Abstract
Free full text

Patient-iPSC-Derived Kidney Organoids Show Functional Validation of a Ciliopathic Renal Phenotype and Reveal Underlying Pathogenetic Mechanisms
Abstract
Despite the increasing diagnostic rate of genomic sequencing, the genetic basis of more than 50% of heritable kidney disease remains unresolved. Kidney organoids differentiated from induced pluripotent stem cells (iPSCs) of individuals affected by inherited renal disease represent a potential, but unvalidated, platform for the functional validation of novel gene variants and investigation of underlying pathogenetic mechanisms. In this study, trio whole-exome sequencing of a prospectively identified nephronophthisis (NPHP) proband and her parents identified compound-heterozygous variants in IFT140, a gene previously associated with NPHP-related ciliopathies. IFT140 plays a key role in retrograde intraflagellar transport, but the precise downstream cellular mechanisms responsible for disease presentation remain unknown. A one-step reprogramming and gene-editing protocol was used to derive both uncorrected proband iPSCs and isogenic gene-corrected iPSCs, which were differentiated to kidney organoids. Proband organoid tubules demonstrated shortened, club-shaped primary cilia, whereas gene correction rescued this phenotype. Differential expression analysis of epithelial cells isolated from organoids suggested downregulation of genes associated with apicobasal polarity, cell-cell junctions, and dynein motor assembly in proband epithelial cells. Matrigel cyst cultures confirmed a polarization defect in proband versus gene-corrected renal epithelium. As such, this study represents a “proof of concept” for using proband-derived iPSCs to model renal disease and illustrates dysfunctional cellular pathways beyond the primary cilium in the setting of IFT140 mutations, which are established for other NPHP genotypes.
Introduction
The kidney is a complex, multicellular organ affected by a wide spectrum of genetic diseases, including congenital abnormalities of the kidney and urinary tract (e.g., OMIM: 610805 and 143400), steroid-resistant nephrotic syndrome (e.g., OMIM: 256300 and 604766), autosomal-dominant polycystic kidney disease (ADPKD [OMIM: 173900]), tubular-electrolyte-processing disorders (e.g., Bartter syndrome [OMIM: 601678]), and stone-forming nephropathies (e.g., cystinuria [OMIM: 220100]). Single-gene disorders account for up to 10% of adult and 50% of pediatric end-stage kidney disease (ESKD).1, 2 The ability of next-generation sequencing techniques to achieve a genetic diagnosis varies significantly between disease groups.3, 4, 5 In addition, the identification of novel gene changes, referred to as variants of unknown significance (VUS), cannot provide a genetic diagnosis without some form of functional evaluation. Modeling of novel VUS has previously been performed in a variety of animal models, including gene knockout or knockdown in cell lines or model organisms from fly through zebrafish to mouse. However, such animal models are limited by interspecies variation in gene structure and function and an inability to account for the possibility of phenotypic variation arising from modifier genes that are present in the affected individual but not in the model organism. Primary cell lines derived from affected individuals overcome these limitations, but an in vitro, three-dimensional (3D), proband-derived renal cellular model has not been previously reported.
Recent advances in the directed differentiation of human iPSCs toward renal lineages could represent a novel disease-modeling platform for functional genomics.6 The generation of iPSCs from human skin or blood samples is now routine.7 Additionally, CRISPR/Cas9-mediated correction of candidate VUS enables the generation of isogenic control iPSC lines, the isolation of the effect of the VUS within the experimental comparison, and the removal of the confounding influences of genomic variation created by a non-isogenic control. In theory, the comparison of organoids generated from proband and matched gene-corrected iPSC lines should isolate the phenotypic effect of the proband’s specific allele. However, kidney organoids currently represent a model of the developing nephron, and the demonstration of a disease phenotype within a proband-derived organoid has not previously been demonstrated.
This study explores the utility of kidney organoids as a disease-modeling platform in the setting of prospective identification of a child with a heritable renal disease. The proband presented with a nephronophthisis (NPHP)-related ciliopathy (NPHP-RC) due to compound-heterozygous mutations in IFT140 (OMIM: 614620). NPHP is a progressive, fibrocystic renal disease histologically characterized by disruption of the tubular basement membrane, tubular dilatation and atrophy, interstitial fibrosis, and an inevitable decline to ESKD. Although rare (the estimated prevalence is 1 in 50,000–80,000 live births),8, 9 NPHP represents the most common monogenic cause of chronic kidney disease in the first three decades of life.10 In spite of the discovery of over 40 associated genes, up to 40% of individuals affected by NPHP-RC remain undiagnosed after targeted exome sequencing.5 Furthermore, the pathogenetic mechanism linking the cilium-centrosome complex to the disease phenotype is unresolved. Proposed mechanisms described in the literature vary and include but are not limited to disturbances in cell adhesion,11 cell polarity,12 cytoskeletal function,13, 14 Wnt signaling,15 Hippo signaling,16 centrosomal function,17 and DNA replication stress.18, 19
A common feature to all genes associated with NPHP-RC is their association with the primary cilium and/or centrosome.10 IFT140 is a core component of the IFT-A complex, which links ciliary maintenance proteins, signaling molecules, and transmembrane receptors to the dynein motor complex in order to descend the ciliary axoneme to the basal body, a process known as retrograde intraflagellar transport (IFT).20 IFT140 dysfunction results in a shortened cilium with an accumulation of IFT cargo distending the ciliary tip.21, 22 In addition, IFT proteins contribute to the selective importation of targeted proteins into ciliary cytoplasm.23, 24, 25 This includes G-protein-coupled receptors important in the negative regulation of hedgehog signaling.26 Beyond these observations, a biological pathway explaining human NPHP disease phenotype is not known.
