urticaria, asthma or anaphylaxis after consumption of the food. Insect bites or stings should generally include urticaria plus angioedema as well as other possible systemic reactions. However, exceptions will always occur. Skin symptoms after ingestion of allergens are quite common, and the same is true for respiratory symptoms. Patients with urticaria might not be the best candidates for skin testing. For some allergens (e.g. from mites) it is difficult to relate clinical symptoms to natural exposure. Also, some individuals with high levels of allergen specific IgE or skin test reactivity are asymptomatic upon natural exposure (Abraham et al., 2007). Therefore, in some cases larger samples might be required to demonstrate the association with clinically relevant allergy.
Sensitization and/or elicitation of an allergic response to the source (allergen extract) may be demonstrated in vitro by basophil activation or in vivo through inhalation, ingestion, intradermal injection, prick-to-prick tests or mucosal contact if performed under institutional review board guidance and following ethical approval.
Positive reactions should be based on criteria such as at least three mm wheal size for allergen extracts greater than the negative SPT control or greater than an established cut-off (e.g. three times the standard deviation of negative control subjects by in vitro IgE binding for ELISA) for extracts or the purified protein.
7. Challenges to the Allergen Nomenclature Sub-Committee
7.1. Revision of allergen names
Due to the risk of confusion and the fact that some names have already been published, the WHO/IUIS Sub-Committee generally takes a conservative position regarding changing allergen names. However, some allergen names have been revised or further specified as the original characterization was incomplete or associations were determined to be wrong based on new evidence.
The seed storage proteins of soybean provide two examples. The Gly m 5 protein is a complex of three beta-conglycinin subunits that were not characterized at the time the allergen name was assigned. One subunit is now known as beta-conglycinin alpha (67kDa), the second as beta-conglycinin alpha (71kDa) and the third as beta-conglycinin beta (50kDa) as described by Holzhauser et al. (2009). The alpha- and alpha- subunits are approximately 82% identical. The sequence of the beta-subunit is only 76% identical to the two alpha subunits. The three subunits fit together to form a trimeric structure. However, different ratios of Gly m 5 subunits form multimeric associations in protein bodies of the soybean (Maruyama et al., 2002). IgE from soybean allergic subjects may bind to one, two or three subunits of Gly m 5. The protein complex was originally named Gly m 5, but following analysis of intact complexes, various combinations of beta-conglycinins were identified to form the multimeric complexes (Holzhauser et al., 2009). Therefore, the individual subunits were finally named as isoallergens Gly m 5.01, Gly m 5.02 and Gly m 5.03 in 2009, with minor variants (e.g. Gly m 5.0301 and Gly m 5.0302).
Similarly, glycinin (Gly m 6) was also named with an understanding that the whole protein has a complex hexameric structure, made of combinations of different subunits (Nielsen et al., 1989). With further gene and protein characterization, it was recognized that at least five genes encode the subunits (Nielsen et al., 1989) and the allergen can be an arrangement of these five proteins which have individual IgE binding properties (Holzhauser et al., 2009), and which were renamed as five isoallergens in 2009.
Bovine milk casein was originally named Bos d 8, but full characterization has demonstrated that it is composed of complexes made up of four primary proteins (Willis et al., 1982). One is now called Bos d 9 (alpha S1 casein) as described by Nagao et al. (1984). The other caseins are defined as Bos d 10 (alpha S2 casein), Bos d 11 (beta casein) and Bos d 12 (kappa casein) with IgE binding defined by Natale et al. (2004). These proteins have high sequence diversity and they form complexes associated with calcium ions involved in a micellar structure. IgE antibodies from cow’s milk allergic subjects may bind to any combination of these subunits. Additional IgE binding from cow’s milk allergic individuals involves beta-lactoglobulin, alpha-lactalbumin and for some, IgG or lactoferrin.
There are examples of homologous allergen domains or whole allergens from grass pollen of related species (Lolium sp, Dactylon sp, and Phleum sp.) with proteins sharing considerable sequence identity which are difficult to assign to either the same or a different group. For example, Lol p 2 and Lol p 3 share approximately 60% identity. Dac g 2 is 60% identical with Lol p 3, and 65% identical to Dac g 3and 65% identical to Phl p 2, which is within the differences in isoforms accepted by the Sub-Committee. However, both grass pollen groups 2 and 3 were kept separate as there is high conservation across species and a marked reduction in sequence identity between group 2 and group 3 proteins among grass pollen. There is also some homology to group 1 beta-expansin proteins of the group 2 and 3 grass pollens though the major group 1 beta-expansin grass pollen allergens including Lol p 1 are much longer proteins and share little IgE cross-reactivity (Devanaboyina et al., 2014).
Two examples exist of allergens that were renamed due to high sequence identities. One is the ragweed allergen Amb a 2 that has a high amino acid sequence identity to group 1, and was therefore was renamed as Amb a 1.0501. The other is a seed storage glycinin allergen in peanut, Ara h 4 that is >90% identical to Ara h 3.0101 and it was therefore renamed as Ara h 3.0201 in 2012.
7.2. Difficulty in naming certain allergens
Some allergens are difficult to name for different reasons.
7.2.1. Complexity
The major cat allergen Fel d 1 (Morgenstern et al., 1991), has a crystal structure similar to that of secretoglobin (Kaiser et al., 2007). This allergen is a tetrameric protein formed by two heterodimers of chain 1 and chain 2, which are encoded by separate genes. Unlike Gly m 5 and Gly m 6 mentioned above, two different proteins are under one allergen name, Fel d 1.0101 (King et al., 1995). The allergen has a unique structure and there is no evidence of IgE cross-reactivity with secretoglobulins of other species (Hilger et al., 2014). A revision of this name in the future is possible but unlikely, given the widely spread use of Fel d 1 in the scientific community.
