Abstract
Background
Pulmonary arterial hypertension (PAH) is a rare disease characterized by pulmonary arteriole remodeling, elevated arterial pressure and resistance, and subsequent heart failure. Compared with adult-onset disease, pediatric-onset PAH is more heterogeneous and often associated with worse prognosis. Although BMPR2 mutations underlie ≈70% of adult familial PAH (FPAH) cases, the genetic basis of PAH in children is less understood.Methods
We performed genetic analysis of 155 pediatric- and 257 adult-onset PAH patients, including both FPAH and sporadic, idiopathic PAH (IPAH). After screening for 2 common PAH risk genes, mutation-negative FPAH and all IPAH cases were evaluated by exome sequencing.Results
We observed similar frequencies of rare, deleterious BMPR2 mutations in pediatric- and adult-onset patients: ≈55% in FPAH and 10% in IPAH patients in both age groups. However, there was significant enrichment of TBX4 mutations in pediatric- compared with adult-onset patients (IPAH: 10/130 pediatric versus 0/178 adult-onset), and TBX4 carriers had younger mean age-of-onset compared with BMPR2 carriers. Mutations in other known PAH risk genes were infrequent in both age groups. Notably, among pediatric IPAH patients without mutations in known risk genes, exome sequencing revealed a 2-fold enrichment of de novo likely gene-damaging and predicted deleterious missense variants.Conclusions
Mutations in known PAH risk genes accounted for ≈70% to 80% of FPAH in both age groups, 21% of pediatric-onset IPAH, and 11% of adult-onset IPAH. Rare, predicted deleterious variants in TBX4 are enriched in pediatric patients and de novo variants in novel genes may explain ≈19% of pediatric-onset IPAH cases.Free full text

Exome Sequencing in Children with Pulmonary Arterial Hypertension Demonstrates Differences Compared to Adults
Abstract
Background
Pulmonary arterial hypertension (PAH) is a rare disease characterized by pulmonary arteriole remodeling, elevated arterial pressure and resistance, and subsequent heart failure. Compared to adult-onset disease, pediatric-onset PAH is more heterogeneous and often associated with worse prognosis. While BMPR2 mutations underlie ~70% of adult familial PAH (FPAH) cases, the genetic basis of PAH in children is less understood.
Methods and Results
We performed genetic analysis of 155 pediatric- and 257 adult-onset PAH patients, including both FPAH and sporadic, idiopathic PAH (IPAH). Following screening for two common PAH risk genes, mutation-negative FPAH and all IPAH cases were evaluated by exome sequencing. We observed similar frequencies of rare, deleterious BMPR2 mutations in pediatric- and adult-onset patients: ~55% in FPAH and 10% in IPAH patients in both age groups. However, there was significant enrichment of TBX4 mutations in pediatric-compared to adult-onset patients (IPAH: 10/130 pediatric- vs 0/178 adult-onset), and TBX4 carriers had younger mean age-of-onset compared to BMPR2 carriers. Mutations in other known PAH risk genes were infrequent in both age groups. Notably, among pediatric IPAH patients without mutations in known risk genes, exome sequencing revealed a 2-fold enrichment of de novo likely gene damaging (LGD) and predicted deleterious missense variants.
Conclusions
Mutations in known PAH risk genes accounted for ~70–80% of FPAH in both age groups, 21% of pediatric-onset IPAH, and 11% of adult-onset IPAH. Rare, predicted deleterious variants in TBX4 are enriched in pediatric patients and de novo variants in novel genes may explain ~19% of pediatric-onset IPAH cases.
Introduction
Pulmonary arterial hypertension (PAH) is a rare disease with high mortality despite advances in treatment. The disease is etiologically heterogeneous, including familial PAH (FPAH), sporadic/idiopathic PAH (IPAH), hereditary PAH (HPAH which includes FPAH and IPAH with identified mutations) and PAH associated with other medical conditions (ie congenital heart disease, connective tissue disorders, portal hypertension and others). PAH manifests in early to mid-life with an estimated prevalence of 4.8–8.1 cases per million for pediatric-onset 1 and 15–50 cases per million for adult-onset 2. Although less prevalent than adult-onset, pediatric-onset disease is more frequently associated with other clinical conditions and less likely to respond to medical treatment. The untreated natural history of IPAH in children is poor with an historical median survival after diagnosis of 10 months compared to 2.8 years for adults 3. Further, the female predominance observed in adult-onset PAH (3.6:1 female:male ratio) is less pronounced in pediatric-onset cases (1.7:1 female:male ratio) 4. Genetics play an important role in the pathogenesis of PAH, including both FPAH and IPAH. However, the genetic basis of pediatric-disease has not been widely-investigated.