By demonstrating an established cellular genotype-phenotype correlation within a proband-derived kidney organoid, this proof-of-concept study validates the use of kidney organoids for disease modeling and future ratification of novel VUS. A simultaneous reprogramming and CRISPR editing protocol27 was used to generate proband and gene-corrected isogenic control iPSCs from the skin fibroblasts of the individual affected by compound-heterozygous IFT140 variants (described above). Both iPSC lines were differentiated to kidney organoids. Within the tubular epithelium of unedited proband organoids, a classical ciliary morphology indicative of retrograde IFT dysfunction was identified. Gene-corrected organoids demonstrated a capacity to resolve this ciliary morphology, thereby verifying the genomic variant as disease causing. Additionally, transcriptional profiling and differential gene expression (DGE) analysis using proband-derived and gene-corrected organoids revealed dysfunctional pathogenetic pathways not previously described in IFT140-deficient disease models but established for other NPHP genes, suggesting that this model could clarify the common pathogenetic mechanisms for this heterogenetic rare disease.
Material and Methods
Genome Sequencing and Analysis
This research was conducted with approval from the human research ethics committees of the Lady Cilento Children’s Hospital (HREC/15/QRCH/126), the University of Queensland (Medical Research Ethics Committee approval 2014000453), and the Royal Brisbane and Women’s Hospital (HREC/14/QRBW/34), including research governance approval at all sites. Written informed consent was obtained from the study participants.
Genomic DNA was isolated from peripheral blood according to standard techniques. The exome of the proband and each of her parents was captured with the Nextera Rapid Capture Exome Kit (Illumina) and then sequenced on an Illumina HiSeq instrument with V4 chemistry (two 125 nt paired-end reads). Read alignment was performed with BWA-mem v.0.7.15; variants were called with the Real Time Genomics integrated analysis tool rtgFamily v.3.6.228 and annotated with SnpEff v.4.1k.29 In-house custom analysis pipelines were used to filter and prioritize variants for autosomal-recessive or de novo disease-causing variants on the basis of the clinical pedigree for the family as previously described.30 Segregation of the candidate variants was confirmed by Sanger sequencing.
Derivation and Correction of Proband iPSCs
Gene-corrected and uncorrected iPSC lines were generated from proband dermal fibroblasts according to a one-step reprogramming and gene-editing protocol as previously described.27, 31 Fibroblasts were harvested with TrypLE (Thermo Fisher) 2 days after passaging and resuspended in Buffer R (Thermo Fisher) at a final concentration of 1 × 107 cells/mL. 100 μL of the cell suspension was added to a tube containing reprogramming plasmids (pEP4E02SET2K, pEP4E02SEN2L, pEP4E02SEM2K, and pSimple-miR302/367), mRNA encoding SpCas9-Gem, a plasmid encoding a single guide RNA that overlaps the proband-specific variant (c.634G>A [p.Gly212Arg]), and a repair template plasmid carrying 942 bp (chr16: 1591716–1592658, UCSC Genome Browser hg38) corresponding to sequence within IFT140. The repair template included a 3 bp synonymous change to act as a Cas9-blocking mutation and to facilitate the identification of gene-corrected iPSC clones by allele-specific PCR. Electroporation was performed in a 100 μL tip with the following conditions: two pulses at 1,400 V for 20 ms. After electroporation, cells were plated on a 6-well Matrigel-coated (BD Biosciences) plate and maintained in fibroblast media until 4 days after transfection and then switched to Essential 7 medium (Essential 8 [E8] medium without TGFβ; Thermo Fisher) supplemented with 100 μM sodium butyrate and changed every other day as described previously.32 Sodium butyrate was removed from the media after the appearance of the first iPSC colonies at around day 10. Gene-corrected iPSCs were identified by allele-specific PCR and confirmed by Sanger sequencing. One gene-corrected clone and one uncorrected iPSC clone were selected and expanded for further analysis. Karyotype was confirmed by Infinium CoreExome-24 DNA microarray (Illumina), and cells tested negative for mycoplasma contamination. Pluripotency of iPSC lines was confirmed by flow cytometry after staining with antibodies to pluripotency markers (EPCAM, CD9, and SSEA4).