7.2.2. Low amino acid identity
Glutathione S-transferases (GST) from cockroach are structurally and functionally related and belong to different GST classes. GSTs attach a glutathione molecule to highly different substrates leading to the high GST diversity regarding function and sequence. The question is whether allergens with a similar biochemical function deserve the same group number, despite highly different sequences. It seems that identical group numbers within a species should generally alert scientists to the possibility of cross-reactivity. Nevertheless, cockroach GSTs from P. americana and B. germanica were assigned to the same group 5, despite their low amino acid identity (15.7% between Bla g 5 and Per a 5; way under the suggested threshold of 67% to be in the same group as isoallergens). More recently the Sub-Committee is trying to reserve the same number for homologous proteins from different species.
7.2.3. Hydrophobic nature of some allergen parts
Oleosins are hydrophobic allergens with an unusual hydrobophic core and few hydrophilic sections responsible for IgE antibody binding. Assignment of names to oleosins was difficult, because the molecular structure was taken into consideration to dissect and compare only the hydrophilic portions of the protein. Other proteins challenging to analyze for the purpose of assigning an allergen name include vitellogenins of chicken (Walsh et al., 1988). The proteins are encoded as very large proteins produced in the liver as polyproteins, transported via blood and taken up as yolk proteins. The process includes proteolysis into independent shorter proteins and one has been listed now as an allergen by WHO/IUIS (Gal d 6) (De Silva et al., 2016). Similar cases were reported earlier for fish roe proteins (Shimizu et al., 2009). The proteins are complex and assignment of IgE binding has often been to the holoprotein.
7.3. Assignment of names to isoallergens or variants (isoforms)
Isoallergens or variants (isoforms) are currently defined based on their amino acid sequence identity, but they might be originated in different ways (Table 1). In some cases, information on whether they are encoded by separate but related genes or the same gene with polymorphisms could assist in the identification. For example, there are 7 gene loci that encode beta expansins in Johnson grass pollen, including Sor h 1. The two allergens Sor h 1.0101 and Sor h 1.0201 are encoded by separate gene loci (Campbell et al., 2015). Similarly, the cockroach allergens Bla g 1.0101 and Bla g 1.0201 are listed as isoallergens due to the high sequence similarity, but recent genomic evidence suggests they are distinct gene products (Fig. 4). Investigators are not required to provide information on gene structure or location, but as this information becomes available it might help to improve the understanding of protein structures and potential name assignments.
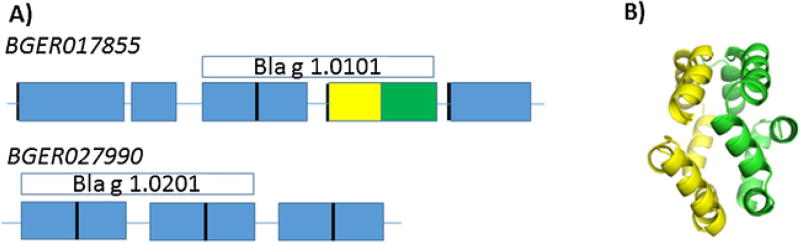
A) Bla g 1 isoallergens are encoded by different genes (in italics), which are composed of fully and partially repeated genetic elements indicated by the boxes. Blue rectangles represent regions with 76–99% sequence identity to either the yellow or green rectangle. The defined allergens at WHO/IUIS only partially cover the full open reading frame. B) The structure of the second half of Bla g 1.0101 was determined (4JRB, Mueller et al., 2013). The yellow and green coloring corresponds to the portion of gene similarly colored in A. The long linkers between structured elements suggest the full protein will resemble beads on a string. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1
Possible origins of isoallergens, variants and other types of allergen heterogeneity.
| Type of heterogeneity | Origin | Characteristics | Known examples in the Allergen Nomenclature database |
|---|---|---|---|
| Dispersed amino acid sequence variability. Named as isoallergens or variants, depending on level of sequence divergence. | Allelic | One isoallergen/variant per haplotype.a | Der f 2; Bet v 1 variants are in part allelic |
| Differential IgE antibody recognition of isoallergens may occur but is less likely for variants. | |||
| Multigene family | Multiple isoallergens/variants, encoded by different genes per haplotype, may be present per organism.a | Bet v 1 | |
| Differential IgE antibody recognition of isoallergens may occur but is less likely for variants. | Ara h 2 | ||
| Size and/or charge distribution | Posttranslational modifications such as addition of carbohydrate, phosphate groups, hydroxy-proline, C-terminal proteolytic cleavage or other moietiesb | Each individual proteins may contain multiple postranslationally modified allergen forms. | No examples |
| Differential IgE antibody recognition of glycoforms may occur. | |||
| Chemical composition | Chemical modification of the allergenb | Modification of the allergen during airborne transport (for aeroallergens), storage and processing (for food allergens), or preparation of extracts, e.g. oxidization, reactions with polyphenols, Maillard reaction during roasting of foods. | No examples |
| Block differences: amino acid sequence differences in segment/s of the protein. | Different protein cleavage | Each individual contains multiple differentially cleaved forms. | Hev b 6.02 and Hev b 6.03 |
| IgE antibody recognition may be affected depending on epitope distribution. | |||
| Alternative mRNA splicing | None yet identified for allergens. | No examples | |
| Modular differences: Different subunit (entire gene product) composition. | Alternative subunit composition | Each individual contains multiple allergen forms, depending on subunit combinations. | Gly m 5, Gly m 6 |
| IgE antibody recognition may be affected depending on epitope distribution. |
New isoallergens will only be accepted in the database if IgE binding has been demonstrated to the isoallergen. New variants will only be accepted if there is justified evidence that the variant is expressed in the organism and tissue causing allergies. The use of PCR based cloning techniques and genome analysis has increased the potential identification of erroneous sequences. The committee would like investigators to sequence multiple clones from independent experiments and to provide accurate information on the source and methods used to determine sequences. Ideally, the existence of the protein in the natural source should be verified by mass spectrometry or N-terminal sequencing with sufficient length to verify the protein sequence matches the nucleic acid sequence. Additional information for inclusion into the database are the characterization of the purified recombinant variant, confirmation of the source and identity of the organism, as well as IgE reactivity with the recombinant protein.