Previous genetic studies of PAH have been conducted primarily in adult cohorts. Germline mutations in the bone morphogenetic protein receptor 2 (BMPR2) gene, encoding a member of the transforming growth factor β (TGFβ) superfamily of receptors, have been identified as the major genetic cause for FPAH (~70% of patients) and, to a lesser extent, IPAH (10–40% of patients) 5. Mutations in other TGFβ family member genes, activin A, receptor type II-like 1 (ACVRL1) and endoglin (ENG) cause hereditary hemorrhagic telangiectasia (HHT) and HPAH 5. Mutations in the BMP receptor type IA and type 1B (BMPR1A and BMPR1B, also called activin receptor-like kinase-6 (ALK6)), caveolin-1 (CAV1), eukaryotic initiation translation factor 2 alpha kinase 4 (EIF2AK4), potassium two-pore-domain channel subfamily K member 3 (KCNK3), SMAD family members 4 and 9 (SMAD4 and SMAD9) and T-box4 (TBX4) have all been identified as less frequent or rare causes of PAH 5–8. Finally, common variants in cerebellin-2 (CBLN2) increase risk for PAH approximately 2-fold 9.
In children, BMPR2 mutations have been evaluated with inconsistent results, but the number of children studied previously was relatively small. One study of 13 children with IPAH failed to identify BMPR2 mutations 10. Another study of 18 pediatric IPAH cases identified four mutations in ACVRL1, ENG or BMPR2 11. A larger, retrospective study reported significantly lower 5-year survival rates among BMPR2 mutation carriers compared to non-carriers in a cohort of 54 children 12. Mutations in other genes including TBX4, BMPR1B and neurogenic locus notch homolog 3 (NOTCH3) have been reported to cause PAH in children 6, 8, 13.
We have previously used exome sequencing to identify CAV1, KCNK3, and EIF2AK4 as novel genetic causes of FPAH, IPAH or pulmonary capillary hemangiomatosis (PCH)/pulmonary veno-occlusive disease (PVOD) 5, 14–16. In the current study, we describe and compare the frequency and spectrum of genes with mutations in pediatric- vs adult-onset PAH using a genomic approach.
Methods
The data, analytic methods, and study materials are available upon request from the corresponding author, for purposes of reproducing the results or replicating the procedure.
Patients
We sought to identify rare inherited and de novo genetic variants causing pediatric- and adult-onset PAH, and to compare the distribution and frequency of genetic mutations in the two age groups. Patients were recruited from pulmonary hypertension centers at Columbia University and Children’s Hospital Colorado. The patients were collected over a 22-year period, from 1993 to 2015, largely recruited from a single center at Columbia University. Patients were diagnosed according to the World Health Organization (WHO) pulmonary hypertension group I classification 17 and included primarily FPAH and IPAH cases. All patients included in the analyses were unrelated. Patients with PAH-CHD, which is much more prevalent among pediatric-onset cases, were excluded to reduce heterogeneity and are the focus of a separate report. The diagnosis of PAH was confirmed by medical record review including right heart catheterization. Pediatric-onset was defined by diagnosis prior to 19 years of age, although subgroup comparison of the rate of de novo mutations considered age-of-onset within the pediatric group (ie. ≤5 vs 5–18 years). Written informed consent (and assent when appropriate) was obtained under a protocol approved by the institutional review board at Columbia University Medical Center and Children’s Hospital Colorado.
FPAH cases were screened for BMPR2 and ACVRL1 mutations by Sanger sequencing and multiplex ligation-dependent probe amplification (MLPA). Then, FPAH cases without mutations in BMPR2 and ACVRL1, and all IPAH cases, were analyzed by exome sequencing (Supplemental Figure 1).
Whole exome sequencing (WES)
WES was performed as a family-based analysis when possible and included 48 pediatric trios (proband and two biological parents), 24 pediatric-/10 adult-onset duos (proband and one biological parent), 60 pediatric-/185 adult-onset singleton probands, 10 pediatric-/13 adult-onset families with multiple affected individuals for a total of 350 probands (Table 1). DNA was extracted from peripheral blood leukocytes and WES was performed in collaboration with the Regeneron Genetics Center (RGC) or the PAH Biobank at Cincinnati Children’s Hospital Medical Center using standard procedures (Supplemental Material).