Cell Culture and Directed Differentiation to Kidney Organoids
Undifferentiated iPSCs were maintained in feeder-free conditions on human embryonic stem cell (hESC)-qualified Matrigel in E8 media. 1 day before differentiation, iPSCs were dissociated from 60%–70% confluence with TrypLE (Thermo Fisher) and plated at 3.5 × 104 cells/cm2 in a 6-well plate on hESC-qualified Matrigel (BD Biosciences) in 2.5 mL E8 media supplemented with Revitacell (Thermo Fisher). The following morning, cells were treated with 8 μM CHIR99021 in APEL2 basal media (STEMCELL Technologies) supplemented with 3.5% Protein Free Hybridoma Media 2 (PFHM2) and Antibiotic-Antimycotic (Life Technologies) for 4 days, followed by FGF9 (200 ng/mL) and heparin (1 μg/mL) for 3 days, during which media were changed every 2 days. At day 7, cells were dissociated with 0.05% trypsin, and 0.25 × 106 cells were spun three times at 300 × g in 1.5 mL tubes. With a large-bore P200 pipette tip, pellets were transferred to a Transwell 0.4 μm pore polyester membrane (CLS3450 Corning). Pellets were cultured over APEL2, 3.5% PFHM2, and Antibiotic-Antimycotic supplemented with 5 μM CHIR 99021 for 1 hr and then over FGF9 (200 ng/mL) and heparin (1 μg/mL) for 5 days. Transwell culture continued without added growth factors for up to 24 days, during which media where changed three times a week.
Immunofluorescence
Organoids were fixed at the desired time point with 2% paraformaldehyde for 20 mins on ice and washed three times with PBS. Organoids were incubated with 10% donkey serum and 0.3% Triton X and PBS overnight at 4°C. Primary antibodies were incubated for 48 hr in the blocking buffer at 4°C. After six washes with 0.3% Triton X and PBS, secondary antibodies were incubated overnight at 4°C. Where applicable, DAPI staining was applied at 1:1,000 for 3 hr. After four washes with PBS, organoids underwent immunofluorescent imaging with a Zeiss LSM 780 confocal microscope. For analysis of ciliary length, three images were captured from two separate experiments. 100 cilia per image were measured in ZEN (v.2.3, Zeiss), and statistical analyses were performed in GraphPad Prism with an unpaired t test and Welch’s correction for unequal variance.
Spheroid culture assays were fixed and stained according to a previously published protocol.33 Matrigel was dissolved, and spheroids were fixed with 4% paraformaldehyde at room temperature for 30 min. Spheroids were washed with DPBS with 1 mM CaCl2 and 1 mM MgCl2 and blocked with a gelatin-based permeabilization buffer (DPBS, 1 mM CaCl2, 1 mM MgCl2, 7 mg/mL gelatin, and 0.5% Triton X) for 1 hr before overnight incubation with primary antibodies in permeabilization buffer at 4°C. Spheroids were washed four times with permeabilization buffer and incubated with secondary antibodies overnight. After incubation with FITC-conjugated phalloidin for 1 hr and DAPI for 10 min, spheroids were washed with DPBS with CaCl2 and MgCl2 three times and mounted with Prolong Diamond Antifade Mountant.
Primary antibodies used included anti-NPHS1 (Bioscientific, AF4263, 1:300), biotinylated Lotus tetragonolobus lectin (LTL; Vector Laboratories, B-1325, 1:300), anti-CDH1 (BD Biosciences, 610181, 1:300), anti-CDH1 (Cell Signaling Technologies, 3195S, 1:300), anti-GATA3 (R&D Systems, AF2605, 1:300), anti-acetylated tubulin (Sigma Aldrich, T6793, 1:500), anti-IFT140 (Proteintech, 17460-1-AP, 1:100), anti-WDR19 (Proteintech, 13647-1-AP, 1:100), anti-IFT88 (Proteintech, 13967-1-AP, 1:100), anti-EPCAM-AF488 (Biolegend, 324210, 1:300), anti-ZO1 (Life Technologies, 40-2300, 1:200), anti-beta catenin (Sigma Aldrich, C2206, 1:200), and Phalloidin-FITC (Sigma-Aldrich, P5282, 1:1,000).
Secondary antibodies included Streptavidin Alexa Fluor 405 conjugate (Life Technologies, S32351, 1:400), Alexa Fluor 488 Donkey anti-mouse IgG (H+L) (Invitrogen, A21202, 1:400), Alexa Fluor 568 Donkey anti-rabbit (H+L) (Invitrogen, A10042, 1:400), Alexa Fluor 647 Donkey anti-sheep (H+L) (Invitrogen, A21448, 1:400), Alexa Fluor 647 Donkey anti-goat IgG (H+L) (Invitrogen, A21447, 1:400), Alexa Fluor 647 Donkey anti-mouse IgG (H+L) (Invitrogen, A31571, 1:400), and Alexa Fluor 568 Donkey anti-mouse IgG (H+L) (Invitrogen, A10037, 1:400).
Ciliary Morphology Assessment
Raw confocal images of cilia were median filtered (radius = 2) and segmented with a Fiji script34 for implementation of the watershed, region-labeling, and volume-filtering (minimum = 200 voxels) functions from the MorpholibJ library.35 A custom Python script was used for processing labeled images to isolate and maximum-project labeled regions representing a single cilium and then randomly select a subset of cilia from the combined dataset for scoring. The program generates blinded and unblinded scoring tables referencing a randomized, multi-image TIFF of individual cilia sampled from both genotypes. Image sets were spiked with automatically generated images (solid grayscale circles) so the scorer could reference their position within a 1,000 image set while scoring. TIFFs were manually scored and referenced back to their genotype with the unblinded table. Proportions were compared with chi-square contingency tests in GraphPad Prism.