7.4. Cross-reactivity might impair the identification of source-specific allergens
Considerations include whether proteins that are homologs of allergenic proteins are allergens themselves, binding IgE and if so, whether patients show symptoms upon exposure. Antibody cross-reactivity can occur with only few amino acids (1–3) in common between homologous proteins (Glesner et al., 2017), but it does not necessarily translate into clinical cross-reactivity. Serum IgE cross-reactivity has been observed between PR-10 related allergens from birch pollen (Bet v 1), apple (Mal d 1) and other plant-derived foods. However, birch pollen is usually the primary sensitizer and elicitor of the allergic immune response, whereas cross-reactive IgE binding to Mal d 1 is often of lower avidity, or may involve a single IgE epitope and therefore may or may not be clinically relevant for individual patients. Similarly, there are patient-specific patterns of IgE cross-reactivity to fish parvalbumins coupled with clinical sensitivity to some fish, but not to other fish species (Kuehn et al., 2014) or to chicken (fish-chicken syndrome) (Kuehn et al., 2016). There are numerous examples of reactivity to homologous allergens from multiple allergen sources that can be explained by IgE cross-reactivity. For example, there is cross-reactivity among lipocalins Can f 6 of dog dander, Fel d 4 of cat dander and Equ c 1 of horse dander (Nilsson et al., 2012). Each of these three allergens can act as primary sensitizer, co-sensitizer or cross-reacting molecule, depending on the exposure profile of the patient. Cross-reactivity with Can f 6 was responsible for a clinically relevant positive specific IgE to dog dander in a horse allergic patient (Jakob et al., 2013). Data on sequence and structural similarity combined with serum IgE binding must be carefully evaluated as demonstrated by the studies of IgE binding and allergy of Ole e 1 (olive pollen allergen), and Fra e 1 (ash pollen allergen) and Pla a 1 (English plantain pollen allergen). The overall structures are similar as could be predicted to some extent by sequence, yet there is a clear gradation of IgE binding among patients, with unique clinical responses (Stemeseder et al., 2017). Thus, sequence or structural information alone is judged insufficient to name a protein as an allergen.
7.5. Is the Sub-Committee naming too many allergens?
The WHO/IUIS Allergen Nomenclature database lists 17 allergens in peanut Arachis hypogaea, 31 in house dust mite Dermatophagoides farinae, 20 in Dermatophagoides pteronyssinus and 23 in Aspergillus fumigatus. Some of those are likely minor allergens or only hypothetical proteins from cDNA (such as Tri a 42 and Tri a 43 in wheat). Some simply have a low IgE binding capacity for some subjects that is only detected when presented at high concentration by in vitro assays, and might not contribute to the IgE binding to the source because the allergen might not be well represented in the allergenic material or allergenic extracts (Casset et al., 2012). Others, like Der p 23, turned out to be significant allergens (Weghofer et al., 2013). In contrast, some allergens such as Der f 22, which shows a low identity (~40%) to Der f 2 and no publications demonstrating evidence of IgE binding or reactivity in the literature for nearly 10 years, probably should not have been accepted by the Sub-Committee. Der f 22 will be re-reviewed and might be removed from the database unless new evidence is uncovered by the Sub-Committee in the future. The Sub-Committee is considering removing other allergens from the database. Some allergens (such as the tomato allergens Sola l 2), bind IgE only through carbohydrates, and this IgE is not clinically relevant. In other cases, binding of trace amounts of different allergen/s to purified natural allergens, can mislead the assessment of a protein as an allergen. In both situations, the use of non-glycosylated recombinant proteins would assist on assessing IgE binding to the desired protein.
7.6. Integration of big data from ‘omics’ research
The application of massively parallel DNA and transcriptome sequencing, proteomics, bioinformatics, mass-spectrometry and protein chemistry processes coupled with sensitive high or multi-allergen array immunoassays is markedly increasing the capacity for data generation and discovery. Through studies of whole allergen source transcriptomics and genomics, it is evident that multiple gene copies exist in a given source for a single protein family that contains allergenic as well as non-allergenic members. Thus not all putative allergen gene sequences actually encode allergens. This may be for a variety of reasons including that some gene loci are pseudogenes. Not all “genes” are transcribed, translated and processed in the relevant tissue at the time when or location where sensitized individuals are exposed. Examples of this include the greater number (six of seven) genes encoding Johnson grass pollen beta-expansin proteins that can be confirmed within the pollen proteome, whilst only two of these gene loci encode IgE reactive proteins isoforms. Similar findings have been observed and/or proposed for the multiple beta-expansin gene loci of rice (Devis et al., 2017) and Sorghum bicolor (Paterson et al., 2009) and profilins of rice (Devis et al., 2017). Other examples are the polcalcin protein family with the allergen Phl p 7, parvalbumins of fish (e.g. Gad m 1) and the PR-10 family, which includes Bet v 1 from birch pollen (Erler et al., 2011).
Currently, there is good knowledge of many protein families that contain allergens (Radauer et al., 2008). Yet many proteins within the families are not known to cause allergy. Although there are numerous publications that attempt to predict allergens using bioinformatics with genomic and transcriptomic data, detection of the proteins in the allergenic source material is essential. Predictive models of allergenicity based on sequence identity are not reliable due to the wide variations in the percentage of amino acid sequence identity that is associated with cross-reactivity, and the lack of knowledge regarding the actual epitopes for these proteins. Other factors are important for allergenicity, such as abundance and structural stability for food allergens or solubility for aeroallergens. Thus the WHO/IUIS Sub-Committee strives to stimulate appropriate characterization of allergenic proteins with demonstration of IgE binding from appropriate human volunteers, allergenicity of these proteins still needs to be demonstrated to prove that a given protein causes allergy.
7.7. Assignment of names before publication
The goal of the WHO/IUIS Allergen Nomenclature Sub-Committee is to assign names to allergens before publication. The purpose is to associate a name with the original publication. However, the danger of this process is that the data used to characterize the allergen have not been evaluated through peer-reviewed publication. In the last four years, the Sub-Committee has been asking for more comprehensive information prior to accepting a candidate as an allergen, and the submission form has been modified accordingly to reflect that and it will likely continue evolving. One goal of this publication is to encourage investigators to meet the need for more relevant information prior to submission.