Table 1
PAH patient population
| Pediatric | Adult | |
|---|---|---|
| Idiopathic, n (%) | 130 (83.9) | 178 (69.3) |
| Familial, n (%) | 25 (16.1) | 79 (30.7) |
|
| ||
| Total, n | 155 | 257 |
| Female:male ratio (n) | 1.6:1 (96:59) | 4.1:1 (207:50)* |
WES Data analysis
The work-flow is outlined in Supplemental Figure 1. We used a previously established bioinformatics procedure to process and analyze WES data. Specifically, we used BWA-MEM (Burrows-Wheeler Aligner) 18 to map and align paired-end reads to the human reference genome (version GRCh37/hg19), Picard MarkDuplicates to identify and flag PCR duplicated reads, GATK 19, 20 HaplotypeCaller (version 3) to call genetic variants and GATK Variant Quality Score Recalibration (VQSR) to estimate accuracy of variant calls. To guard against technical artifacts, we excluded variants that met any of the following conditions: minimum read depth ≤8 reads, allele balance ≤20% 21, genotype quality <30, mappability <1 (based on 150 bp reads), genomicSuperDups segmental duplication similarity ≥95%, or VQSR >99.7. All candidate variants were confirmed with Sanger sequencing and tested for disease segregation when family DNA samples were available.
We used ANNOVAR 22 to annotate the variants and aggregate information of allele frequency and in silico predictions of deleteriousness. We used population allele frequencies from internal databases and public data from the Exome Aggregation Consortium (ExAC) 23 and Genome Aggregation Database (gnomAD) to define rare variants with a population allele frequency ≤0.1%. We used multiple bioinformatics prediction algorithms including PolyPhen 2, Mutation Taster, SIFT, PROVEAN, metaSVM 24 and Combined Annotation Dependent Depletion (CADD) 25 to predict variant deleteriousness but ultimately focused on metaSVM (damaging) to define damaging missense variants (D-Mis) for enrichment analyses. We identified de novo variants as described previously, using trios composed of proband and unaffected biological parents, and manually inspected all candidate de novo variants using the Integrative Genomics Viewer (IGV) 26 to reduce the false-positive rate. Finally, we inferred copy-number variants from WES data using the CLAMMS algorithm 27.
Identification of pathogenic/likely pathogenic variants in established PAH risk genes and novel candidate risk genes
Variants identified in known PAH risk genes were classified as pathogenic, likely pathogenic, or of uncertain significance according to the American College of Medical Genetics and Genomics (ACMG) guidelines 28. We compared deleterious rare or de novo variants with mutations reported in the literature and in genetic databases (Online Mendelian Inheritance in Man database, Human Genome Mutation Database 29 and ClinVar 30). Variants in established PAH genes of uncertain significance by ACMG guidelines and variants in novel candidate risk genes were classified based on predictions of deleteriousness. We defined three levels of deleterious variants: 1) “high,” likely-gene-disrupting (LGD) variants (including premature stopgain, frameshift indels, canonical splicing variants, and deletion of exons) and missense variants predicted to be damaging by MetaSVM and CADD (phred-scale) score ≥15; 2) “medium,” missense variants predicted to be damaging by MetaSVM or CADD score ≥15; and 3) “low,” all others.
Statistical analysis
To compare the frequency of rare deleterious variants between pediatric- and adult-onset PAH cases in known risk genes, we used a binomial test, with a null hypothesis that the frequency of patients carrying rare deleterious variants in a known risk gene (BMPR2 or TBX4) or gene set (ACVRL1, BMPR1A, BMPR1B, CAV1, EIF2AK4, ENG, KCNK3, SMAD4 and SMAD9) is the same between pediatric- and adult-onset patients.
To assess the enrichment of rare variants in known risk genes in cases, we obtained WES data of 2,426 unaffected parents from a congenital heart disease study by the Pediatric Cardiac Genetics Consortium (PCGC) 31 as controls, and selected subjects (cases and controls) of European ancestry using Principle Components Analysis (PCA) implemented in PLINK version 1.9 32, 33. We used an exact binomial test to test the significance of enrichment of rare variants in cases compared to controls, with a null hypothesis that the number of rare variants observed in cases follows a binomial distribution, given the total number of such variants in cases and controls, and a rate determined by the number of cases over the total number of cases and controls.
To estimate the burden of de novo variants in cases, we used previously calibrated background mutation rate 34, 35 to calculate the expected number of de novo variants of various types (e.g. LGD, missense, or synonymous) by chance in a number of cases in all genes or a gene set. For a certain type of de novo variant in a gene set, if the observed number in cases is m1, the expected number is m0, then we estimated the enrichment rate by (m1/m0), and test the significance by an exact Poisson test using m0 as the expectation.
We used fisher.test, binom.test, and poisson.test functions in R to perform statistical tests described above.
