Abstract
Free full text

An in vivo model of functional and vascularized human brain organoids
Abstract
Differentiation of human pluripotent stem cells to small brain-like structures known as brain organoids offers an unprecedented opportunity to model human brain development and disease. To provide a vascularized and functional in vivo model of brain organoids, we established a method for transplanting human brain organoids into the adult mouse brain. Organoid grafts showed progressive neuronal differentiation and maturation, gliogenesis, integration of microglia, and growth of axons to multiple regions of the host brain. In vivo two-photon imaging demonstrated functional neuronal networks and blood vessels in the grafts. Finally, in vivo extracellular recording combined with optogenetics revealed intragraft neuronal activity and suggested graft-to-host functional synaptic connectivity. This combination of human neural organoids and an in vivo physiological environment in the animal brain may facilitate disease modeling under physiological conditions.
The development of adequate model systems to study the human brain in health and disease has proved challenging. Animal models, particularly rodents, and cell culture systems, such as those using human pluripotent stem cell (hPSC)-derived neural lineages1, have provided tremendous insights into the development, function, and dysfunction of the human brain, but do not capture its full complexity2,3. Three-dimensional (3D) neural tissues generated from hPSCs, termed brain organoids, have enormous potential for investigating aspects of human brain development and mental disorders4–11. Brain organoids contain multiple cell types, have neuronal functionality in vitro that recapitulates some features of the development and maturation of complex neuronal tissue, and are more representative of in vivo physiology than two-dimensional cell cultures. While brain organoids have been successfully applied to study both human brain development and complex human diseases, limitations that preclude their broader application remain8,12–16.
Current brain organoid systems lack the microenvironment, neuronal circuits, vascular circulation, and immune system that exist in vivo. Vascularization is particularly necessary for oxygen penetration, nutrient supply, and efficient neural progenitor differentiation17,18. The lack of oxygen penetration renders the center of the organoid necrotic, which can interfere with its normal development and potential neuronal migration routes19. Integrating a vascular structure that allows adequate delivery of oxygen and nutrients is necessary to generate large organoids that are not limited in size by diffusion. Although co-cultures with vascular cells or tissue engineering may be capable of forming tubular networks in organoids and other 3D structures18,20, the issues of nutrient supply and authentic blood microenvironment remain. Thus, an alternative approach may be engraftment into an animal host. While such engraftment has been successful for a wide variety of solid tissues and for both normal and cancer organoids21–30, to our knowledge, such an in vivo model system for vascularized human brain organoids has not yet been reported.
Here, we developed an efficient in vivo engraftment model of hPSC-derived brain organoids within a physiological tissue environment. The grafted organoids readily integrated into the mouse brain, exhibited progressive neuronal temporal differentiation patterns, developed a functional vasculature system, and displayed unprecedented axonal outgrowth to generate mature and functional human brain tissues in vivo that responded to physiological stimuli triggered by anesthesia. Finally, we used optogenetics to show functional synaptic connectivity between grafted organoids and the host brain.
RESULTS
Intracerebral grafting of brain organoids
Intracerebral implantation in rodents supports the long-term survival and vascularization of fetal and adult mouse tissue21,31,32. Therefore, we hypothesized that intracerebral implantation of cerebral organoids could create a permissive environment for their development and long-term survival. We first transduced human embryonic stem cells (hESCs) with a lentivirus vector encoding EGFP to generate a GFP hESC line. GFP+ cerebral organoids were generated as described previously (Fig. 1a and Supplementary Fig. 1a,b)8,33. Organoids were grown in culture for 40–50 d, unless stated otherwise, before intracerebral implantation into a cavity made in the retrosplenial cortex of NOD-SCID (nonobese diabetic- severe combined immunodeficient) (Prkdcscid)-immunodeficient mice (Fig. 1a). Only organoids that passed the quality criteria described previously33—clearing of embryoid body borders, formation of radially organized neuroepithelium before Matrigel embedding, and outgrowth and development of defined buds in Matrigel without massive cyst formation or premature differentiation—were selected for implantation. In addition, we confirmed the generation of polarized, radially organized, ventricular zone–like regions (VZ-L), and dorsal forebrain identity in the organoids (Supplementary Fig. 1c,d).
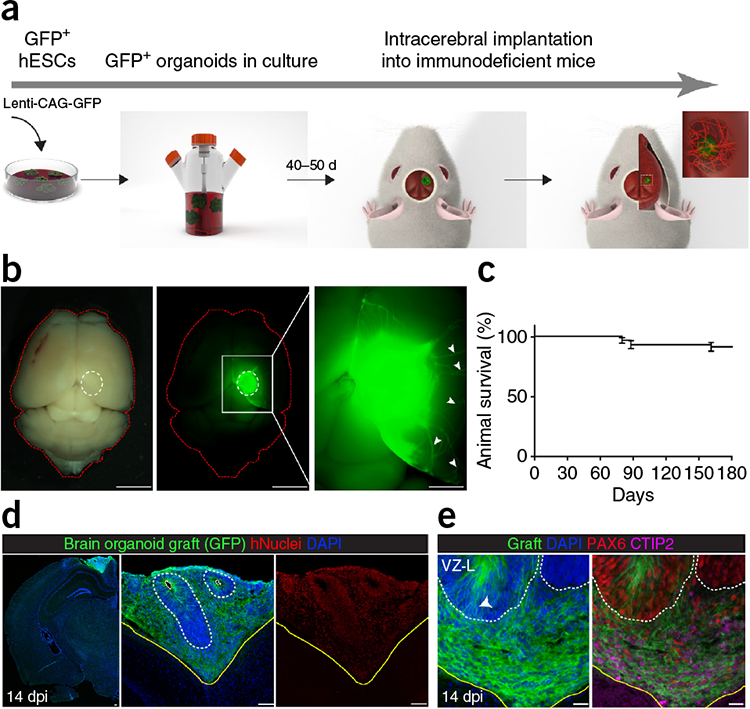
Intracerebral grafting of brain organoids into mouse brain. (a) Illustration of the experimental procedure for generation of GFP+ organoids from hESCs and intracerebral implantation into immunodeficient mouse brain. (b) Whole-mount dorsal view image of mouse brain grafted with a GFP+ cerebral organoid and harvested at 50 dpi; the graft is outlined in white. Right, magnified graft displaying neurite outgrowth from the organoid toward the host brain (arrowheads). (c) Kaplan–Meier survival curve for overall survival of mice after engraftment with brain organoids. (91.8% survival beyond 180 dpi, n = 61 mice from ten experiments). (d) GFP+ organoids were grafted into mouse brain and harvested at the indicated dpi. Coronal sections were analyzed using immunofluorescence and confocal microscopy. Immunofluorescence staining for GFP and human nuclear antigen (hNuclei), demonstrates that the implant survived well and distributed throughout the lesion cavity at 14 dpi. Left image shows confocal stitched tile scan; image was vertically inverted. White solid lines indicate apical/ventricular surface. Dotted white line indicates the radial glia VZ-L regions. Yellow lines indicate the graft-host border. (e) Graft immunostained for GFP, dorsal telencephalic progenitor marker PAX6 and the deep-layer subcortical neuron marker CTIP2. Radially organized cells (arrowhead, left panel) represent the PAX6+ VZ-L region (dotted white lines). n = 4 animals in b, and n = 3 animals in d,e. Nuclei were counterstained with DAPI. Scale bars: 1 μm in b, 100 μm in d, and 20 μm in e.
Approximately 92% of grafted animals survived beyond 180 d (Fig. 1c). To test organoid graft survival, we examined GFP expression in the whole-mount brain after dissection. At 0.5–3 months post-implantation (PI), we observed robust integration and survival of the graft (80% ± 4%, n = 28 mice), with GFP+ processes extruding from the graft into the recipient brain (Fig. 1b,d and Supplementary Fig. 2). Only brains with clearly identifiable grafts were subsequently analyzed.
Organoid grafts exhibit progressive differentiation
To examine the differentiation potential of grafted organoids, we analyzed mouse brains at 0.5 to ~8 months. Human cells were first identified based on their expression of GFP, human nuclear antigen (hNuclei), and anti-human mitochondria antibody. At 14 and 90 d post-implantation (dpi), grafts exhibited robust integration and survival and were distributed throughout most of the implantation cavity (Fig. 1d and Supplementary Fig. 2a,b). A hallmark of cerebral organoids is the relatively dominant formation of forebrain identity, with well-defined neuroepithelial ventricular zones and cortical-like neuronal layers8,12 (Supplementary Fig. 1c,d). At 14 dpi, grafts showed segregation of well-defined SOX2+ VZ-L regions with typical radial morphology (Fig. 1d,e and Fig. 2a). The dorsal marker PAX6 was expressed in radial glia progenitors in the VZ-L regions, and CTIP2, a marker of an early-born deep neuronal cortical layer, was expressed in the cortical plate–like area (Fig. 1e). The combination of PAX6 and CTIP2 expression domains in the graft indicates their dorso-cortical identity. These results suggested that the regional and morphological identities of grafted organoids were preserved in vivo.