Journals cannot rely on reviewers to make sure that correct allergen nomenclature is used in publications unless it is emphasized by the editorial office and forms part of the “Instructions to Authors”. Allergen names are sometimes published that do not comply with the official WHO/IUIS nomenclature, causing inconsistencies in the literature. Currently, journals (especially journals that are not only allergy focused) still sometimes do not publish names accepted by the Sub-Committee. To help improve the correct assignment and use of allergen names, the WHO/IUIS Allergen Nomenclature Sub-Committee has contacted in 2017 the editorial offices of the main journals that publish allergen studies, to request the implementation of a policy requiring the use of the official allergen nomenclature in their publications. There has been a positive response from the journals and most of them have already implemented such a change of the instructions to authors.
Some allergens not recognized and listed by the WHO/IUIS have been published in journals indexed in PubMed. Even more commonly, genomics, transcriptomics and proteomics projects are assigning proteins in sequence databases (e.g. GenBank) with “allergen-like” names or even listing proteins or genes with a WHO/IUIS-like name, however, without having provided evidence-based data on protein allergenicity. Authors are encouraged to submit such allergens to the Sub-Committee for its consideration and having an official name assigned.
8. Other allergen sequence and structure databases
Several publicly available allergen databases exist with different purposes, strengths and weaknesses (reviewed by Radauer, 2017). The WHO/IUIS allergen nomenclature database provides a systematic and official allergen nomenclature for many allergens. Allergome (www.allergome.org) comprises a large amount of partially filtered information and references of allergens, public references for the proteins and links to many reference sources. AllergenOnline (www.allergenonline.org) is a peer-reviewed risk assessment tool for food safety and includes allergens from all sources (Goodman et al., 2016). AllergenOnline is updated annually with reviews of individual proteins and data of IgE binding and clinical responses after publication of data on the allergens and allergenicity. The COMPARE database (comparedatabase.org) hosted by the Health and Environmental Sciences Institute was derived from the AllergenOnline.org database (>99% of entries in COMPARE) and shows the same “year adopted” for entries as in the AllergenOnline database. It does not yet have a sequence search system, but is intended to eventually be a risk assessment tool similar to AllergenOnline. The Structural Database of Allergenic Proteins database (SDAP, fermi.utmb.edu) is linked to a structural database source and is intended as a risk assessment tool. The Immune Epitope Database (IEDB, www.iedb.org) provides information on T-cell and B-cell epitopes as well as MHC binding proteins of many proteins including pathogens, viruses, bacteria and some allergens. The AllFam database (www.meduniwien.ac.at/allfam) focuses on protein families of allergens based on information from publications, from WHO/IUIS, AllergenOnline and Pfam databases. Users of these databases should understand the information and intent of each of these databases and their limitations.
9. Future directions and new initiatives
The Sub-Committee has had recognition and some financial support from the International Union of Immunological Societies (IUIS) for a number of years. In 2016 the Sub-Committee gained additional support from the European Academy of Allergy and Clinical Immunology (EAACI) that recognized it as one of its committees. In 2017, support was gained from the American Academy of Allergy, Asthma and Immunology (AAAAI). The time and effort needed to maintain the database and for members to provide careful review of submissions is considerable. All members act on a voluntary basis, without financial compensation.
Allergen database hosting. From 2007 through 2017 the WHO/IUIS database was hosted without charge in the Department of Food Science & Technology, on a Food Allergy Research and Resource Program server at the University of Nebraska-Lincoln (USA). The Who/IUIS database was transferred to the EAACI computer server in November 2017 and the cost of maintaining the database will be shared by the IUIS, EAACI and AAAAI. The WHO/IUIS Allergen Nomenclature Sub-Committee is now an official committee in the EAACI organization.
The Sub-ommittee is reviewing and improving available information and tools on the database to help researchers compare their newly identified candidate allergens in the context of the database. The Sub-ommittee welcomes suggestions that may be submitted to the hairman or Sub-ommittee members.
As advances are made in technology of gene sequencing, transcriptomics and proteomics, more questionable allergen candidates are being submitted to the WHO/IUIS Sub-Committee and other sequence databases. The Sub-Committee encourages that scientists revise and remove “unproven” allergens from protein databases (GenBank Protein, and UniProt). However, it is important to note that as a committee on nomenclature, the mission is to provide a framework for consistent allergen identification and not to pass judgement on whether an allergen is a major, mid-tier, or minor allergen. The scientific discussion about the merits of a particular allergen is simply facilitated by accurate nomenclature to identify the molecule in question.
How the Allergen Nomenclature Sub-Committee responds to and evaluates the increasing numbers of putative allergens will be an increasing challenge. Not all putative allergen-like sequences are demonstrably allergenic. Processes and criteria for assessing allergenicity might need to be continually evaluated and revised. Research and critical thinking will need to address the capacity to assimilate many different data sources into useful and clinically relevant information that aids the diagnosis and treatment of allergic patients.
A full online review process analogous to the ones for manuscripts can be envisioned. Name assignments to allergen candidates are performed by the Chair of the Sub-Committee and two to three reviewers. Currently the WORD file submissions are maintained on the server. Once an online entry form is developed, the submissions should be handled with less chance of manual errors, although the committee will review all entries before they become public.