Results
There were 155 pediatric PAH patients diagnosed prior to age 19 (25 FPAH, 130 IPAH) and 257 adult-onset patients (79 FPAH, 178 IPAH). The female to male ratio was 1.6:1 (96/59) in the pediatric-onset and 4.1:1 (207/50) in the adult-onset groups (Table 1), and the difference is statistically significant (Fisher’s exact test p-value=4.9e-05). We performed targeted genetic analysis of BMPR2 (Sanger sequencing and MLPA) and ACVRL1 (sequencing) in 104 FPAH cases, and identified pathogenic or likely pathogenic variants in 63 cases according to ACMG guidelines. Next, WES was performed for the 42 BMPR2/ACVRL1 mutation-negative FPAH and all 308 IPAH patients, for a total of 350 samples (Supplemental Figure 1). The full set of rare pathogenic, likely pathogenic, or predicted deleterious variants in established PAH genes are provided in Supplemental Table 1 (pediatric-onset) and Supplemental Table 2 (adult-onset).
Similar BMPR2 mutation frequencies in pediatric- and adult-onset PAH patients
The combined analysis of Sanger sequencing/MLPA/WES resulted in the identification of 84 rare, predicted deleterious BMPR2 mutations in FPAH/IPAH probands (Supplemental Tables 1 and 2). Altogether, there were 58 LGD variants, 25 D-Mis variants, and 1 in-frame deletion of seven amino acids. Fourteen of the 84 mutations are novel and all are depicted in Supplemental Figure 2. Similar BMPR2 mutation frequencies were observed in pediatric- and adult-onset FPAH (14/25, 56% pediatric and 43/79, 54.4% adult) and IPAH (13/130, 10% pediatric and 14/178, 7.9% adult) patients (Table 2). However, there was a difference in the distribution of LGD and D-Mis variant types between the two age groups: LGD variants occurred more frequently in patients with older age of onset (31.3 ± 14.0 years, mean ± SD) compared to D-Mis variants (19.3 ± 12.5 years) (P =0.0006, Mann–Whitney U test). The distribution of variant locations was similar with almost all of the D-Mis variants in both age groups located within the conserved activin or protein kinase domains and most of the LGD variants located in the first half of the protein that contains the two domains (Supplemental Figure 2).
Table 2
Enrichment of rare genetic variants in known PAH genes* in pediatric- and adult-onset patients of European ancestry.
| Mutation type† | Observed (cases) | Observed (controls, n=1319) | Enrichment rate | p-value | |
|---|---|---|---|---|---|
| Pediatric-onset (n=88) | SYN | 4 | 30 | 2.00 | 0.16 |
| LGD | 3 | 2 | 22.48 | 2.22E-03 | |
| MIS | 9 | 69 | 1.96 | 0.06 | |
| D-Mis | 5 | 29 | 2.58 | 0.06 | |
|
| |||||
| Adult-onset (n=160) | SYN | 5 | 30 | 1.37 | 0.42 |
| LGD | 8 | 2 | 32.98 | 6.90E-07 | |
| MIS | 10 | 69 | 1.19 | 0.59 | |
| D-Mis | 2 | 29 | 0.57 | 0.77 | |
Substantial contribution of TBX4 mutations to pediatric-onset PAH
We identified 13 likely pathogenic/predicted highly deleterious TBX4 variants in 13 probands (12 pediatric- and 1 adult-onset) (Supplemental Tables 1 and 2). The variants included 9 LGD and 3 D-Mis variants, including one intragenic deletion encompassing exons 3–9. All of these variants are novel and most reside within the conserved T-box domain (Figure 1). Based on the 12 probands with at least one parental sample available, we determined the inheritance pattern of a subset of variants: 1 de novo, 4 inherited from mothers without PAH, and 4 inherited from fathers without PAH. The observed inheritance of TBX4 variants from an unaffected parent in at least 8 of 13 cases is consistent with incomplete penetrance that is also observed for BMPR2 mutation carriers36, suggesting that the effect of any single risk variant on development of disease is likely dependent on genetic background, somatic mutations, environmental effects or some combination thereof. Notably, pediatric-onset IPAH cases were significantly enriched for TBX4 mutations compared to adult-onset patients (10/130, 7.7% versus 0/178, p-value <0.0001) (Figure 2A). Although a much smaller sample size, there was a similar trend for FPAH cases (Figure 2A). Additionally, TBX4 carriers had a 20-year younger age-of-onset (7.9 ± 9.0, mean ± SD; range 0.6–33 years) compared to BMPR2 carriers (28.2 ± 15.4; range 1.5–72 years) (p <0.0001) (Figure 2B).
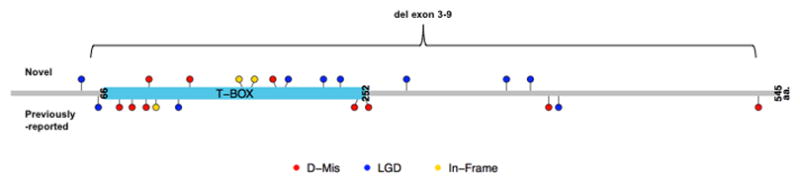
Novel mutations identified by WES are indicated above the protein schematic. Previously-reported mutations not identified in this study are indicated below the schematic. D-Mis, damaging missense mutations predicted by MetaSVM (red), LGD, likely gene damaging (blue) and In-frame insertion/deletions (yellow).