Survival and differentiation of grafted organoids. Organoids were grafted and harvested at the indicated dpi, coronal sections were analyzed using immunofluorescence and confocal microscopy. (a) Double immunofluorescence staining for SOX2 and NeuN (left), and GFP and SMI312 (right). At 14 dpi, the graft expresses both the NPCs marker SOX2 and the mature neuronal marker NeuN but shows low numbers of SMI312-positive processes. At 50 dpi, the graft retains a lower expression domain of SOX2, NeuN+ cells, and the SMI312-positive area increase. White solid lines indicate the apical/ventricular surface. Dotted white lines indicate the radial glia VZ-L regions. Yellow lines indicate the graft-host border. (b) Quantification of the percentage (mean ± s.e.m.) of SOX2+/DAPI+ and of NeuN+/DAPI+ cells in the graft at the indicated grafting time point. P values were calculated using one-way ANOVA with post hoc Tukey’s test between group comparisons (F(2, 6) = 18.81; P = 0.0026 for NeuN, F(2, 6) = 24.3; P < 0.0001 for SOX2), n = 3 independent animals per group. Asterisks indicate pair-wise comparisons with 14 dpi. (c) Comparative quantification of the percentage of SOX2+/DAPI+ and of NeuN+/DAPI+ cells in the graft compared with stage-matched organoids in culture at the indicated grafting or culture time point. Data are presented as mean ± s.e.m., unpaired two-tailed t-test. For SOX2, 53 day vs. 14 dpi (t = 3.059, df = 8, P = 0.0156) and 102 d vs. 50 dpi (t = 1.617, df = 9, P = 0.1369, not significant). For NeuN, 53 d vs. 14 dpi (t = 1.208, df = 8, P < 0.2617, not significant) and 102 d vs. 50 dpi (t = 12.15, df = 9, P < 0.0001). Day 53 (n = 7 organoids), day 102 (n = 8 organoids) from three independent patches. n = 3 animals for 14 dpi and 50 dpi. (d) Immunofluorescence staining for GFP and human-specific GFAP (hGFAP) at the indicated time points showing astrocyte differentiation in the graft with increased abundance over time. Right panel is a higher magnification of the boxed area. (e) Immunostaining for oligodendrocyte marker Olig2 in the graft at 50 and 90 dpi. (f) Human graft contains Iba1+ microglia that do not co-localize with GFP. (g) Double immunofluorescence staining for presynaptic marker Synapsin (Syn) and the postsynaptic marker PSD95 at 50 dpi, showing a co-association between pre- and post-synaptic compartments and the formation of synaptic connections in the graft. The box indicates the region of magnification from the left panel; arrowheads indicate direct contact between a pre- and post-synapses puncta. Image shows a single plane confocal-Z-section. Nuclei were counterstained with DAPI. Scale bars: 100 μm in a, 50 μm in d–f, 5 μm in f (right panel) and g. *P < 0.05,
**P < 0.01, ****P < 0.0001. n.s., not significant.
At their early developmental stage in culture, cerebral organoids consist of both neural progenitor cells (NPCs) and mature neuronal cells; at later stages NPCs undergo neurogenesis and mature to neurons8,12,13,33. To examine whether similar progressive maturation occurred over time in vivo, we used the NPC marker SOX2 and the mature neuronal markers NeuN, MAP2, and SMI312. By 14 dpi, grafted organoids expressed SOX2 and NeuN, with relatively low presence of SMI312+ regions (Fig. 2a). At 50 dpi and 90 dpi, NeuN+ cells were enriched at the expense of SOX2, and prominent expression of the neuronal markers SMI312 and MAP2 was observed (Fig. 2a,b and Supplementary Fig. 3b–d. Organoid grafts survived up to 233 dpi, the latest time point tested in our study, and showed persistent expression of NeuN and SOX2 (Supplementary Fig. 3e,f). Comparison of differentiation dynamics of grafted organoids and nearly stage-matched cerebral organoids cultured in vitro revealed relatively higher numbers of NeuN+ cells at 50 dpi compared to 102-day in vitro organoids (41.76% ± 1.817;10.94% ± 1.346, respectively), with no significant differences at earlier stages (Fig. 2c), suggesting that the in vivo environment enhanced cellular maturation and/or survival of organoids.
We next examined the level of gliogenesis within organoid grafts. Consistent with previous reports, we rarely observed the mature astrocyte marker GFAP in cultured organoids before the engraftment stage8,12,13,34 (Supplementary Fig. 3a). In grafted organoids, there were few human-specific GFAP+ cells at 14 dpi and a substantial increase at later time points (Fig. 2d). Glial astrocyte identity was confirmed by staining for S100β (Supplementary Fig. 3b). At 50, 90, and 233 dpi, the majority of SOX2+ cells were not organized in tight, regionally restricted expression domains resembling stem cell niches, as observed at 14 dpi and in cultured organoids (Fig. 2a, Supplementary Fig. 1d and 3c,e), but were sparsely distributed, raising the possibility that these were SOX2-expressing astrocytes35 rather than NPCs. Co-localization of SOX2 and GFAP expression (Supplementary Fig. 3b) suggested an astrocyte fate. Furthermore, OLIG2 staining at middle and late stages revealed the presence of oligodendrocytes in the grafts (Fig. 2e and Supplementary Fig. 3g). However, when staining for myelin basic protein (MBP), we did not detect signs of myelination.
Immunostaining for the presynaptic marker Synapsin I and the post-synaptic marker PSD95 showed not only intragraft punctate staining of these markers, but also multiple Synapsin and PSD95 colocalized puncta, suggesting synaptic connectivity in the grafts (Fig. 2g). Immunostaining for the microglia marker Iba1 revealed abundant migration of Iba+ cells from the host throughout the graft at 90 and 233 dpi with typical ramified morphology (Fig. 2f and Supplementary Fig. 3g). Iba1+ cells were GFP negative, suggesting that they were of host origin, consistent with the lack of microglia cells in ungrafted organoids at least at day 38 of culture (Supplementary Fig. 3a).
Collectively, these results demonstrate that grafting of cerebral organoids supports progressive differentiation of neuronal and glial lineages and infiltration of brain immune cells into the graft.
Extensive axonal growth of engrafted organoids
We next investigated axonal outgrowth of cerebral organoids into the host brain. By 90 dpi, high levels of GFP+ axons were growing out of the graft sites, with axon density progressively diminishing as a function of distance from the graft (Supplementary Fig. 4). Ventrally innervated bundles of human axons were detected in both hemispheres, with the ipsilateral hemisphere containing the highest number of fibers. Most axons grew out of organoids as a bundle through the cortical layers and the corpus callosum (Fig. 3a,b and Supplementary Fig. 4a,b). GFP+ human axons also traversed the amygdalar nucleus and the striatum. Human axons with lower fiber density were also observed ipsilaterally in the hippocampus, hypothalamus and thalamus, and in contralateral regions of the host brain. Notably, axonal growth was found not only in the grafted regions but also more rostrally (Supplementary Fig. 4a,b). These results demonstrate that grafted organoids can generate long-distance axonal projections to distant targets in the host brain.