10. Conclusions
The WHO/IUIS Allergen Nomenclature Sub-Committee serves as an expert professional group that reviews supporting evidence of allergenicity of proteins, assigns names and manages an official list of recognized allergens. The Sub-Committee also clarifies the understanding of qualities of allergen molecules and focuses current thinking about the criteria for allergenicity of proteins from molecular, biochemical, in vitro immunoassay-based methods as well as clinical perspectives. Investigators are encouraged to follow the criteria set by the Sub-Committee to provide evidence-based data on allergenicity prior to submitting an allergen. Ultimately, whether a protein is an allergen for a particular patient depends on the level of sensitisation, exposure and clinical allergy status. As the introduction of allergen components has had major impact on analytical sensitivity, specificity and allergy diagnosis (Matricardi et al., 2016), allergen nomenclature will remain and become increasingly important in the context of the application of the wide array of “omics” technologies in allergy research. In the future, the WHO/IUIS Allergen Nomenclature Sub-Committee will continue to provide expertise and criteria to evaluate evidence of the specificity of IgE binding for candidate allergens. These criteria will most likely evolve as additional data are generated from new technologies. Recently, there has been an explosion of “Big Data”, from various “omics” techniques that will impact on the submissions to the database. The continued activities of the WHO/IUIS Allergen Nomenclature Sub-Committee are an important component of the review process to assign names to allergens before peer-reviewed publication. These activities help scientists and reviewers to focus on relevant data supporting characterization of allergens and IgE binding, and stimulate more rigorous review of clinical implications applied to allergens. The assignment of reliable names to allergens is useful for pharmaceutical companies making diagnostic and therapeutic materials for patients, helps provide information as the medical field of allergology continues to move from using crude extracts to the use of individual allergens. However, database users should remember that a WHO/IUIS name designation relies on data provided by the scientist submitting the candidate allergens, before peer-review publication. The Sub-Committee contributes to improve continued interactions of scientists and clinicians in the effort to improve diagnoses, therapy and risk avoidance for those who have specific allergies.
Acknowledgments
The authors thank the IUIS, EAACI and AAAAI organizations for travel grants and administrative costs to the Sub-Committee.
Footnotes
Uncited references
Bennich et al. (1968), Bradford (1898), Hilger et al. (2012), Krombach et al. (2004), Mose et al. (2017), Noon et al. (1911), Ring and Gutermuth (2011), Stanworth (1993) and Vogel et al. (2005).
References
- Abraham CM, Ownby DR, Peterson EL, Wegienka G, Zoratti EM, Williams LK, Joseph CL, Johnson CC. The relationship between seroatopy and symptoms of either allergic rhinitis or asthma. J. Allergy Clin. Immunol. 2007;119(5):1099–1104. [Abstract] [Google Scholar]
- Baer H, Anderson MC, Hale R, Gleich GJ. The heat stability of short ragweed pollen extract and the importance of individual allergens in skin reactivity. J. Allergy Clin. Immunol. 1980;66:281–285. [Abstract] [Google Scholar]
- Beyer K, Grabenhenrich L, Hartl M, Beder A, Kalb B, Ziegert M, Finger A, Harandi N, Schlags R, Gappa M, Puzzo L, Roblitz H, Millner-Uhlemann M, Busing S, Ott H, Lange L, Niggemann B. Predictive values of component specific IgE for the outcome of peanut and hazelnut food challenges in children. Allergy. 2015;70(1):90–98. In: Questions on the History of Medicine from the L, pp. [Abstract] [Google Scholar]
- Breiteneder H, Chapman MD. Allergen nomenclature. In: Lockey RF, Ledford DK, editors. Allergens and Allergen Immunotherapy: Subcutaneous, Lingual and Oral. CRC Press, Taylor and Francis Group; Boca Raton, Florida, USA: 2014. pp. 37–49. [Google Scholar]
- Campbell BC, Gilding EK, Timbrell V, Guru P, Loo D, Zennaro D, Mari A, Solley G, Hill MM, Godwin ID, Davies JM. Total transcriptome, proteome, and allergome of Johnson grass pollen, which is important for allergic rhinitis in subtropical regions. J. Allergy Clin. Immunol. 2015;135(1):133–142. [Abstract] [Google Scholar]
- Casset A, Mari A, Purohit A, Resch Y, Weghofer M, Ferrara R, Thomas WR, Alessandri C, Chen KW, de Blay F, Valenta R, Vrtala S. Varying allergen composition and content affects the in vivo allergenic activity of commercial Dermatophagoides pteronyssinus extracts. Int. Arch. Allergy Immunol. 2012;159(3):253–262. [Europe PMC free article] [Abstract] [Google Scholar]
- Chapman MD, Pomes A, Breiteneder H, Ferreira F. Nomenclature and structural biology of allergens. J. Allergy Clin. Immunol. 2007;119:414–420. [Abstract] [Google Scholar]
- Chapman MD. Allergen nomenclature. In: Lockey RF, Bukantz SC, Bousquet J, editors. Allergens and Allergen Immunotherapy. Marcel Decker, Inc.; New York, USA: 2004. pp. 51–64. [Google Scholar]