RNA Sequencing
RNA from the epithelial fraction of whole kidney organoids was extracted with the PureLink RNA Mini Kit (Invitrogen), including treatment with the PureLink DNase Set (Invitrogen), and sequencing libraries were prepared according to Illumina’s TruSeq Stranded Total RNA protocol with RiboZero rRNA depletion. In total, six samples (three from each iPSC clone) were sequenced with an Illumina NextSeq 500 sequencer at the Translational Genomics Unit of Murdoch Children’s Research Institute. The data were submitted to the Gene Expression Omnibus (GEO) (accession number GEO: GSE107230). The raw reads were quality and adaptor trimmed with Trimmomatic v.0.3536 and mapped to the human genome (hg38) with STAR v.2.5.2a37 in the two-pass mapping mode. Reads were summarized over genes with featureCounts38 and GENCODE v.20 annotation. Genes that had expression levels of at least one count per million in at least three samples were kept for further analysis. The data were TMM normalized39 and voom transformed.40 DGE was initially identified with robust paired moderated t tests in the limma R Bioconductor package41 with a false-discovery rate cutoff of less than 5%. This identified 1,097 downregulated and 1,244 upregulated genes in the corrected iPSC versus uncorrected iPSC lines. Testing for enrichment of the Broad Institute’s Hallmark gene sets (see Web Resources) was performed with CAMERA.42 To further refine the list of differentially expressed genes, we removed 570 genes from the DGE analysis that we had previously identified as being highly variable from experiment to experiment and therefore likely false positives (Table S3) and restricted our focus to significantly up- and downregulated DGE (adjusted p < 0.01, n = 954). Gene-set enrichment analysis of these 308 genes was performed with the ToppGene Suite.43 Analysis of downregulated DGE (log fold change < −0.7, n = 308) was performed with an adjusted p value cutoff < 0.05. Histograms and heatmaps were generated with GraphPad Prism (v.7.01). Protein interactomes were generated with STRING v.10.5,44 and nodes were recolored from GraphPad Prism heatmaps in Adobe Illustrator (v.2015.2.1).
cDNA was generated from RNA samples with the GoScript Reverse Transcription Kit (Promega), and qPCR validation of selected differentially expressed genes was performed with the SensiFAST SYBR Lo-ROX Kit (BIOLINE) and QuantStudio5 Real-Time PCR System (Thermo Fisher). Oligonucleotide primers are listed in Table S7.
Magnetic-Activated Cell Sorting (MACS) and Cyst Culture
12 whole organoids were dissociated in 2.0 mL TrypLE Select (Thermo Fisher) for 12 min and inactivated by dilution with chilled MACS buffer solution (PBS, 2 mM EDTA, and 0.5% bovine serum albumin). The cell solution was passed sequentially through a 70, 40, and 30 μm sieve. Cells were counted, centrifuged at 300 × g for 5 min, and resuspended in 300 μL of MACS buffer and 100 μL of EPCAM+ (CD326) Microbeads (Miltenyi Biotech) for 30 min. Cells were rinsed with MACS buffer and centrifuged twice before resuspension in 500 μL of MACS buffer and passage through an MS MACS column according to the manufacturer’s protocol (Miltenyi Biotec). The EPCAM+ fraction was eluted gently from the column in chilled Renal Epithelial Cell Growth Medium 2 (RECGM2, Promocell). 1.0 × 105 cells in 100 μL of RECGM2 were mixed with 100 μL Matrigel GFR (BD Biosciences) and plated immediately into an 8-well chamber slide. The cell-Matrigel suspension was incubated at 37°C for 30 min before the addition of 200 μL RECGM2 supplemented with 1× Revitacell overnight. RECGM2 without Revitacell was refreshed daily until spheroids were visible by bright-field microscopy. After fixation and immunofluorescent staining (as above), spheroids were imaged with a Zeiss LSM 780 confocal microscope. 100 consecutive spheroids were assessed per condition, and proportions were compared statistically in GraphPad Prism (v.7.01) with Fisher’s exact test. Cilia per nucleus were manually counted in ten epithelial cysts imaged at high resolution in Fiji with a Cell Counter plugin. Mean cilia per nucleus expressed as a percentage for each clone were compared in GraphPad Prism with an unpaired t test and Welch’s correction for unequal variance.
Simple Western Blot
1.0 × 106 EPCAM+ cells were centrifuged in cold MACS buffer and then lysed in RIPA buffer with protease inhibitor (1:500) for 15 min on ice. Lysates were vortexed for 45 s and centrifuged at 13.3 × g for 10 min at 4°C. The supernatant was collected and diluted 1:5 with 1× Sample Buffer (ProteinSimple). Protein quantification was performed with a 66–440 kDa plate in a ProteinSimple Wes Capillary Western Blot analyzer according to the manufacturer’s instructions.
Results
Identification of Compound-Heterozygous IFT140 Variants Reclassifies Phenotype to Mainzer-Saldino Syndrome
The proband was diagnosed with retinitis pigmentosa (OMIM: 617781) at 1 year of age (Figure 1A) and with ESKD at 6 years of age, suggesting a phenotype of Senior Løken syndrome (SLS [OMIM: 266900]). SLS is a NPHP-RC syndrome describing the co-occurrence of retinal and renal ciliopathy. Renal ultrasound identified two kidneys that were appropriately sized for her age and displayed increased renal cortical echogenicity and loss of corticomedullary differentiation without any visible cysts (Figure 1B). Renal biopsy was not performed because it was deemed unlikely to contribute any diagnostic features beyond those of advanced, end-stage kidney fibrosis. Neither the proband’s non-consanguineous parents nor any of her three siblings showed signs of disease, and there was no extended family history of renal or other inherited disease (Figure 1C).