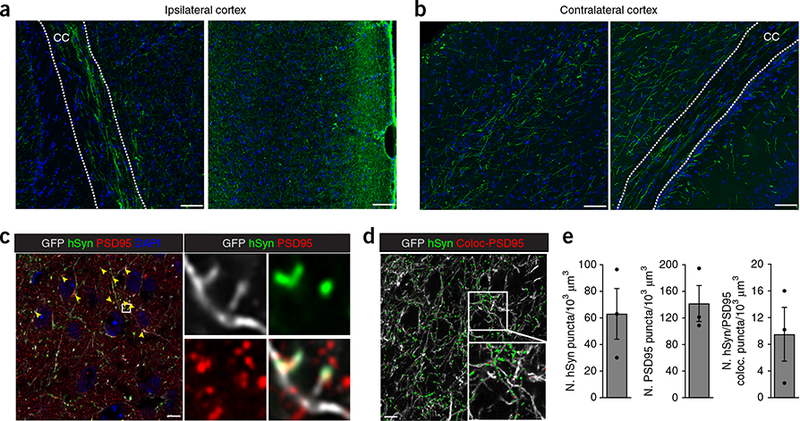
Organoid graft sends axonal projections with synaptic connectivity. (a,b) Confocal images of brain sections stained for GFP and obtained from ipsilateral (a) and contralateral cortex (b) of 90-dpi grafted mouse brain. Images show robust integration of GFP+ organoids and very large numbers of axons extending into the cortical regions and corpus callosum (CC) of the host brain. (c) Single focal plane confocal image of immunostaining for GFP, human-specific Synaptophysin (hSyn), and PSD95 in the host cortex at 90 dpi. Colocalization (arrowheads) and close association of hSyn and PSD95 indicate synaptic connectivity between the organoid axons and the host brain. Right panels show magnification of the boxed region in the left panel. (d) GFP signal with Z-stack 3D reconstruction of hSyn and colocalized only PSD95 puncta. (e) Quantification of the number of hSyn, PSD95, and hSyn/PSD95 colocalized puncta normalized to area and obtained from ipsilateral host cortical regions of 90-dpi grafted mouse brains. Data are presented as mean ± s.e.m. n = 3 grafted animals. Nuclei were counterstained with DAPI. Scale bars: 50 μm in a,b and 5 μm in c,d.
Examination of synaptic connectivity between human axons and the host brain at 90 dpi showed a dense network of human presynaptic marker Synaptophysin (hSyn)-positive structures colocalized with GFP+ fibers in the host cortex (Fig. 3c), indicating maturation and, most likely, functionality of graft axons. Co-staining with PSD95, which recognizes human and mouse synapses, identified individual synapses between graft axons and the host brain (Fig. 3c–e), suggesting that human axons established synaptic connectivity with neurons in the host brain.
Vascularization and high viability of engrafted organoids
We next used a cranial glass window, which facilitated optical tracing of blood vessel growth, to test whether grafted organoids had a functional vascularization system (Fig. 4a). Blood vessels started to invade the graft area by 7–10 dpi and became extensively vascularized by 14 dpi (Fig. 4b), with successful vascularization occurring in 85.4% ± 6.4 of grafted organoids (Supplementary Fig. 5c). Vascularization was essential for graft survival, as organoid implants that failed to vascularize displayed little or no fluorescent signal (data not shown). Immunostaining for the human and mouse endothelial cell markers Endoglin and CD31 demonstrated the growth of blood vessels in the grafts at 14 and 90 dpi, but not at 5 dpi, suggesting that organoid vascularization occurred between 5 and 14 dpi (Fig. 4c and Supplementary Fig. 5a). Moreover, CD31+ blood vessels did not coincide with staining for human-specific CD31, indicating that the vascular network in the grafts was derived from the host (Supplementary Fig. 5b). These results demonstrate growth of the vascular network in grafts and suggest that successful vascularization is essential for graft survival.
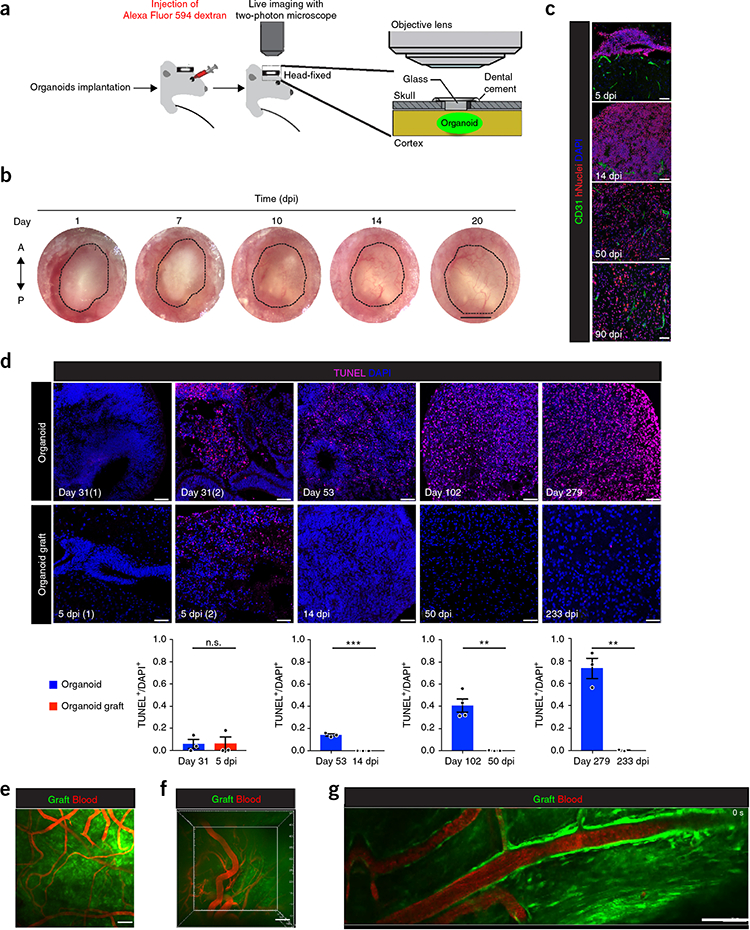
Functional vasculature and decreased apoptosis in grafts. (a) Illustration of the imaging approach for live two-photon microscopy imaging of the organoid graft. (b) Serial macroscopic tracking of grafts showing dynamics of blood vessel perfusion by the recipient vascular system. Dotted area indicates the graft. (c) Grafts co-immunostained for the endothelial markers CD31 and hNuclei at the indicated time points. (d) Organoids have an elevated degree of cell death that is rescued after grafting. Top, TUNEL staining of grafted organoids and stage-matched cultured organoids at the indicated stage. Left panels show staining obtained from two different organoids and organoid grafts at day 31 and 5 dpi, respectively. Bottom, quantification of TUNEL+/DAPI+ cells in grafted organoids and nearly stage-matched cultured organoids of indicated ages. Values are represented as mean ± s.e.m., (n = 3, except for 102-d organoid, n = 4, and 233 dpi n = 2); unpaired two-tailed t-test was used to compare mean difference between each group. Day 31 vs. 5 dpi (t = 0.03656, df = 4, P = 0.9726, not significant), day 53 vs. 14 dpi (t = 14.67, df = 4, P = 0.0001), day 102 vs. 50 dpi (t = 5.943, df = 5, P = 0.0019), and day 279 vs. 233 dpi (t = 6.267, df = 3, P = 0.0082). (e–g) In vivo two-photon imaging of blood vessels via dextran infusion as viewed through the cranial window. Organoids were implanted and TexasRed-dextran was injected at different time points of post-implantation (30 dpi in e, 120 dpi in f). (e) maximum projection of a 300-μm stack taken in a 30-dpi graft (Supplementary Video 2). (f) top view of a three-dimensional reconstruction of a 500-μm Z section in the organoids from a 120-dpi grafted animal (Supplementary Video 3). (g) Single z-plane obtained from 120-dpi graft and acquired at 141-μm depth below the organoid surface, showing blood flow in the vasculature network (Supplementary Video 4). Nuclei were counterstained with DAPI. Scale bars: 1 mm in b, 50 μm in c,d, and 100 μm in e–g. **P < 0.01, ***P < 0.001.
We next sought to understand how the in vivo brain environment and vascularization affected the growth and viability of organoids. Surface size tracing from day 0 to day 14 showed an overall reduction in graft size before vascularization, followed by a relative increase in the graft surface area that coincided with the timing of vascularization (Supplementary Fig. 5d,e). We compared cell death in the graft before and after vascularization to that of nearly stage-matched organoids cultured in vitro. In cultured organoids, apoptotic cell death was widespread at day 279 compared to day 31 (Fig. 4d). In contrast, grafts showed either limited or no detectable cell death at 14, 50, and 233 dpi in vivo. Notably, we observed low, yet variable, apoptotic signal in 31-d cultured organoids and at 5 dpi before overt vascularization occurred (two out of three of the grafts exhibited no detectable TUNEL, and one had a low TUNEL signal). These findings suggest that grafting and host-mediated vascularization promote viability of cerebral organoids that would otherwise display progressive cell death in vitro.
We studied whether implanted organoids could be imaged in awake grafted mice using in vivo two-photon scanning fluorescence microscopy. We imaged a clear GFP signal up to 500 μm in depth (Supplementary Fig. 5f and Supplementary Video 1). After infusing the grafted animal intravenously with a high-molecular-weight dextran dye, we imaged the graft under a two-photon microscope through a cranial window (Fig. 4a). Simultaneous live imaging of dextran and GFP showed blood vessel growth with active blood flow, demonstrating the development of functional vasculature networks inside the grafts (Fig. 4e–g, Supplementary Fig. 5g and Supplementary Videos 2 and 3).