- Cohen SG, Zelaya-Quesada M. Portier, Richet, and the discovery of anaphylaxis: a centennial. J. Allergy Clin. Immunol. 2002;110(2):331–336. [Abstract] [Google Scholar]
- Commins SP, Platts-Mills TA. Tick bites and red meat allergy. Curr. Opin. Allergy Clin. Immunol. 2013;13(4):354–359. [Europe PMC free article] [Abstract] [Google Scholar]
- Commins SP, Satinover SM, Hosen J, Mozena J, Borish L, Lewis BD, Woodfolk JA, Platts-Mills TA. Delayed anaphylaxis, angioedema, or urticaria after consumption of red meat in patients with IgE antibodies specific for galactose-alpha-1,3-galactose. J. Allergy Clin. Immunol. 2009;123:426–433. [Europe PMC free article] [Abstract] [Google Scholar]
- De Silva C, Dhanapala P, Doran T, Tang ML, Suphioglu C. Molecular and immunological analysis of hen’s egg yolk allergens with a focus on YGP42 (Gal d 6) Mol. Immunol. 2016;71:152–160. [Abstract] [Google Scholar]
- Deak PE, Vrabel MR, Pizzuti VJ, Kizeitepe T, Bilgicer B. Nanoallergens: a multivalent platform for studying and evaluating potency of allergen epitopes in cellular degranulation. Exp. Biol. Med. 2016;241:996–1006. [Europe PMC free article] [Abstract] [Google Scholar]
- Devanaboyina SC, Cornelius C, Lupinek C, Fauland K, Dall’Antonia F, Nandy A, Hagen S, Flicker S, Valenta R, Keller W. High-resolution crystal structure and IgE recognition of the major grass pollen allergen Phl p 3. Allergy. 2014;69:1617–1628. [Europe PMC free article] [Abstract] [Google Scholar]
- Devis DL, Davies JM, Zhang D. Molecular features of grass allergens and development of biotechnological approaches for allergy prevention. Biotechnol. Adv. 2017;35(5):545–556. [Abstract] [Google Scholar]
- Ebisawa M, Brostedt P, Sjolander S, Sato S, Borres MP, Ito K. Gly m 2S albumin is a major allergen with a high diagnostic value in soybean-allergic children. J. Allergy Clin. Immunol. 2013;132(4):976–978. [Abstract] [Google Scholar]
- Erler A, Hawranek T, Krückemeier L, Asam C, Egger M, Ferreira F, Briza P. Proteomic profiling of birch (Betula verrucosa) pollen extracts from different origins. Proteomics. 2011;11(8):1486–1498. [Abstract] [Google Scholar]
- Foetisch K, Westphal S, Lauer I, Retzek M, Altmann F, Kolarich D, Scheurer S, Vieths S. Biological activity of IgE specific for cross-reactive carbohydrate determinants. J. Allergy Clin. Immunol. 2003;111(4):889–896. [Abstract] [Google Scholar]
- Freidhoff LR, Ehrlich-Kautzky E, Grant JH, Meyers DA, Marsh DG. A study of the human immune response to Lolium perenne (Rye) pollen and its components, Lol p I and Lol p II (Rye I and Rye II) J. Allergy Clin. Immunol. 1986;78:1190–1201. [Abstract] [Google Scholar]
- Glesner J, Vailes LD, Schlachter C, Mank N, Minor W, Osinski T, Chruszcz M, Chapman MD, Pomés A. Antigenic Determinants of Der p 1: Specificity and cross-reactivity associated with IgE antibody recognition. J. Immunol. 2017;198(3):1334–1344. [Europe PMC free article] [Abstract] [Google Scholar]
- Goodman RE, Ebisawa M, Ferreira F, Sampson HA, van Ree R, Vieths S, Baumert JL, Bohle B, Lalithambika S, Wise J, Taylor SL. AllergenOnline: a peer-reviewed, curated allergen database to assess novel food proteins for potential cross-reactivity. Mol. Nutr. Food Res. 2016;60(5):1183–1198. [Abstract] [Google Scholar]
- Hamsten C, Tran TAT, Starkhammar M, Brauner A, Commins SP, Platts-Mills TAE, van Hage M. Red meat allergy in Sweden: association with tick sensitization and B-negative blood groups. J. Allergy Clin. Immunol. 2013;132(6):1431–1434. [Europe PMC free article] [Abstract] [Google Scholar]
- Hamsten C, Starkhammar M, Tran TA, Johansson M, Bengtsson U, Ahlén G, Sällberg M, Grönlund H, van Hage M. Identification of galactose-alpha-1,3-galactose in the gastrointestinal tract of the tick Ixodes ricinus; possible relationship with red meat allergy. Allergy. 2013;68:549–552. [Abstract] [Google Scholar]
- Hilger C, Kler S, Arumugam K, Revets D, Muller CP, Charpentier C, Lehners C, Morisset M, Hentges F. Identification and isolation of a Fel d 1-like molecule as a major rabbit allergen. J. Allergy Clin. Immunol. 2014;133(3):759–766. [Abstract] [Google Scholar]
- Holzhauser T, Wackermann O, Ballmer-Weber B, Bindslev-Jensen C, Scibilia J, Perono-Garoffo L, Utsumi S, Poulsen LK, Vieths S. Soybean (Glycine max) allergy in Europe: gly m 5 (beta-conglycinin) and Gly m 6 (glycinin) are potential diagnostic markers for severe allergic reactions to soy. J. Allergy Clin. Immunol. 2009;123:452–458. [Abstract] [Google Scholar]
- Jakob T, Hilger C, Hentges F. Clinical relevance of sensitization to cross-reactive lipocalin Can f 6. Allergy. 2013;68(5):690–691. [Abstract] [Google Scholar]
- Kaiser L, Velickovic TC, Badia-Martinez D, Adedoyin J, Thunberg S, Hallén D, Berndt K, Grönlund H, Gafvelin G, van Hage M, Achour A. Structural characterization of the tetrameric form of the major cat allergen Fel d 1. J. Mol. Biol. 2007;370(4):714–727. [Abstract] [Google Scholar]
- King TP, Hoffman D, Løwenstein H, Marsh DG, Platts-Mills TA, Thomas WR. Allergen nomenclature: WHO/IUIS Allergen Nomenclature Subcommittee. Int. Arch. Allergy Immunol. 1994;105:224–233. [Abstract] [Google Scholar]
- King TP, Hoffman D, Løwenstein H, Marsh DG, Platts-Mills TAE, Thomas W. Allergen Nomenclature. Allergy. 1995;50:765–774. [Abstract] [Google Scholar]