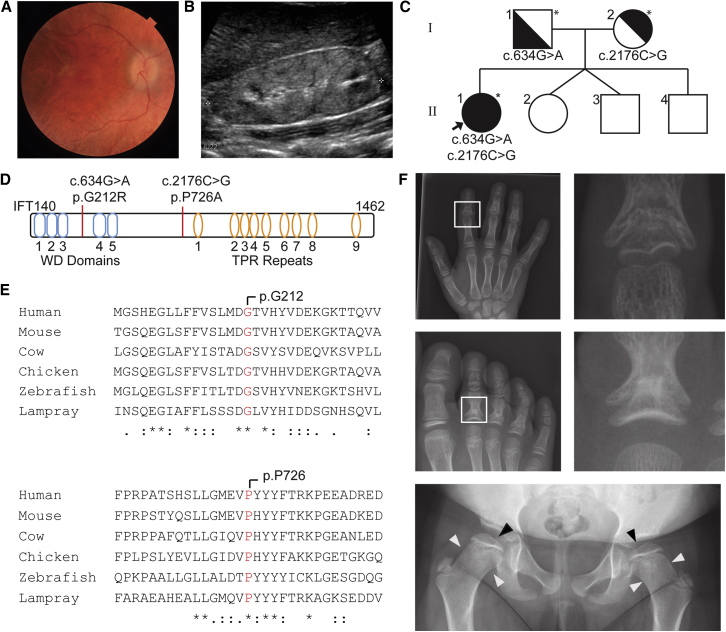
Whole-Exome Sequencing (WES) Leads to Revision of Clinical Phenotype
(A) Proband retinal photograph demonstrating retinitis pigmentosa.
(B) Renal ultrasound demonstrating echogenic kidney with loss of corticomedullary differentiation forms the basis of the eponymous diagnosis of SLS before WES.
(C) Trio WES identifies compound-heterozygous IFT140 variants (asterisks indicate sequenced individuals).
(D) IFT140 structure with variant loci identified.
(E) Conservation of both amino acid loci across vertebrate species.
(F) After the identification of IFT140 variants, a skeletal survey found cone-shaped epiphyses in the phalanges of the hand and foot (white boxes), flattened femoral heads (black arrowhead), and widened femoral neck (white arrowheads) consistent with MSS.
Trio exome sequencing analysis identified compound-heterozygous, single-base-pair IFT140 variants, confirmed by Sanger sequencing, to exist in trans: c.634G>A (p.Gly212Arg) (GenBank: NM_014714.3) and c.2176C>G (p.Pro726Ala) (GenBank: NM_014714.3) (Figure 1D). Both sites are highly conserved throughout the vertebrate lineage (Figure 1E) and are predicted to be damaging by PolyPhen, MutationTaster, and CADD. Both c.634G>A and c.2176C>G have low population allelic frequencies of 4.1 × 10−6 and 5.4 × 10−5, respectively (Table S1). The c.634G>A variant is located at the −1 position of the splice donor site of exon 6. This variant has been previously described in individuals with Mainzer-Saldino syndrome (MSS) and Jeune’s asphyxiating thoracic dystrophy (JATD) (both OMIM: 266290) and validated by expression of FLAG-tagged transcripts in immortalized human retinal pigmented epithelial (RPE) cells, suggesting mislocalization of the protein away from the primary cilium.45 The nucleotide c.G634 is totally conserved in all vertebrates, and c.634G>A is predicted to disrupt the activity of the donor splice site.46 The c.2176C>G variant has not previously been described.
Compound-heterozygous mutations in IFT140 have not previously been associated with SLS but have been reported as a cause of JATD and MSS, which are distinguished by classical skeletal phenotypes.45, 47, 48 A subsequent skeletal survey in the proband identified flattened femoral heads with widened necks as well as cone-shaped phalangeal epiphyses (Figure 1F), revising her phenotype to MSS.
Simultaneous Reprogramming and Gene Correction of Proband Fibroblasts
A previously described one-step protocol that combines both an episomal-based reprogramming strategy and CRISPR/Cas9-mediated gene editing27 was used to generate clonal uncorrected and gene-corrected iPSC lines within a single experiment (Figure 2A). The Cas9 repair template included correction of the c.634G>A variant as well as an additional synonymous 3 bp change to facilitate the identification of corrected clones and prevent Cas9-mediated cleavage after successful homology-directed repair (Figure 2A). Gene correction was identified in 1 of 44 screened iPSC clones (2.3%). One uncorrected proband (henceforth referred to as PR) and the isogenic, gene-corrected clone (GC) were carried forward to further experiments. Pluripotency was confirmed by culture morphology and staining for common pluripotency markers and subsequent flow cytometry (Figures S1A and S1B).