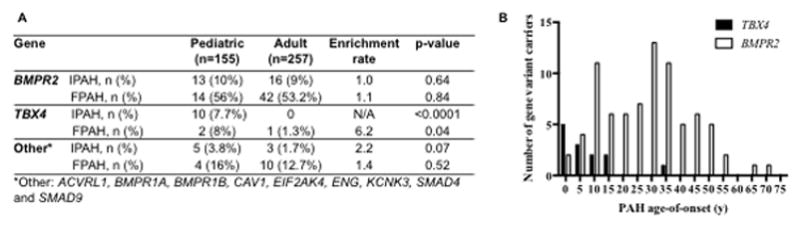
A, Enrichment of rare, predicted deleterious variants in TBX4, but not other known risk genes, in pediatric-onset cases. P-values were calculated by binomial tests. B, Younger age-of-disease onset for TBX4 variant carriers compared to BMPR2 variant carriers (P <0.0001, Mann-Whitney U test)
Rare genetic causes of PAH
We identified a small number of patients with rare deleterious mutations in recently identified genes for PAH, PAH associated with hereditary hemorrhagic telangiectasia (PAH-HHT), and genes involved in the BMP and TGFβ signaling pathway (Supplemental Tables 1 and 2). Two pediatric-onset patients were observed to carry KCNK3 mutations: a novel paternally-inherited two amino acid insertion (c.641_642insGCAGAC:p.214insQT) (IPAH) and a previously-reported heterozygous missense variant (c.G544A:p.E182K) 15 (FPAH). A novel heterozygous frameshift mutation in CAV1 (c.471delC; p.D157fs) was found in an adult patient with a pediatric-onset daughter who died at 9 years old. Compound heterozygous LGD variants in EIF2AK4 (c.C3766T:p.R1256X and c.1150dupG:p.S383fs) were identified in a familial case with PCH. A previously-reported homozygous nonsense mutation in EIF2AK4 (c.C1387T:p.R463X) 37 was present in an adult FPAH patient whose brother had PAH and died at age 33 and was not available for testing.
In our total PAH cohort, 9 patients had a diagnosis of PAH-HHT. ACVRL1 and ENG are known risk genes for PAH-HHT and targeted Sanger sequencing/WES identified 3 pediatric- and 5 adult-onset patients with mutations in ACVRL1 as well as 1 adult patient with an ENG mutation (Supplemental Tables 1 and 2). Interestingly, one patient with isolated PAH without HHT carried a paternally-inherited missense variant in ACVRL1 (c.C1199T:p.A400V) as well as a paternally-inherited missense variant in BMPR2 (c.A1509C:p.E503D).
Finally, three candidate pathogenic variants were identified in genes in the BMP and TGFβ signaling pathway. A deleterious missense variant and a nonsense variant were identified in SMAD9 and a deleterious missense variant in BMPR1B (Supplemental Tables 1 and 2). In total, the rare genetic cause mutations had similar frequencies in pediatric and adult PAH cases: 3.8% (5/130) of IPAH cases and 16% (4/25) of FPAH pediatric-onset patients and 1.7% (3/178) of IPAH and 12.7% (10/79) of FPAH adult-onset cases (Table 2).
Excess of rare deleterious variants in known PAH risk genes due to BMPR2 and TBX4
We then evaluated the burden of rare variants in all 11 established PAH-associated risk genes from WES data. We used 1319 unaffected parents from a congenital heart disease cohort in the Pediatric Cardiac Genetics Consortium (PCGC) 31 as population controls for allele frequency estimates. To avoid spurious association due to population stratification, we inferred ethnicity of all subjects using principal components analysis 33, and selected 88 pediatric-onset and 160 adult-onset PAH cases of European ancestry for this analysis. We observed a significant burden of rare LGD variants in both the pediatric- (enrichment rate=22, p-value=0.002) and adult-onset cases (enrichment rate=33, p-value=6.9e-7) in this established PAH gene set (Table 3). There was also a trend toward enrichment of rare, predicted deleterious missense variants in pediatric-onset cases (odds ratio=2.6, p-value=0.06), but not in adult-onset cases. The enrichment was not observed when BMPR2 and TBX4 were removed from the analysis (Supplemental Table 3).