- Kuehn A, Swoboda I, Arumugam K, Hilger C, Hentges F. Fish allergens at a glance: variable allergenicity of parvalbumins, the major fish allergens. Front. Immunol. 2014;5:179. https://doi.org/10.3389/fimmu2014.00179. [Europe PMC free article] [Abstract] [Google Scholar]
- Kuehn A, Codreanu-Morel F, Lehners-Weber C, Doyen V, Gomez-André SA, Bienvenu F, Fischer J, Ballardini N, van Hage M, Perotin JM, Silcret-Grieu S, Chabane H, Hentges F, Ollert M, Hilger C, Morisset M. Cross-reactivity to fish and chicken meat – a new clinical syndrome. Allergy. 2016;71(12):1772–1781. [Abstract] [Google Scholar]
- Mari A, Ooievaar de Heer P, Scala E, Giani M, Pirrotta L, Zuidmeer L, Bethell D, van Ree R. Evaluation by double-blind placebo-controlled oral-challenge of the clinical relevance of IgE antibodies against plant glycans. Allergy. 2008;63(7):891–896. [Abstract] [Google Scholar]
- Marsh DG, Belin L, Bruce A, Lichtenstein LM, Hussain R. Rapidly released allergens from short ragweed pollen. J. Allergy Clin. Immunol. 1981;67(3):206–216. [Abstract] [Google Scholar]
- Marsh DG, Goodfriend L, King TP, Lowenstein H, Platts-Mills TA. Allergen nomenclature. Bull. World Health Organ. 1986;64(5):767–774. [Europe PMC free article] [Abstract] [Google Scholar]
- Maruyama N, Ramlan M, Salleh M, Takahashi K, Yagasaki K, Goto H, Hontani N, Nakagawa S, Utsumi S. Structure-physicochemical function relationships of soybean beta-conglycinin heterotrimers. J. Agric. Food Chem. 2002;50:4323–4326. [Abstract] [Google Scholar]
- Matricardi PM, Kleine-Tebbe J, Hoffmann HJ, Valenta R, Hilger C, Hofmaier S, Aalberse RC, Agache I, Asero R, Ballmer-Weber B, Barber D, Beyer K, Biedermann T, Bilò MB, Blank S, Bohle B, Bosshard PP, Breiteneder H, Brough HA, Caraballo L, Caubet JC, Crameri R, Davies JM, Douladiris N, Ebisawa M, Eigenmann PA, Fernandez-Rivas M, Ferreira F, Gadermaier G, Glatz M, Hamilton RG, Hawranek T, Hellings P, Hoffmann-Sommergruber K, Jakob T, Jappe U, Jutel M, Kamath SD, Knol EF, Korosec P, Kuehn A, Lack G, Lopata AL, Mäkelä M, Morisset M, Niederberger V, Nowak-Węgrzyn AH, Papadopoulos NG, Pastorello EA, Pauli G, Platts-Mills T, Posa D, Poulsen LK, Raulf M, Sastre J, Scala E, Schmid JM, Schmid-Grendelmeier P, van Hage M, van Ree R, Vieths S, Weber R, Wickman M, Muraro A, Ollert M. EAACI molecular allergology user's guide. Pediatr. Allergy Immunol. 2016;27(Suppl. 23):1–250. [Abstract] [Google Scholar]
- Morgenstern JP, Griffith IJ, Brauer AW, Rogers BL, Bond JF, Chapman MD, Kuo MC. Amino acid seqeunce of Fel dI, the major allergen of the domestic cat: protein seqeunce analysis and cDNA cloning. Proc. Natl. Acad. Sci. U. S. A. 1991;88(21):9690–9694. [Europe PMC free article] [Abstract] [Google Scholar]
- Mueller GA, Pedersen LC, Lih FB, Glesner J, Moon AF, Chapman MD, Tomer KB, London RE, Pomés A. The novel structure of the cockroach allergen Bla g 1 has implications for allergenicity and exposure assessment. J. Allergy Clin. Immunol. 2013;132(6):1420–1426. [Europe PMC free article] [Abstract] [Google Scholar]
- Nagao M, Maki M, Sasaki R, Chiba R. Isolation and sequence analysis of bovine alpha-s1-casein cDNA clone. Agric. Biol. Chem. 1984;48:1663–1667. [Google Scholar]
- Natale M, Bisson C, Monti G, Peltran A, Garoffo LP, Valentini S, Fabris C, Bertino E, Coscia A, Conti A. Cow’s milk allergens identification by two-dimensional immunoblotting and mass spectrometry. Mol. Nutr. Food Res. 2004;48(5):363–369. [Abstract] [Google Scholar]
- Nielsen NC, Dickenson CD, Cho TJ, Thanh VH, Scallon BJ, Fischer RL, Sims TL, Drews GN, Goldberg RB. Characterization of the glycinin gene family in soybean. Plant Cell. 1989;1:313–328. [Abstract] [Google Scholar]
- Nilsson OB, Binnmyr J, Zoltowska A, Saarne T, van Hage M, Grönlund H. Characterization of the dog lipocalin allergen Can f 6: the role in cross-reactivity with cat and horse. Allergy. 2012;67(6):751–757. hay feverL. [Abstract] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, Schmutz J, Spannagl M, Tang H, Wang X, Wicher T, Bharti AK, Chapman J, Feltus FA, Fowik U, Grigoriev IV, Lyons E, Maher CA, Martis M, Narechania A, Otillar RP, Penning BW, Salamov AA, Wang Y, Zhang L, Carpita NC, Freeling M, Gingle AR, Hash CT, Keller B, Klein P, Kresovich S, McCann MC, Ming R, Peterson DG, Mehboob-ur-Rahman Ware D, Westhoff P, Mayer KF, Messing J, Rokhsar DS. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. [Abstract] [Google Scholar]
- Pomés A, Chruszcz M, Gustchina A, Minor W, Mueller GA, Pedersen LC, Wlodawer A, Chapman MD. 100 Years later: celebrating the contributions of x-ray crystallography to allergy and clinical immunology. J. Allergy Clin. Immunol. 2015;136(1):29–37. [Europe PMC free article] [Abstract] [Google Scholar]
- Radauer C, Bublin M, Wagner S, Mari A, Breiteneder H. Allergens are distributed into few protein families and possess a restricted number of biochemical functions. J. Allergy Clin. Immunol. 2008;121:847–852. [Abstract] [Google Scholar]
- Radauer C, Nandy A, Ferreira F, Goodman RE, Larsen JN, Lidholm J, Pomes A, Raulf-Heimsoth M, Rozynek P, Thomas WR, Breiteneder H. Update of the WHO/IUIS Allergen Nomenclature Database based on analysis of allergen sequences. Allergy. 2014;69(4):413–419. [Abstract] [Google Scholar]