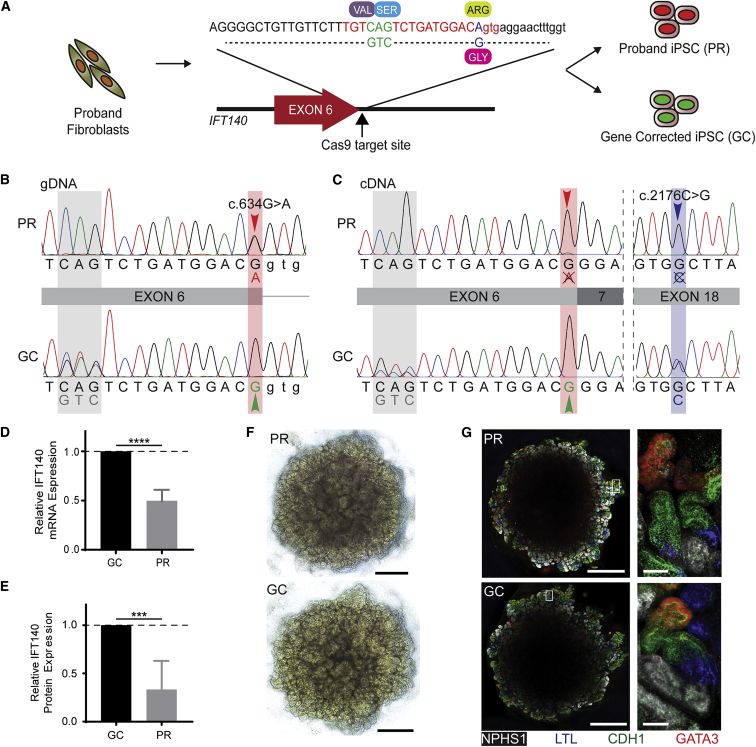
Gene Correction, Transcript Analysis, and Differentiation to Kidney Organoid
(A) The gene-correction template for the c.634G>A variant contains the corrected 1 bp substitution (blue) and a synonymous downstream 3 bp substitution (green) for ease of identification by Sanger sequencing. Successfully reprogrammed clones were screened for unsuccessful (red nuclei, PR) or successful (green nuclei, GC) gene correction.
(B) DNA sequencing demonstrates the c.634G>A variant in the PR clone (red arrowhead) and successful correction of this variant in the GC clone (green arrowhead), as well as the synonymous 3 bp change in this allele (gray shading).
(C) Sanger-sequenced RNA (copy DNA) transcripts suggest that the c.634G>A allele is not detectable as a stable transcript, indicated by a single chromatogram peak at both the c.634G>A locus (red arrowhead) and the c.2176C>G variant (blue arrowhead) in the PR clone. The double chromatogram peak at the synonymous 3 bp substitution (gray) and the c. 2176C>G variant locus (not gene-corrected; blue) in the GC clone indicates that gene correction has rescued this transcripts.
(D) qPCR identifies 50% relative mRNA expression in PR iPSCs compared with control cells (data represent the mean + 95% CI; p = 0.0001).
(E) Capillary western blot reveals 43% relative IFT140 expression in PR differentiated renal epithelial cells compared with GC cells (data represent the mean + 95% CI; p < 0.001).
(F) Bright-field images of kidney organoids on day 25 of culture (day 18 of aggregate culture). Scale bars, 1 mm.
(G) Immunofluorescence images of whole organoids demonstrate balanced expression of glomerular precursors (NPHS1, white), proximal tubule, (LTL, blue), distal tubule (CDH1, green), and collecting duct (CDH1, green; GATA3, red). Scale bars, 1 mm (low-magnification images) and 50 μm (high-magnification images).
Gene Correction of iPSCs Rescues Unstable mRNA Transcript
The proband-specific variants were confirmed after Sanger sequencing of PCR amplicons spanning exon 6 or exon 18 of the IFT140 locus (Figure 2C, red arrowhead), whereas loss of the c.634G>A variant and gain of the synonymous 3 bp change were confirmed in gene-corrected iPSCs.
Total RNA was extracted, and RT-PCR was performed to amplify IFT140 transcripts from GC and PR iPSCs, which were then sequenced. This analysis revealed that only the c.2176C>G allele was detectable at the transcriptional level in PR iPSCs (Figure 2C), indicating that the c.634G>A variant results in defective splicing of exon 6 and that the incorrectly spliced transcripts are potentially targeted for degradation. Hence, only one IFT140 allele is likely to be transcribed and translated. In contrast, transcription from both alleles was detected in GC iPSCs, as evidenced by the presence of the synonymous 3 bp change adjacent to the corrected variant (Figure 2C). This was also supported by the finding of 53% IFT140 mRNA expression and 33% quantitative protein in the PR clone relative to the GC clone (Figures 2D and 2E).
Kidney Organoid Tubules Demonstrate Ciliary Morphology Consistent with Defective Retrograde IFT
Multicellular kidney organoids were differentiated from both PR and GC iPSC lines (Figure 2F) through adaptation of a protocol previously published by Takasato et al.49 to one that used fully defined serum- and feeder-free culture conditions (Figure S1C). Organoids produced from both lines were validated as reaching a renal endpoint by the contiguous immunofluorescent localization of nephrin (NPHS1, staining glomerular precursors), LTL (staining proximal tubule), cadherin 1 (CDH1, staining distal tubule), and co-immunofluorescence for CDH1 and GATA3 (collecting duct) (Figure 2G).