Two-photon imaging of neuronal activity in engrafted organoids
We explored the functionality of grafted organoids by longitudinally measuring neuronal network activity in vivo using two-photon calcium imaging. Grafts displaying good GFP fluorescence were injected with an AAV8-CaMKII-jRGECO1a viral vector36, which allows specific expression of a red fluorescent calcium sensor mainly in excitatory neurons (Fig. 5a). After allowing at least a 2-week period for protein expression, we acquired several recordings of calcium activity from awake animals (Fig. 5b and Supplementary Videos 4,5,6). Imaging data were analyzed by calculating relative changes in fluorescence (ΔF/F) in regions of interest corresponding to neurons in the grafts. Grafted organoids had robust, recurrent, spontaneous rhythmic calcium transients interspersed with periods of no activity. The activity of the neuropil and cells was highly synchronized, and all identified cells in each field of view took part in every burst (Fig. 5c,d). We imaged engrafted animals at nearly 1-month intervals and were able to identify the same population of neurons over different imaging sessions. Spontaneous transient elevations of intracellular calcium were detected from the same regions and, in some cases, the same single neurons after more than 3 months of imaging (Fig. 5e,f). In summary, our in vivo two-photon calcium imaging demonstrated synchronized neural activity in the organoid grafted neurons.
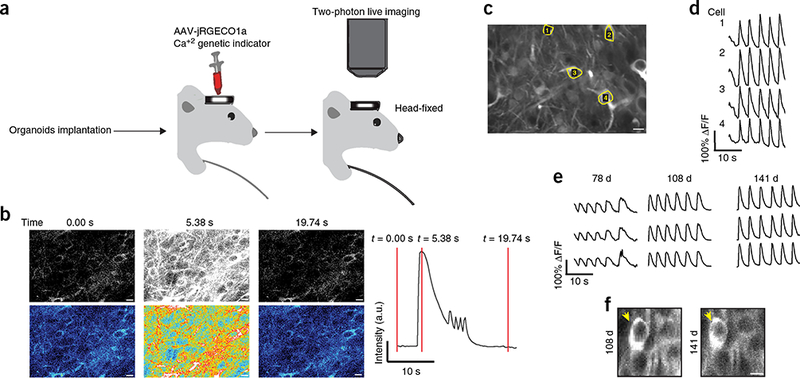
Two-photon imaging reveals neuronal activity in the graft. (a) illustration of our imaging system for Ca2+ imaging to monitor neuronal activity of the organoid graft using a two-photon microscope. (b) Representative images (from three frames) of two-photon Ca2+ imaging shown in grayscale (upper) and pseudo-color (lower). The images illustrate epochs of low (left and right panels) and high (middle) Ca2+ concentration in the graft during a Ca2+ transient at 78 dpi. The fluorescence intensity of the calcium sensor is markedly higher during periods of neuronal activity. Graph is a plot of full-frame average intensity during a Ca2+ transient, and red lines indicate time points corresponding to the frames in the left panels. (c) Typical field of view of neurons expressing jRGECO1a in the graft at 168 dpi. (d) Representative ΔF/F calcium traces of cells from the field of view depicted in c. Note the rhythmic and perfectly synchronized activity. (e) Representative ΔF/F traces from the same mouse at different time points after implantation. (f) The same cells in the graft can be imaged over different imaging sessions. n = 3 grafted animals in b–e, n = 1 grafted animal in f. Scale bar, 10 μm in b,c.
Electrophysiological recording of neuronal activity in engrafted organoids
To determine whether organoid grafts exhibited electrophysiological activity, we conducted extracellular recording using multielectrode arrays37 at various time points after grafting (Fig. 6a–e and Supplementary Figs. 6 and 7). For example, we successfully recorded action potentials (spikes) from two sites of the mid-stage 50-dpi-grafted organoids at different dorsal-ventral depths (DV) (Supplementary Fig. 6a–c) and from four sites of the late-stage 115-dpi graft (Fig. 6d,e). In contrast to the late-stage graft, which displayed large numbers of isolated active neurons (three to eight neurons per recording site) at multiple depths, both low numbers of firing neurons (two to five neurons per recording site) and locations were observed in the mid-stage graft (Fig. 6d,e and Supplementary Fig. 6a–c). This observation indicates relatively advanced maturity at 115 dpi and is consistent with the temporal progressive neuronal maturation we observed in grafts (Fig. 2). Firing rates of these neurons gradually increased after removal of isoflurane anesthesia and sharply decreased with reintroduction, indicating suppression of neuronal activity by anesthesia. Thus, our data demonstrate neuronal activity and state-dependent firing changes of neurons in organoids that respond to external environmental stimuli.
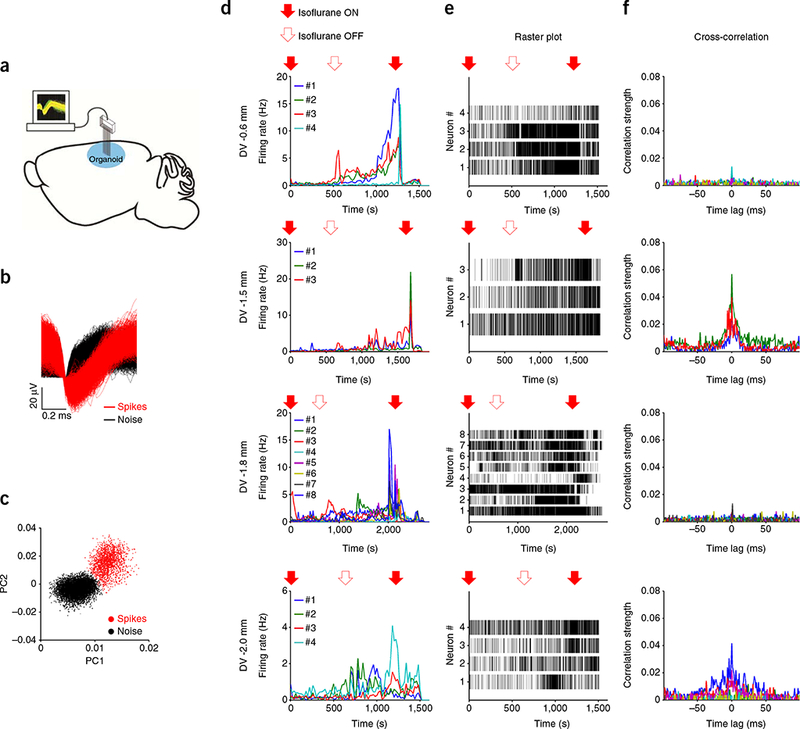
In vivo electrophysiological recording in the graft. (a) Illustration of in vivo multielectrode array recording of the organoid graft. (b) Waveforms of action potentials (red) from neuron 3 shown in d,e, third row (DV −1.8 mm). (c) Principal component analysis (PCA) of waveforms from the same neuron in b. (d) Firing rate changes of single neurons (color-coded) obtained from a 115-dpi grafted organoid at four different DV depths (from top to bottom: −0.6 mm, −1.5 mm, −1.8 mm, and −2.0 mm) from the organoid surface. Each line (color-coded) indicates the firing rate of an individual neuron. Arrows on the top denote the time isoflurane was turned ON (filled arrows) and OFF (empty arrows). (e) Spike raster plots from the neuronsin d at four different depths (from top to bottom: −0.6 mm, −1.5 mm, −1.8 mm, and −2.0 mm). Each vertical bar indicates a single spike. (f) Cross-correlation of neuron pairs shown in d. At depths of −1.5 mm and −2.0 mm, neuron pairs show strong correlated activity. n = 3 grafted animals.
To assess neuronal activity and interactions among neurons at the population level, we calculated the cross-correlation for neuron pairs in the same recording site. While neuron pairs in 50-dpi grafts showed little or no correlated activities, neuron pairs in 115-dpi organoids at depths of −1.5 mm and −2.0 mm showed strong correlation within ±50 ms, suggesting synchronized firings of neurons in the late stage (Fig. 6f and Supplementary Fig. 6c). Overall, these results showed that late-stage grafts were accompanied by a more active and coordinated neuronal network than early-stage grafts, suggesting functional maturation at the late stage of grafted organoids.