Table 3
Enrichment of rare de novo mutations in pediatric-onset IPAH trios (n=36).
| Mutation type* | Observed | Expected by chance | Enrichment | p-value |
|---|---|---|---|---|
| SYN | 11 | 11.12 | 0.99 | 1 |
| LGD | 6 | 3.42 | 1.75 | 0.46 |
| MIS | 29 | 24.58 | 1.18 | 0.36 |
| D-Mis | 11 | 4.68 | 2.35 | 0.009 |
| LGD&D-Mis | 17 | 8.04 | 2.11 | 0.004 |
Burden of de novo variants in pediatric-onset patients
To further explore the genetic underpinnings of PAH, we performed a genome-wide burden test for de novo variants. Forty-six pediatric-onset cases for which we had ascertained parental DNA were available for trio WES. We were unable to perform parallel trio analyses in adult patients due to the lack of available parental DNA samples. Excluding the cases with rare deleterious variants in the 11 known risk genes, 36 pediatric-onset IPAH cases were used to compare the frequency of de novo variants in cases with expectation from the background mutation rate 34, 35. We detected a 2.35-fold enrichment (p=0.01) of de novo D-Mis variants in the pediatric IPAH cases (Table 3). Subgroup analysis revealed that very early-onset cases (≤5 years of age) were enriched for LGD variants (6/16 patients) compared to older-onset cases (>5–18 years, 0/18 patients) (p =0.01) (Supplemental Table 4). A list of all de novo mutations/genes identified in this study, excluding synonymous variants, is provided in Supplemental Table 5. As expected, eight of these patients harbored deleterious de novo mutations in two or more genes. Overall, the data suggest that ~25% of pediatric-onset IPAH without mutations in known PAH genes, or 19% of all pediatric-onset IPAH cases, may be explained by de novo LGD or deleterious missense variants in novel genes.
For patients without rare, deleterious variants in known PAH risk genes, we also performed a genome-wide burden test for rare, deleterious variants excluding the set of 11 known risk genes. We again used the unaffected individuals from the PCGC 31 as controls and selected 66 pediatric- and 120 adult-onset PAH cases of European ancestry for this analysis. We observed no enrichment of rare, deleterious variants in either pediatric (Supplemental Table 6) or adult (Supplemental Table 7) cohorts.
Discussion
In a cohort of 412 PAH patients, we identified new rare, deleterious mutations in known PAH genes, candidate causal de novo variants in novel genes, and compared the results of genetic analyses in pediatric- vs adult-onset disease. Fourteen novel mutations were identified in the major PAH risk gene, BMPR2, 13 in TBX4, and 12 in other known PAH risk genes. Most of the mutations are located in conserved protein functional domains. Using a genome-wide de novo screen, we identified 6 LGD and 10 deleterious missense variants in novel candidate disease genes. While the frequencies of BMPR2 mutations were similar for pediatric- and adult-onset PAH cases, the frequency of TBX4 mutations among pediatric-onset patients was significantly greater than that of adult-onset patients, with significantly younger age-of-onset. Furthermore, we identified an enrichment of LGD and deleterious missense de novo variants in pediatric IPAH patients without mutations in known PAH risk genes.
BMPR2 and other genes in the TGFβ pathway in PAH
The TGFβ superfamily includes the TGFβ /BMP ligands, receptors, accessory proteins, activins, and downstream signaling mediators SMADs and NOTCH3 38. In this study, we identified rare, deleterious mutations in known PAH risk genes BMPR2, BMPR1B, ACVRL1, ENG, and SMAD9 for a total yield of 48 variants in pediatric-onset (20/25, 80% FPAH and 28/130, 21.5% IPAH) and 74 in adult-onset (53/79, 67.2% FPAH and 19/178, 10.7% IPAH) patients. Additionally, we identified rare, deleterious variants in other candidate genes in this pathway: BMPR1A, ACVR2B, GDF2, SMAD6, TGFB, and TGFBR2. BMPR2 mutations accounted for ~55% of FPAH cases in both children and adults. Although the contribution of BMPR2 mutations to PAH in this study was slightly lower than in previous reports, it is likely that patients with clinically-identified BMPR2 mutations chose not to enroll in this genetic study. Exploration of the novel candidate risk genes in additional pediatric and adult PAH patients, along with functional characterization of potentially pathogenic variants, will help further our understanding of this pathway and how its dysregulation contributes to the pathophysiology of PAH.