Previous IFT140-null mouse models demonstrated shortened primary cilia with swollen ciliary tips in developing limb buds.22 It should be noted that dual IFT140 knockout resulted in embryonic lethality.22 Primary cilia stained with acetylated tubulin and ARL13B were clearly identified in the tubular epithelium of iPSC-derived kidney organoids by immunofluorescence (Figures 3A and 3D). Automated image processing was used to generate a random series of images of individual cilia from CDH1+ regions of kidney organoids (n = 900 cilia) for blinded scoring (Figure 3A). A clubbed morphology similar to that seen in the IFT140-null mouse was evident in the majority of PR cilia (59%; 95% confidence interval [CI] = 54%–63%), whereas a similar majority of GC cilia (60%; 95% CI = 56%–65%) demonstrated wild-type morphology (Figure 3B). PR cilia were significantly shorter than GC cilia, and the difference between PR and GC mean ciliary length increased with time in aggregate culture (Figure 3C). Ciliary IFT140 localization within club-shaped cilia in GATA3+ regions of organoid tubules demonstrated increased IFT140 staining of the swollen ciliary axoneme. Other IFT-A and IFT-B proteins demonstrated a similar intensification of fluorescence in the ciliary tip, consistent with defective retrograde IFT (Figure 3D).
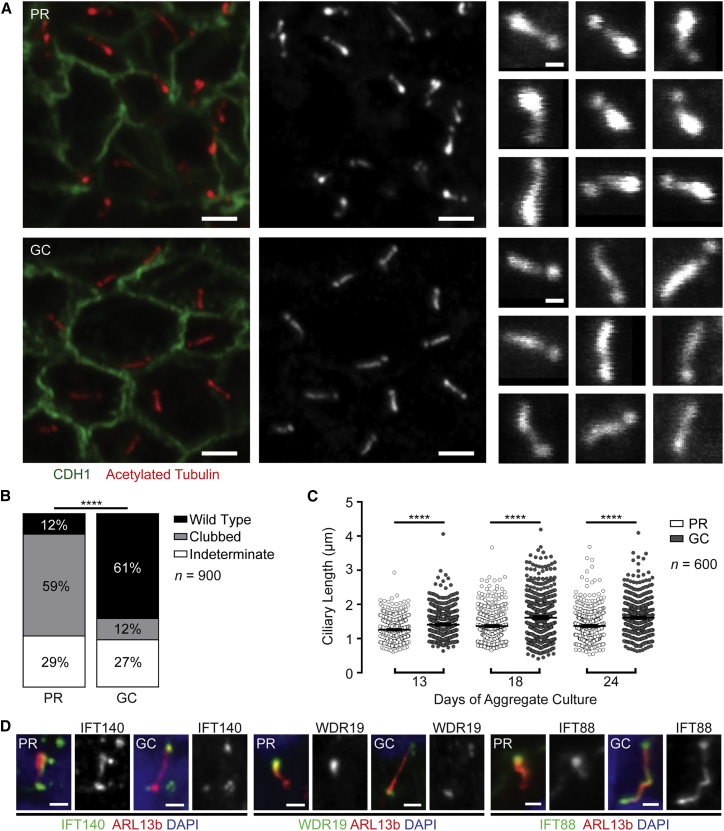
Proband Organoid Tubules Demonstrate Abnormal Cilia, which Are Rescued by Gene Correction
(A) Cilia of CDH1+ tubular epithelial cells demonstrate classical morphology associated with defective retrograde IFT: short with swollen ciliary tips (left and middle panels; scale bars, 2 μm). Individual cilia were isolated from these images (right panels; scale bars, 500 nm) and shuffled for blinded scoring of morphology.
(B) Quantitative output of blinded cilia-morphology analysis demonstrates predominantly clubbed cilia in PR organoids (59%) and predominant wild-type morphology in gene-corrected cilia (61%) (n = 900; p < 0.0001, Chi-square test).
(C) PR cilia were shorter than GC cilia at every culture time point, such that an increasing difference was noted with time in culture (n = 600 cilia per condition; p < 0.0001, Welch’s t test; error bars represent the mean + 95% CI).
(D) IFT components IFT140, WDR19, and IFT88 were all found to accumulate in the PR ciliary tip. Gene-corrected organoids displayed a wild-type distribution (scale bar, 1 μm).
Transcriptional Profiling Identifies Dysfunctional Cellular Processes Resulting from c.634G>A
The isolated transcriptional effects of the c.634G>A variant were investigated by comparison of the bulk RNA sequencing (RNA-seq) profiles (accession number GEO: GSE107230) of the epithelial (EPCAM+) cell fractions isolated from iPSC-derived kidney organoids by MACS (Figure 4A). According to co-immunofluorescence analysis, this EPCAM+ fraction included proximal tubule (LTL+) and collecting duct structures (GATA3+) as well as an epithelial structure resembling Bowman’s capsule surrounding visceral epithelial cells (Figures S2A and S2B). Validation of EPCAM+ purification by MACS was confirmed by flow cytometry (Figure S2C). Replicates clustered appropriately on a principle-component analysis with clear separation between PR and GC transcription profiles upon unbiased clustering (Figures 4B and 4C).