Optogenetics revealed graft-to-host functional connectivity
To investigate functional connectivity between organoid neurons and the host brain, we established optogenetic control of grafts by injecting a tdTomato+ graft with AAV expressing Channelrhodopsin2-EYFP under the control of human synapsin promoter (AAV9-ChR2-eYFP). ChR2::YFP was expressed in both the grafts and their tdTomato+ axonal projections (Supplementary Fig. 8a,b). To functionally verify ChR2 expression, we activated a ChR2-transduced graft, or uninjected negative control, while simultaneously recording its activity using in vivo extracellular recording. An electrode array bundled with optic fiber was implanted in the graft, and the graft was stimulated with blue laser while recording simultaneously. One sample neuron showed robust laser-evoked responses to three stimulation patterns as 1 s constant light, 50 Hz, and 20 Hz; no responses were observed in the uninjected control (Supplementary Fig. 9a–d). The response latency to laser onset was less than 5 ms and no significant response change was observed before or after the period of light illumination. These results indicated that graft neurons expressing ChR2 could be optogenetically activated by laser stimulation.
Next, we stimulated the ChR2-expressing graft while recording in host brain regions to search for potential neural activity changes in the host (Supplementary Fig. 10). We succeeded in recording local field potential (LFP) changes in host brain (fiber location: DV −1.7 mm; array location: AP −2.54 mm, ML −1.5 mm, DV −2.2 mm) (Supplementary Fig. 10e–h). In this example, the graft was activated with a 20-Hz stimulation pattern for 1 s, and laser onset was at 0 s. LFP signal from one electrode showed a negative peak right after laser onset (Supplementary Fig. 10e). The peak latency from laser onset was 20 ms (Supplementary Fig. 10e, inset). This peak was followed by 20-Hz oscillation lasting for 1 s (Supplementary Fig. 10e,f). These results showed that this host brain region received excitatory inputs when stimulating the graft, which indicated functional connectivity from the graft to the host brain. Notably, another electrode in the same brain region detected a positive peak after laser onset, accompanied by a 20-Hz oscillation (Supplementary Fig. 10g,h). The response latency to laser onset was 20 ms (Supplementary Fig. 10g, inset). No responses were observed when recording from either a more dorsal position in the same animal or from a similar array position in the uninjected control (Supplementary Fig. 10a–c, i–k). This inhibition observed in the host brain region evoked by stimulation was likely caused by integration through the local neural network, which again strongly supports the existence of functional connectivity between the graft and the host. Taken together, these results demonstrate the utility of optogenetics in dissecting functional interaction between the organoid and the host brain and suggest functional integration of the organoids’ neurons in host synaptic circuits.
In light of this finding of functional integration, we examined the behavior of grafted animals. We tested the spatial learning abilities of grafted mice and age-matched, NOD-SCID ungrafted controls using the Barnes maze. During training, no group differences were observed in the latency to locate the target location; nor was a group difference observed in the latency to locate the target during the probe test (Supplementary Fig. 11a,b). Analysis of errors made during training revealed that, although grafted mice made fewer errors on training day 1, this effect disappeared by day 2 (Supplementary Fig. 11c). However, the pattern of search errors during the probe trial did differ by group, with NOD-SCID controls searching primarily in the area adjacent to the target location whereas grafted mice showed little spatial preference for the target (Supplementary Fig. 11d–f). Together, these data suggest that both groups showed improvement with training, and no significant difference in learning ability was detected between grafted and ungrafted animals; however, mice with organoid implants did not perform as well as controls when tested for spatial memory, possibly due to the lesion made for organoid implants.
DISCUSSION
Brain organoids hold great potential for the investigation of aspects of human brain development and mental disorders. A limitation of organoid systems, however, is that they lack a vascular system and the complex physiological context of the intact human brain, such as, the interactions among cells, tissue, organ, and organism. In this study, we demonstrate that human brain organoids are amenable to transplantation in the rodent brain, showing integration, viability, long-term survival, vascularization, functional neuronal activity, and synaptic connectivity of the grafted organoid and the host. We also establish research tools that enable recording and optogenetic manipulation of neuronal activity with high spatial and temporal resolution within brain–organoid chimeras.
In the 1970s, transplantation of solid rodent tissue intracerebrally on a rich vascularized surface overlaying the anterior colliculus enabled a high degree of revascularization and survival of the implant21,31,32. We adapted this classic knowledge for transplantation of brain organoids derived from hESCs. Our results show extensive infiltration of the host vasculature within a few days post-transplantation, with no sign of human endothelial cells in the graft (Fig. 4 and Supplementary Fig. 5a). Although the majority of grafted organoids survived transplants that did not undergo vascularization failed to survive. Previous reports have demonstrated engraftment of several organoid systems in vasculature-rich areas such as the kidney capsule or colon23,24,26; however, successful engraftment and maturation of human intestinal tissues required the presence of a mesenchymal niche, and lack of a mesenchymal component resulted in failure of engraftment24. Moreover, while liver organoids were transplanted in the brain with quite high success25, co-culture with endothelial cells was required for vascularization and engraftment, highlighting the importance of the vascular bed. It is reasonable to assume that the survival of our organoid grafts was also promoted by adjacency to the brain ventricle, which allows circulation of cerebrospinal fluid through the graft cavity. We conclude that a vascular bed region in the brain aids successful engraftment of organoids, without the need for mesenchymal and endothelial cells.
Our data show that transplantation of cerebral organoids results in integration, progressive maturation and neuronal differentiation, synaptogenesis in the graft, and gliogenesis, thus sharing features with the dynamics of normal development and neurogenesis, and is consistent with organoids cultured in vitro. Transplanted embryonic neurons have been shown to integrate with great specificity into neocortical circuits in the adult brain38. We observed human axonal growth to distant targets and various regions in the host brain, and evidence of functional synaptic connectivity between the graft and the host brain. Further research is needed to address questions about the specificity of the axonal projections and the precise synaptogenesis mapping.
Although they express mature markers and contain functional synapses, in vitro organoids generated with current systems appear to represent an embryonic developmental stage3,8,34. Our Ca2+ imaging data demonstrated that the neurons in grafted organoids have synchronized activity, rather than sparse, isolated activity, suggesting an active neuronal network in the graft. Moreover, a study of the pattern of neuronal activity in mouse cortex showed that, as early as postnatal day 5, the activity was highly synchronous in local clusters of neurons39. At the end of the second postnatal week, neocortical networks underwent a transition to a much more desynchronized state that lacked a clear spatial structure39. Our observation of a similar pattern of Ca2+ imaging and neuronal activity in the grafted organoids suggests that, at least from a functional perspective, the organoids resemble late embryonic or early postnatal tissue. On the other hand, our extracellular multielectrode recordings, which have higher temporal resolution than Ca2+ imaging, indicated both higher numbers of active neurons and a partial switch from non-correlative to correlative neuronal activity during the course of transplantation, suggesting progressive neuronal maturation and generation of functional neuronal networks in the graft (Fig. 6 and Supplementary Fig. 6). A recent study found a similar progressive functional maturation and emergence of a neuronal network in cultured cerebral organoids using extracellular multielectrode recording13. Determination of whether organoid transplantation enhances functional maturation of neural circuits compared to cultured organoids, and of the corresponding human development time point(s) of grafted organoids, requires further research. In addition, it would be interesting to test whether transplantation of other types of brain organoids, or further modifications, could promote full maturation.
We expect the experimental system presented here to serve as a useful tool for a number of biomedical applications. Transplantation has been used to study the survival, integration, axonal projection pattern, and synaptic connectivity of embryonic and hPSC-derived neurons cultured in two-dimensional (2D)38,40–44, and to investigate the feasibility of using hPSC-derived neurons to treat central nervous system disorders1,45–50. However, the inability of these 2D models to recapitulate the in vivo-like cytoarchitectural organization and synaptic connections of the brain has impeded their use in precise disease modeling or drug screening applications.
In contrast to neural 2D culture, cultured organoids recapitulate several key features of human brain organogenesis and cytoarchitecture, making them attractive models for studies of certain aspects of brain development and disease3,10,15,19,51. However, in vitro studies are limited for investigating cell–tissue interactions, patterns of axonal projections, synaptic connectivity, and cross-interaction with the brain immune system. Transplantation of brain organoids could enable assessment of differentiation, axonal growth, and synaptic integration of neural organoids derived from patient-specific iPSCs under physiological conditions in vivo, facilitating mechanistic studies into the pathogenesis of neurodevelopmental, neuropsychiatric, and neurodegenerative disorders. Moreover, it could elucidate how neural progenitors and/or mature neural cells interact with their endogenous environments and vascular niches.
Preclinical animal models generally have a poor track record for accurately predicting which drugs will work in humans, as up to 80% of drugs that pass preclinical tests fail in humans52,53. Our approach might be helpful for preclinical drug testing of patient-derived brain organoids under physiological conditions.