TBX4 and pediatric PAH
TBX4 is a transcription factor in the T-box gene family expressed in the atrium of the heart, the limbs, and the mesenchyme of the lung and trachea. TBX4, jointly with TBX5, has been shown to interact with FGF10 during lung growth and branching 39. TBX4 was first suggested as a candidate PAH risk gene because of its localization within chromosome 17q microdeletions of patients presenting with severe developmental delays sometimes associated with pulmonary hypertension40, 41. Then, Kerstjens-Frederikse and colleagues performed sequencing of TBX2 and TBX4, both located within the deletions, in PAH patients and identified 3 rare TBX4 mutations and 3 novel 17q22q23.2 microdeletions in 20 pediatric patients as well as one rare TBX4 mutation among 49 adults; no mutations in TBX2 were identified8. Heterozygous loss of function mutations in TBX4 have been reported to cause small patella syndrome (SPS; MIM #147891) 42 and five of the TBX4 variant carriers reported by Kerstjens-Frederikse were retrospectively found to have clinical features of SPS8. More recent studies have identified 3/40 TBX4 mutation carriers in a children’s PAH cohort43 and 3/136 adult carriers in a Spanish PAH cohort44. Our study is the largest to date, confirming a role for rare, predicted deleterious TBX4 variants in PAH, especially pediatric-onset disease. Of the 13 PAH patients carrying LGD or D-Mis TBX4 variants, only one was diagnosed with SPS; however, no radiologic records of knees, pelvis or feet were available for retrospective assessment of the others. The clinical phenotypes of the TBX4 mutation carriers ranged from mild/moderate PAH to persistent, progressive PAH unresponsive to vasodilators and fatal or requiring lung transplantation. Notably, we observed a significant enrichment of rare, predicted deleterious TBX4 variants among pediatric- compared to adult-onset patients. Further, TBX4 variant carriers had a 20-year earlier mean age-of-onset compared to BMPR2 carriers.
Gender bias of PAH presentation is modified by age of disease onset
In studies carried out primarily in adult cohorts, PAH has been shown to occur approximately 3-fold more frequently in females than males. However, a previous study of pediatric PAH reported that females represented only ~60% of patients 4. In our study of both pediatric- and adult-onset patients, the female gender bias was significantly lower in pediatric compared to adult patients (Table 1). Genetically, BMPR2, ACVRL1, and ENG mutations occurred more frequently in females, while TBX4 mutations, which exhibited enrichment in the pediatric-onset patients, occurred with equal frequency among both genders. Mutations in other known PAH genes occurred less frequently and effects of age and gender could not be determined.
Pediatric- vs adult-onset PAH genetics
A comparison of the genetic causes of pediatric- and adult-onset PAH as suggested from our study is depicted in Figure 3. While the contribution of BMPR2 mutations was similar between both groups of patients, mutations in TBX4 made a significantly greater contribution to pediatric- vs adult-onset PAH patients. The contribution of other known PAH risk genes was rare in both age groups. Analysis of de novo mutations in pediatric IPAH patients and their unaffected relatives revealed an enrichment of D-Mis de novo mutations, while vary early age-of-onset patients had an enrichment of LGD de novo mutations. Of note, a pediatric-onset patient who died in childhood carried a D-Mis mutation in MAPK6 (mitogen-activated protein kinase 6). MAPK6 encodes ERK3 (extracellular signal-regulated kinase 3) and mice carrying null alleles exhibit intrauterine pulmonary hypoplasia and early neonatal death 45. AMOT (angiomotin) encodes an angiostatin-binding protein involved in embryonic endothelial cell migration and tube formation as well as endothelial cell tight junctions and angiogenesis 46–48; the frameshift insertion mutation carried by pediatric-onset patient JM0004 (diagnosed by 1 year of age) would result in the loss of 2 coiled-coil motifs, a PDZ protein-interaction domain as well as the angiostatin-binding motif. KEAP1 (Kelch-like ECH associated protein 1), encoding an E3 ubiquitin ligase, regulates cellular responses to oxidative stress through interactions with NRF2. Disruption of KEAP1-NRF2 interaction ameliorated oxidative stress and inhibited apoptosis in murine vascular cells 49, and endothelial-specific deletion of NRF2 reduced endothelial cell sprouting in vivo 50. NUCB1, nucleobindin 1, encodes a protein with multifunctional domains, and the frameshift variant at amino acid 189 would disrupt a DNA-binding site and cause loss of two Ca+-binding sites and a leucine zipper motif 51. These and other de novo variants identified in this study may provide novel insights into the pathophysiology underlying PAH.
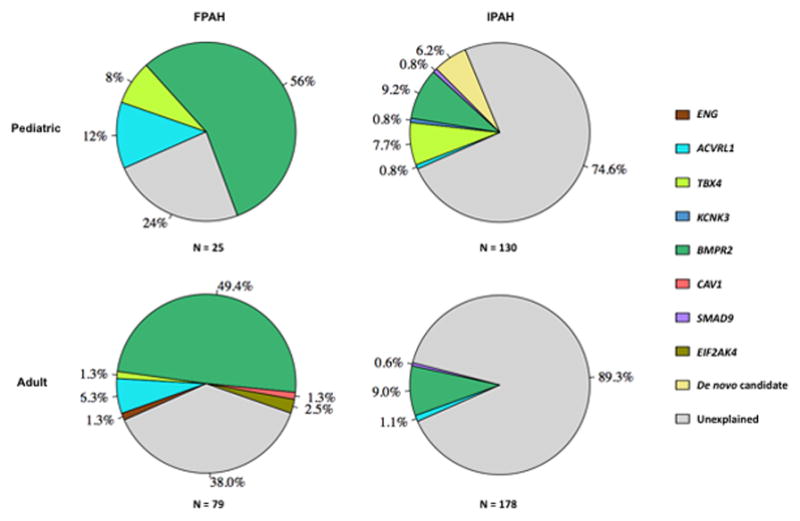
The percentage of total rare, deleterious mutations identified by Sanger sequencing or WES attributable to known PAH risk genes, de novo candidates, or unexplained is depicted as pie charts for pediatric- and adult-onset FPAH and IPAH.