- Radauer C. Navigating through the jungle of allergens: features and applications of allergen databases. Int. Arch. Allergy Immunol. 2017;173(1):1–11. [Abstract] [Google Scholar]
- Sastre J, Landivar ME, Ruiz-Garcia M, Andregnette-Rosigno MV, Mahillo I. How molecular diagnosis can change allergen-specific immunotherapy prescription in a complex pollen area. Allergy. 2012;67(5):709–711. [Abstract] [Google Scholar]
- Schwager C, Kull S, Behrends J, Rockendorf N, Schocker F, Frey A, Homann A, Becker WM, Jappe U. Peanut oleosins associated with severe peanut allergy-importance of lipophilic allergens for comprehensive allergy diagnosis. J. Allergy Clin. Immunol. 2017;140(5):1331–1338. [Abstract] [Google Scholar]
- Seyfarth F, Miguel D, Schliemann S, Hipler UC. Diagnostic precision of component-resolved vs. extract-based in vitro diagnosis of hymenoptera venom allergy: effects on clinical management. J. Dtsch. Dermatol. Ges. 2017;15(5):507–515. [Abstract] [Google Scholar]
- Shimizu Y, Nakamura A, Kishimura H, Hara A, Watanabe K, Saeki H. Major allergen and its IgE cross-reactivity among salmonoid fish roe allergy. J. Agric. Food Chem. 2009;57(6):2314–2319. [Abstract] [Google Scholar]
- Stemeseder T, Freier R, Wildner S, Fuchs JE, Briza P, Lang R, Batanero E, Lidholm J, Liedl KR, Campo P, Hawranek T, Villalba M, Brandstetter H, Ferreira F, Gadermaier G. Crystal structure of Pla l 1 reveals both structural similarity and allergenic divergence within the Ole e 1-like protein family. J. Allergy Clin. Immunol. 2017;140(1):277–280. [Europe PMC free article] [Abstract] [Google Scholar]
- Taylor G, Walker J. Charles Harrison Blackley, 1820–1900. Clin. Exp. Allergy. 1973;3:103–108. [Abstract] [Google Scholar]
- Van der Veen MJ, van Ree R, Aalberse RC, Akkerdaas J, Koppelman SJ, Jansen HM, van der Zee JS. Poor biologic activity of cross-reactive IgE directed to carbohydrate determinants of glycoproteins. J. Allergy Clin. Immunol. 1997;100(3):327–334. [Abstract] [Google Scholar]
- Walsh BJ, Barnett D, Burley RW, Elliott C, Hill DJ, Howeden ME. New allergens from hen’s egg white and egg yolk. In vitro study of ovomucin, apovitellenin I and VI, and phosvitin. Int. Arch. Allergy Appl. Immunol. 1988;87(1):81–86. [Abstract] [Google Scholar]
- Weghofer M, Grote M, Resch Y, Casset A, Kneidinger M, Kopec J, Thomas WR, Fernandez-Caldas E, Kabesch M, Ferrara R, Mari A, Purohit A, Pauli G, Horak F, Keller W, Valent P, Valenta R, Vrtala S. Identificaction of Der p 23, a peritrophin-like protein, as a new major Dermatophagoides pteronyssinus allergen associated with the peritrophic matrix of mite fecal pellets. J. Immunol. 2013;190(7):3059–3067. [Europe PMC free article] [Abstract] [Google Scholar]
- Willis IM, Stewart AF, Caputo A, Thompson AR, Mackinlay AG. Construction and identification by partial nucleotide sequence and analysis of bovine casein and beta-lactoblobulin cDNA clones. DNA. 1982;1(4):375–386. [Abstract] [Google Scholar]
- Yunginger JW, Gleich GJ. Measurement of ragweed antigen E by double antibody radioimmunoassay. J. Allergy Clin. Immunol. 1972;50(6):326–337. [Abstract] [Google Scholar]
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.molimm.2018.03.003
Article citations
Exploring Fish Parvalbumins through Allergen Names and Gene Identities.
Genes (Basel), 15(10):1337, 18 Oct 2024
Cited by: 0 articles | PMID: 39457461 | PMCID: PMC11507022
Review Free full text in Europe PMC
Allergen Testing: A Review of the Indications, Procedures, and Limitations in Ocular Allergy.
Clin Rev Allergy Immunol, 14 Sep 2024
Cited by: 0 articles | PMID: 39276294
Review
IgE-Mediated Legume Allergy: A Pediatric Perspective.
J Pers Med, 14(9):898, 25 Aug 2024
Cited by: 0 articles | PMID: 39338152 | PMCID: PMC11433522
Review Free full text in Europe PMC
Identification of cross-reactive IgE-binding proteins from Philippine allergenic grass pollen extracts.
Asia Pac Allergy, 14(3):108-117, 05 Aug 2024
Cited by: 0 articles | PMID: 39220572 | PMCID: PMC11365694
Bioaerosol Exposures and Respiratory Diseases in Cannabis Workers.
Curr Allergy Asthma Rep, 24(7):395-406, 15 Jun 2024
Cited by: 0 articles | PMID: 38878249 | PMCID: PMC11233357
Review Free full text in Europe PMC
Go to all (69) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe
-
(1 citation)
PDBe - 4JRBView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Update of the WHO/IUIS Allergen Nomenclature Database based on analysis of allergen sequences.
Allergy, 69(4):413-419, 01 Apr 2014
Cited by: 82 articles | PMID: 24738154
Review
Keeping Allergen Names Clear and Defined.
Front Immunol, 10:2600, 19 Nov 2019
Cited by: 11 articles | PMID: 31798576 | PMCID: PMC6878850
Review Free full text in Europe PMC
ALLERDB database and integrated bioinformatic tools for assessment of allergenicity and allergic cross-reactivity.
Cell Immunol, 244(2):90-96, 01 Dec 2006
Cited by: 5 articles | PMID: 17467675
Newly defined allergens in the WHO/IUIS Allergen Nomenclature Database during 01/2019-03/2021.
Allergy, 76(11):3359-3373, 05 Aug 2021
Cited by: 17 articles | PMID: 34310736 | PMCID: PMC9290965
Review Free full text in Europe PMC
Funding
Funders who supported this work.
AAAAI organizations
EAACI
IUIS
Intramural NIH HHS (2)
Grant ID: ZIA ES102906-07
Grant ID: Z99 ES999999