Finally, a wide variety of 3D organoid systems have been transplanted in vivo24–30,54,55, in some cases to repair and rescue tissue damage23,25,55–57, indicating that organoids may have potential for cell therapy. Our finding that human brain organoids can integrate and form functional circuits in the mouse brain suggests that organoids could provide an alternative to pure populations of a particular cell type, especially for the treatment of complex brain disorders or injuries. A central challenge will be to determine whether brain organoids can restore specific lost, degenerated, or damaged brain regions upon integration.
METHODS
Methods, including statements of data availability and any associated accession codes and references, are available in the online version of the paper.
ONLINE METHODS
Animals
All animal experiments described in this study were approved by the Institutional Animal Care and Use Committee (IACUC) at the Salk Institute for Biological Studies (12–00022), and the University of California San Diego (S12201), and were conducted in accordance with the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals. Male and female mice were group-housed when possible with up to five mice per cage, under standard conditions, and kept on a 12 h light/dark cycle (lights on at 6:00 a.m.) with ad libitum access to food and water. Immune-deficient NOD-SCID mice, aged 5–6 weeks, were purchased from Jackson Laboratories (JAX Stock: 001303) or were generated in our lab by breeding. In the majority of the experiments, female mice 6–10 weeks of age were used and the feasibility of the approach was validated in males.
Human embryonic stem cell culture.
Human embryonic stem cells (hESCs) H9 (Wisconsin International Stem Cell (WISC) Bank, WiCell Research Institute, WA09 cells)58 were cultured according to WiCell protocols (www.wicell.org/home/support/stem-cell-protocols/stem-cell-protocols.cmsx) on MEF feeders (Applied StemCell, Cat ASF-1114) in hESC medium containing DMEM/F12, 20% knockout serum replacement, 1% non-essential amino acids, 2 mM GlutaMAX, 0.1% (55 μM) 2-Mercaptoethanol (all from Invitrogen) and 10 ng/ml bFGF (Humanzyme), or on Matrigel (Corning, hESC-Qualified) in mTeSR1 (homemade by Salk STEM Core). Cultures were maintained in 6-well tissue culture plates and passaged at a ratio of 1:3 or 1:6 every 4–6 d by 1 mg/ml collagenase type IV (Invitrogen) for 30 min at 37 °C and mechanical scraping with a 5-ml glass pipette. All human pluripotent stem cells were maintained below passage 50 and confirmed negative for mycoplasma. Cells were confirmed to be karyo-typically normal. All experiments involving cells from human subjects were performed in compliance with the institutional Embryonic Stem Cell Research Oversight (ESCRO) committee.
Generation of cerebral organoids.
Cerebral organoids were generated and processed for analysis as described previously8,33 with minor modification. Briefly, hESCs were washed with Dulbecco’s phosphate-buffered saline (D-PBS, Invitrogen) and dissociated with EDTA followed by Accutase to generate single cells. A total of 1 × 104 cells were then seeded into each well of an ultra-low-attachment 96-well plate (Nunc) to form single embryoid bodies (EBs) in medium containing DMEM/F12, 20% KSR, 3% FBS, 2 mM GlutaMAX, 1% non-essential amino acids, 50 nM b-mercaptoethanol and 4 ng/ml bFGF. ROCK inhibitor Y27632 (40 uM) was included for the first 24 h. EBs were maintained in 96-well plates for 2 d, and then the medium was changed to the same medium without FGF, and the cells were incubated for another 3–4 d. Medium then was changed to neural induction medium (NIM) consisting of DMEM/F12, 1× N2 supplement, 1% non-essential amino acids, 2 mM GlutaMAX and 1 μg/ml heparin (Sigma). On days 10–12, EBs were embedded in 30-μl droplets of Matrigel and were allowed to gel at 37 °C for 20–30 min. Embedded EBs were subsequently cultured in cerebral differentiation medium (CDM) consisting of 50% DMEM/F12, 50% Neurobasal, 0.5× N2 supplement, 1× B27 (without RA) supplement, 2 mM GlutaMAX, 2.8 ng/ml human insulin (Sigma), 0.5% non-essential amino acids, and 55 μM b-mercaptoethanol. Droplets were cultured in a stationary condition in 6-cm suspension culture dishes for 4 d, and then transferred to spinning flasks (Corning) containing culture with CDM +RA media. A total of ~40 organoids were added to each spinning flask, and medium was changed once every week or when the medium turned dark yellow, for the duration of the culture process. Matrigel was removed mechanically at day 20–22. For some experiments, an orbital shaker (65–80 r.p.m.) was used instead of the spinning flask. Only organoids that passed quality control—namely, clearing of embryoid body borders, formation of radially organized neuroepithelium before Matrigel embedding, and outgrowth and development of defined buds in Matrigel without massive cyst formation or premature differentiation—were selected for transplantation. In addition, organoids were examined by immunostaining to confirm the generation of polarized ventricle zone-like regions and dorsal forebrain identity in the organoids batch.
Immunofluorescence staining and histological processing.
For organoids, residual Matrigel was manually removed from organoids and three washes were performed by placing organoids into mesh inserts in wells containing PBS. Organoids were then fixed in 4% PFA in 0.1 M phosphate buffer (PB, pH 7.2–7.5) with gentle shaking on ice, or at 4 °C, for 45 – 60 min; they were then washed three times in PBS and cryoprotected in 15% sucrose in PB for 4–5 h followed by 30% sucrose overnight at 4 °C. Three to five organoids then were embedded together in blocks in tissue-freezing medium (General Data), frozen on dry ice, and stored at −70 °C. 20-μm cryosections of organoids were obtained using a cryostat (Leica), mounted on Superfrost plus slides (Thermo Scientific, Menzel-Glaser), dried at room temperature (RT) for 30 min and stored up to 12 months at −70 °C. For each section set, every two sections were collected onto a slide to obtain a total of ten slides. This collection method was repeated with the same ten slides until 6–8 sections were mounted per slide and an entire serially sectioned collection set was obtained representing the whole organoid. Slides were dried for 30 min at RT and stored at −80 °C.
Immunofluorescence staining was performed as previously described59. Slides were thawed, dried for 5–10 min in a 37 °C incubator, and rehydrated in PBS for 5 min, and blocked and permeabilized in blocking solution (5% normal donkey serum, 0.1% Triton X-100 in PBS). The tissue area and slide edges were outlined using a hydrophobic PAP pen, then slides were incubated with ~500 ml blocking/permeabilization solution (0.1% Triton-X, 5% normal donkey serum in PBS) in a humidified chamber for 1 h at RT. Slides were incubated in the appropriate primary antibody diluted in blocking solution under parafilm at 4 °C overnight. Sections were washed three times (5 min each) in PBT (0.05% Tween in PBS), incubated with appropriate fluorochrome-conjugated secondary antibodies diluted in blocking solution at RT for 1 h in the dark, washed twice in PBS, counterstained with DAPI (1 μg/ml, D9542; Sigma) for 5 min, rinsed twice in PBS and mounted with Shandon Immu-Mount (Thermo Scientific, 9990412) ProLong gold for high-magnification imaging (Invitrogen, P36934). All images were collected on a Zeiss LSM880 confocal microscope and processed with Zen software, ImageJ software (NIH), and Adobe Photoshop CS4 (Adobe Systems, San Jose, CA). A list of the primary and secondary antibodies used in this study is provided as Supplementary Table 1. Some primary antibodies were used with two different dilutions for either tissue (T) or organoids (O). Cell death was detected using TUNEL assay (C10247, Invitrogen) following the manufacturer’s protocol.
Mouse brain tissue collection.
Mice were anesthetized with a lethal dose of ketamine and xylazine (130 mg/kg, 15 mg/kg; i.p.) and perfused transcardially with 0.9% NaCl or with 0.1 M PBS followed by fresh 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer (pH 7.2–7.5). Brains were dissected and postfixed in 4% PFA overnight. Brains were cryoprotected for 72 h in 30% sucrose with 0.05% NaN3 at 4 °C. Brains were sectioned coronally on a cryostat at 20 mm thickness into 0.1 M PB and processed for immunostaining as described above.
Lentivirus production and generation of GFP hESCs.
pCSC-CAG-GFP and pBOB-CAG-tdTomato were obtained from the lab of Inder Verma at the Salk Institute (Addgene 12337). VSVG-coated lentivirus particles were produced in HEK-293T cells. Briefly, HEK293 cells were transfected with a mixture of lentiviral construct and third-generation packaging plasmids using Polyethylenimine (PEI) (Cat 23966, Polysciences, Inc.). Culture medium was changed 4–5 h after and collected 72 h after transfection. Virus-containing medium was filtered through 0.45-μm filter concentrated via ultracentrifugation and stored at high concentration at −80 °C before usage.