In summary, differences in the genetic basis of PAH in children compared to adults include an enrichment of TBX4 mutations, likely contributing to the earlier onset of disease. Notably, de novo mutations account for a significant fraction of PAH in children and could provide an important strategy to identify novel PAH genes in this patient group. The genetic assessment of larger pediatric study cohorts could provide an important opportunity to identify novel genes, elucidate the pathogenic mechanisms of PAH and provide targets for future therapies.
Acknowledgments
We thank the families for their generous contribution. Robyn Barst and Jane Morse were critical members of the team to enroll and clinically characterize patients. Patricia Lanzano provided oversight of the biorepository at Columbia University. Philip Allen provided assistance with variant curation. Children’s Hospital Colorado patient samples were enrolled and sequenced as part of the National Biological Sample and Data Repository for PAH (PAH Biobank).
Sources of Funding: Funding support was provided by NHLBI HL060056 (WKC), HL105333 (DDI, WCN, MWP, KAL), HL114753 (DDI) as well as NIH/NCATS Colorado Clinical and Translational Science Award UL1 TR001082 (DDI), The Frederick and Margaret L. Weyerhaeuser Foundation (DDI) and The Jayden de Luca Foundation (DDI).
Footnotes
Disclosures: CG-J, AKK, JGR, JDO, AB and FD are full time employees of Regeneron Pharmaceuticals Inc. and receive stock options as part of compensation.
References
Full text links
Read article at publisher's site: https://doi.org/10.1161/circgen.117.001887
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc5896781?pdf=render
Citations & impact
Impact metrics
Article citations
The causes of pulmonary hypertension and the benefits of aerobic exercise for pulmonary hypertension from an integrated perspective.
Front Physiol, 15:1461519, 17 Oct 2024
Cited by: 0 articles | PMID: 39483752 | PMCID: PMC11525220
Review Free full text in Europe PMC
Seeing pulmonary hypertension through a paediatric lens: a viewpoint.
Eur Respir J, 63(6):2301518, 20 Jun 2024
Cited by: 0 articles | PMID: 38575157 | PMCID: PMC11187317
Frameshift variants in C10orf71 cause dilated cardiomyopathy in human, mouse, and organoid models.
J Clin Invest, 134(12):e177172, 17 Jun 2024
Cited by: 0 articles | PMID: 38950288
Homozygous missense variants in YKT6 result in loss of function and are associated with developmental delay, with or without severe infantile liver disease and risk for hepatocellular carcinoma.
Genet Med, 26(7):101125, 21 Mar 2024
Cited by: 0 articles | PMID: 38522068
Defining the clinical validity of genes reported to cause pulmonary arterial hypertension.
Genet Med, 25(11):100925, 05 Jul 2023
Cited by: 12 articles | PMID: 37422716 | PMCID: PMC10766870
Go to all (66) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Diseases
- (1 citation) OMIM - 147891
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genes that drive the pathobiology of pediatric pulmonary arterial hypertension.
Pediatr Pulmonol, 56(3):614-620, 09 Jan 2020
Cited by: 9 articles | PMID: 31917901 | PMCID: PMC7343584
Review Free full text in Europe PMC
Rare variant analysis of 4241 pulmonary arterial hypertension cases from an international consortium implicates FBLN2, PDGFD, and rare de novo variants in PAH.
Genome Med, 13(1):80, 10 May 2021
Cited by: 34 articles | PMID: 33971972 | PMCID: PMC8112021
Genetic analyses in a cohort of children with pulmonary hypertension.
Eur Respir J, 48(4):1118-1126, 01 Sep 2016
Cited by: 48 articles | PMID: 27587546
Funding
Funders who supported this work.
NCATS NIH HHS (1)
Grant ID: UL1 TR001082
NHLBI NIH HHS (4)
Grant ID: R01 HL114753
Grant ID: R01 HL060056
Grant ID: R24 HL105333
Grant ID: R01 HL136748
NIGMS NIH HHS (1)
Grant ID: R01 GM120609