To transduce hESCs with lentivirus, hESCs cultured on Matrigel were first dissociated using collagenase type IV and further dissociated by drawing small aggregates in and out of pipette briefly. hESCs were plated as small colonies onto Matrigel-coated dishes and fed mTeSR medium containing 10 μM ROCK inhibitor Y27632; the next day lentivirus particles were included in the media. When 80–90% confluency was reached, hESCs were subsequently FACS-sorted for GFP expression and replated at high density on Matrigel-coated dishes and fed mTeSR medium containing 10-μM ROCK inhibitor Y27632. Expression of lentiviral transgenes was evaluated using GFP fluorescence.
AAV viral injection to the organoid graft.
AAV8-CaMKII-JRGECO1a36 and AAV8-CaMKII-GCaMP6M (titer was 3.89 × 1012 GC/mL) were obtained from the Salk Virus Core. Mice were anesthetized with isoflurane (1–2% via a nose cone) and placed in a stereotaxic frame for virus injection. A ~0.5- to 1-mm hole was drilled through the cranial window and virus solution was delivered through stereotaxic surgery to the grafted organoids at three different Z locations using a microinjector (Nanoject II, Drummond Science). The mice were returned to their cages and allowed to recover. Mice were kept for at least 10 d before imaging.
Dextran injection.
For dextran injection, Texas-Red 70,000 MW Dextran (D1830, Invitrogen) was diluted to 5% w/v (25 mg in 0.5 ml saline 0.9 NaCl). The mice were anesthetized and 50 μl dextran was injected retro-orbitally using a 27-G needle. Mice were imaged immediately.
Surgical procedures: intracerebral implantation of cerebral organoids.
Cerebral organoids were cultured for 40–50 d, or 31 d for 5- and 40-dpi experiments, before implantation. Only organoids that passed the quality control criteria mentioned above were selected. Mice were anesthetized with 5% (induction) and 1–2% (maintenance) isoflurane in oxygen. Dexamethazone (2.5 mg/kg) was injected subcutaneously to minimize edema. Each animal was fixed in a stereotactic frame and body temperature was maintained at 37 °C using a water-circulation heating pad (Gaymar Industries). The skin above the skull was cut with a small incision. A ~3-mm diameter craniotomy was performed by drilling into the skull; the underlying dura mater was removed and a cavity was first made by aspiration with a blunt-tip needle attached to a vacuum line. The aspirative lesion was made unilaterally in the region of the retrosplenial cortex by removing the tissue overlaying the anterior colliculus. The vascular bed of the choroidal fissure was exposed and the transplant was placed on the pial vessels overlying the collicles just caudal to the hippocampus. Sterile saline was used to irrigate the lesion and keep it free of blood throughout the surgery, and a piece of Gelfoam (Pfizer) was used to slow the bleeding and absorb the excess blood. The implanted organoid was then covered with a 5-mm cover slip to create a cranial window and was sealed with adhesive glue as described previously60,61. The wound was closed with sutures or, alternatively, the implant was held in place with dental cement and a small titanium bar (9.5 × 3.1 × 1.3 mm) was attached to the skull to secure the animal to the microscope stage. Following completion of the surgery, carprofen (5 mg/kg, i.p., 100 μl of 1 mg/ml) and buprenorphine (0.1 mg/kg, subcutaneous 100 μl 0.01 mg/ml) were administrated for inflammation and analgesic relief. The mice were then allowed to recover in a temperature-controlled cage and were returned to their home cages. Only brains with clearly identifiable GFP+ organoid grafts (indicating graft survival) were subsequently analyzed.
Two- photon imaging.
Morphology and blood flow imaging were done under isoflurane anesthesia (1% isoflurane in O2, vol/vol) and mice were head-fixed to the microscope stage via a titanium bar implant while resting on a 37 °C electrical heat blanket (Harvard Instrument). For imaging of Ca2+ activity, mice were head-fixed to the microscope using a custom-made titanium implant and allowed to run on a cylindrical treadmill62.
All in vivo imaging was done with a 2-photon laser scanning microscope (MOM, Sutter Instruments) using a femtosecond-pulsed laser (Chameleon Ultra II, Coherent) tuned to 910-nm or a 1,055-nm femtosecond-pulsed laser (Fidelity 2, Coherent) and a 16× water immersion objective (0.8 NA, Nikon). Images were acquired using the ScanImage software63, which was written in MATLAB (MathWorks). Calcium imaged data were acquired at 3.9 Hz (512 × 128 pixels) and analyzed in MATLAB by extracting fluorescent intensity data from regions-of-interest corresponding to visualized cells expressing the calcium sensor, as previously described39. Briefly, to correct for slight x-y drift, we aligned movies using a cross-correlation–based, subpixel image registration routine. Fluorescence traces were low-pass filtered and averaged to define a baseline level, and fluorescent changes normalized this baseline (ΔF/F)39. Timestamping of movies, AVI file conversion, and color look-up table selection were done using ImageJ Plots of average full-frame intensity were done in Matlab.
In vivo electrophysiology.
The in vivo neuronal recording procedure was performed as detailed before37. Briefly, before recording, mice were lightly anesthetized using isoflurane (4% induction; 1–2% sustained) and placed in a stereotactic frame. For electrophysiological recording, we used electrode arrays (Innovative Neurophysiology Inc.) of 16 tungsten contacts (2 × 8), with each electrode being 35 μm in diameter. Electrodes were spaced 150 μm apart in the same row and 200 μm apart between two rows. Total length of electrodes was 5 mm. The array was incrementally lowered into the organoid graft and silver grounding wire was attached to the scalp.
Neural activity was recorded using the MAP system (Plexon Inc., TX). The spike activities were initially sorted online with a build-in algorithm (Plexon Inc., TX). Only spikes with stereotypical waveforms clearly distinguished from noise and relatively high signal-to-noise ratio were tagged and saved for further analysis. After the recording session, the recorded spikes were further isolated into individual units by an offline sorting software (Offline Sorter, Plexon Inc.). Each individual unit displayed a clear refractory period in the inter-spike interval histogram, with no spikes during the refractory period (larger than 1.3 ms).
Spike sorting.
Recording processing was performed as described previously37,64. Briefly, the raw signals detected by a single electrode were first high-pass filtered with a cutoff of approximately 200–300 Hz, which yielded the continuous spike signal sampled at the same 40-kHz rate as the original signal using the OmniPlex Neural Data Acquisition System (Plexon Inc., TX). A sample section of the original signal is shown in Supplementary Figure 6f. Spikes with magnitude exceeding the threshold were tagged and recorded as waveforms (red arrows). Then we applied PCA analysis to these waveforms to discriminate the neuronal spikes from noise. On the PC plane, waveforms showing distinct shapes are classified into separate clusters.
Cross-correlation.
The strength of correlated firings between neurons was estimated using a cross-correlation function65. The cross-correlation function is defined as:
where It denotes the activity of neuron I at time t. Jt+t0 denotes the activity of neuron J at time t + t0. CIJ (t0) represents the correlation between neuron I and J at time lag t0. The temporal resolution of time lag was 1 ms. T is the total length of the spike train. In the calculation, the correlation strength was normalized against the firing rates of the neuron pair, which eliminated the influence of firing rate on correlation estimation.
Optogenetic stimulation.
For optogenetic stimulation experiments, 1 μl of concentrated AAV encoding Channelrhodopsin-2 (AAV9-ChR2-EYFP, titer 1.07 × 1013 GC/mL) was injected into the center of tdTomato-expressing organoid graft at 150 dpi. Two weeks later, the injected, and uninjected control, organoid graft was recorded using electrode arrays with an optic fiber directly attached. The fiber was ~200 μm from the electrode tips and capable of delivering laser light. Uninjected or injected organoid graft were stimulated by 473-nm laser (5 mW, Laserglow Technologies) with three stimulation patterns as 1 s constant light, 50 Hz (50 pulses in 1 s) and 20 Hz (20 pulses in 1 s). The bundle of array and fiber was slowly lowered down into the organoid. The depth was adjusted within the range of 0 to −2.0 mm for the optimization of placement. For the local field potential (LFP) recordings, the array and the optic fiber were placed in the mouse brain region and in the organoid graft, respectively (fiber location: DV −1.7 mm; array location: AP −2.54 mm, ML −1.5 mm, DV −2.2 mm) and the same stimulation patterns were delivered. Note that in extracellular recording, the negative and positive voltage change represent excitation and inhibition, respectively. All experiments were performed while animals were under anesthesia.
Barnes maze.
Organoid-implanted mice and age-matched NOD-SCID controls (~5-month-old animals;100–116 dpi) were tested for spatial learning ability using the Barnes maze (n = 7 mice per group). The maze was 91 cm in diameter with 20 holes spaced evenly around the perimeter, one of which led into a darkened escape box. Prior to training, all mice completed a single habituation trial. Mice were placed in an opaque cylinder in the center of the maze for 30 s to disorient them to spatial cues in the room; then the cylinder was removed and mice were gently guided to the escape location, where they remained for 2 min to familiarize them with the escape box. For training, all mice received three trials per day for 3 d. Each trial consisted of 30 s in the disorientation cylinder, followed by 3 min of time to locate the escape box. Mice that did not successfully escape within the time limit were gently guided to the correct location. After escaping, mice were left in the escape box for 20 s. The maze surface and escape box were cleaned with Sani-Cloth wipes after each trial. A probe test was performed on day 4, 24 h after the final training day. The escape box was covered, and mice were allowed to explore the maze for 60 s for a single trial before being removed. The latency to locate the correct location was recorded during both training and probe phases. Errors (head dips or nose pokes above incorrect locations) were scored by a blinded observer from video recordings.
Quantification and statistical analysis.
To determine the percentage of NeuN+ and SOX2+ cells in the organoid grafts, whole-brain sections of implanted brains stained with DAPI and the appropriate immunohistochemistry markers were imaged and used for quantification. The number of DAPI+ cells expressing various markers was manually counted using the “Cell Counter” plugin of Fiji. For each grafted brain, at least three regions of interest (ROI) were outlined using the “ROI tools.” The numbers of DAPI nuclei and cells expressing each marker from three sections and three grafting experiments per age group were included in the analysis. The cell counts from each grafted organoid were summed, and the percentage of DAPI+ cells expressing each marker was calculated. The percentage was averaged across three grafting experiments for each time point. Staining with DAPI and GFP was used to identify all cells and to determine the location of the graft. Moreover, human identity of the counted cells was confirmed by staining sequential sections with the hNuclei antibody.
Quantification of growth rate of the brain organoid after transplantation.
Brain organoids were monitored from time 0 through day 13 or 14 under the stereomicroscope. Images were collected every day, and organoid surface areas were measured using Fiji. Normalized total organoid surface area (normalized to day 0, 100%) was calculated and averaged from at least three experiments (day 0–5 n = 9, day 6 n = 8, days 7 and 8 n = 6, day 9 n = 5, day 13 n = 4, day 14 n = 3).
Quantification of NeuN and SOX2 for comparison between grafted and ungrafted organoids (Fig. 2) was performed using automatic particle counting in Fiji, by first setting the threshold to obtain a binary image and remove background, then the tissue was traced in the DAPI channel and particles number were measured for all channels. Cell counts were measured and averaged relative to DAPI.
Quantification of vascularization success rate was performed by macroscopic monitoring of blood vessel outgrowth after 14 dpi. The success rate was calculated by first averaging each experiment separately, and then average success rate from the ten independent experiments.
Barnes maze analysis.
Mean latency and errors for each mouse were analyzed in GraphPad Prism 7. Training performance was analyzed by two-way ANOVA, with group as a between-subject factor and day as a within-subject factor. Sidak’s test was used post hoc for multiple comparisons. Latency to locate the target during the probe trial was analyzed by the Mann–Whitney test. Tests were two-tailed.
Statistics.
Graphs and statistical analyses were generated using Prism 7 (GraphPad). For comparing multiple groups, a one-way ANOVA with post hoc Tukey’s test was used (Figs. 2d,e). Unpaired Student’s t-test (two-tailed) was used to compare two groups. Data are presented as a mean ± s.e.m. (standard error of mean). Experiments were not randomized, and the investigator was not blinded. The experiments were repeated at least three times for each experiment, unless stated otherwise.
Ethics statement.
All animal experiments described in this study were approved by the Institutional Animal Care and Use Committee (IACUC) at the Salk Institute for Biological Studies (12–00022), and the University of California San Diego (S12201), and were conducted in accordance with the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals. All experiments involving cells from human subjects were performed in compliance with the institutional Embryonic Stem Cell Research Oversight committee (ESCRO).
Life Sciences Reporting Summary.
Further information on experimental design is available in the Life Sciences Reporting Summary.
Data availability statement.
The authors declare that the data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Suppl
Suppl. 2
Suppl. 3
Video 1
Video 2
Video 3
Video 4
Video 5
Video 6
ACKNOWLEDGMENTS
We thank members of the Gage laboratory for helpful discussions; S. Schafer for the pCSC-CAG-GFP lentivirus and I. Verma for the pBOB-CAG-Td-Tomato construct. We also thank M. Shtrahman for assistance and two-photon imaging expertise, L. Moore, S. Baktvar, S. Kim, B. Miller, C. Lim, and I. Guimont for their technical assistance, M.L. Gage for editorial comments, V. Mertens for illustrations, I. Farhy-Tselnicker and J. Klug for technical advice, and M. Shtrahman, and T. Toda for critical reading and comments on the manuscripts. We thank U. Manor and the Waitt Advanced Biophotonics Core, K. Diffenderfer and the Salk Stem Cell Core, C. O’Connor and C. Fitzpatrick and the FACS Core, and the Salk Institute for generously providing critical infrastructural and financial support. We apologize to those whose work was not cited owing to space limitations. This work was supported by the NIH (U19 MH106434, U01 MH106882), The Paul G, Allen Family Foundation, Bob and Mary Jane Engman, The Leona M, and Harry B, Helmsley Charitable Trust Grant (2012-PG-MED), Annette C, Merle-Smith, The G, Harold and Leila Y, Mathers Foundation, JPB Foundation, Dolby Family Ventures for F.H.G. and NIH grants (R01NS083815, R01AG047669) for X.J. S.F. was funded by CIRM Bridges to Stem Cell Research Internship Program. A.A.M. received funding from the EMBO Postdoctoral Long-term Fellowship (ALTF 1214–2014, EMBO fellowship is co-funded also by the European Commission FP7-Marie Curie Actions, LTFCOFUND2013, GA-2013–609409), and is currently supported by the Human Frontiers Science Program (HFSP Long-Term Fellowship- LT001074/2015).
Footnotes
COMPETING INTERESTS
The author declares no competing interests.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nbt.4127
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc6331203?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/nbt.4127
Article citations
Organoids in the oral and maxillofacial region: present and future.
Int J Oral Sci, 16(1):61, 01 Nov 2024
Cited by: 0 articles | PMID: 39482304 | PMCID: PMC11528035
Review Free full text in Europe PMC
A tumorigenicity evaluation platform for cell therapies based on brain organoids.
Transl Neurodegener, 13(1):53, 29 Oct 2024
Cited by: 0 articles | PMID: 39472972 | PMCID: PMC11520457
Engineering human immune organoids for translational immunology.
Bioact Mater, 44:164-183, 18 Oct 2024
Cited by: 0 articles | PMID: 39474315 | PMCID: PMC11513670
Review Free full text in Europe PMC
Human midbrain organoids: a powerful tool for advanced Parkinson's disease modeling and therapy exploration.
NPJ Parkinsons Dis, 10(1):189, 20 Oct 2024
Cited by: 0 articles | PMID: 39428415 | PMCID: PMC11491477
Review Free full text in Europe PMC
A bibliometric analysis of cerebral small vessel disease.
Front Aging Neurosci, 16:1400844, 07 Oct 2024
Cited by: 0 articles | PMID: 39435188 | PMCID: PMC11492496
Go to all (550) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Brain organoids get vascularized.
Nat Biotechnol, 36(5):407-408, 01 May 2018
Cited by: 24 articles | PMID: 29734310
Bioengineering tissue morphogenesis and function in human neural organoids.
Semin Cell Dev Biol, 111:52-59, 12 Jun 2020
Cited by: 14 articles | PMID: 32540123 | PMCID: PMC8755418
Review Free full text in Europe PMC
Toward the formation of neural circuits in human brain organoids.
Curr Opin Cell Biol, 61:86-91, 16 Aug 2019
Cited by: 11 articles | PMID: 31425932
Review
Applications of brain organoids in neurodevelopment and neurological diseases.
J Biomed Sci, 28(1):30, 22 Apr 2021
Cited by: 34 articles | PMID: 33888112 | PMCID: PMC8063318
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIA NIH HHS (1)
Grant ID: RF1 AG047669
NIMH NIH HHS (2)
Grant ID: U19 MH106434
Grant ID: U01 MH106882
NINDS NIH HHS (1)
Grant ID: R01 NS083815