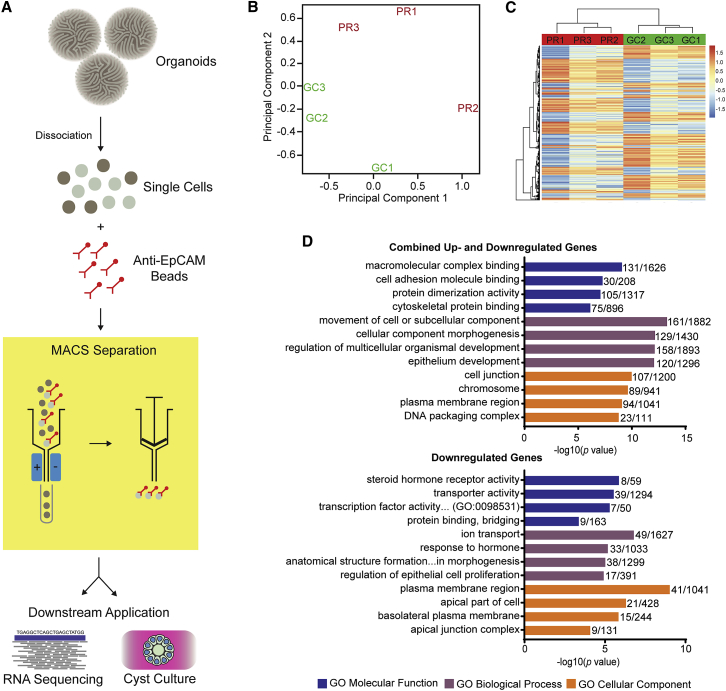
RNA-Seq of Epithelial Sub-fractions of Organoids Demonstrates Dysfunctional Cellular Processes Resulting from IFT140 Variants
(A) Diagrammatic representation of epithelial MACS from dissociated organoids. Dissociated organoids were incubated with magnetic beads conjugated with EPCAM antibody and passed through a magnetic field, trapping epithelial cells that could be gently eluted for downstream application after removal of the magnetic field.
(B) Principal-component analysis of primary RNA-seq data demonstrates clustering of PR and GC replicates.
(C) Heatmap demonstrates consistency of gene expression across replicates.
(D) Top GO terms output from ToppGene gene-list enrichment of set 1 (combined up- and downregulated genes with significant differential expression, adjusted p < 0.01; top graph) and set 2 (genes with log fold change > 0.7 favoring GC and significant differential expression < 0.05; bottom graph). Bars represent the −log10(p value), and the adjacent fraction represents the number of genes within the dataset (numerator) and the number of genes within the GO gene list (denominator). Similar GO terms with a near-identical DGE representation and GO term were filtered out to avoid repetition. Colors are as follows: blue, GO molecular function; purple, GO biological process; and orange, GO cellular component.
For the purposes of gene enrichment analysis, the primary DGE dataset (Table S2) was filtered for genes that have previously exhibited high transcriptional variability between repeated differentiation experiments of a human wild-type iPSC cell line (CRL1502, clone C32).50 This transcriptional “noise” is batch associated and likely to result from slight variations in relative maturation between individual organoids. Any of the top 1,000 most variable genes within the CRL1502 dataset were excluded from further analysis (n = 570; Table S3).
Gene enrichment analysis of the remaining genes with significant DGE (adjusted p value < 0.01, n = 954; Table S4) returned gene ontology (GO) terms illustrating disturbances in cell adhesion and cytoskeletal interaction pathways (Figure 4D). This analysis returned one single human phenotype term, “vitreoretinal degeneration” (HP:0000655), consistent with the proband phenotype (Figure S3A) and inclusive of CRB1 (OMIM: 604210), a critical component of the highly conserved Crumbs polarity complex, which is strongly associated with retinitis pigmentosa (OMIM: 600105) and Leber’s congenital amaurosis (OMIM: 613835).51 Cell polarity complexes are highly conserved, critical regulators of epithelial polarity and modulate asymmetrical cytoskeletal organization within the cell and cell-junction integrity in order to preserve apicobasal expression of transmembrane receptors and transporters.52, 53 Differential expression of Crumbs complex genes within the primary RNA-seq dataset suggested downregulation in PR renal epithelia compared with GC epithelia (Figure S3B). Transcriptional differences within the associated PAR polarity complex were less consistent between replicates with the exception of CDC42 (OMIM: 116952), which was downregulated in PR (Figure S3C). Of interest, animal knockout models of Cdc42−/− develop a NPHP-like renal histology and demonstrate loss of photoreceptor cilia.54 CDC42 was also a central node within a STRING protein interaction network of DGE highlighted within the “adherens Junction” term (GO:0005912; Figures S4A and S4B). EGFR was also a significantly downregulated central interacting gene product identified by the STRING protein interactome. EGF signaling is strongly associated with the proliferative epithelial phenotype seen in ADPKD cysts, and inhibition of this signaling pathway reduces cyst progression in animal models (reviewed in Harskamp et al.55). The observation of CDK1 (int:CDK1) and CDK2 (int:CKD2) in the top five interacting gene products is of interest given the increasing evidence of benefit in both animal and cellular nephronophthisis models treated with CDK inhibitors.56, 57, 58, 59
IFT-related ciliopathy is often associated with increases in canonical Wnt, Shh, and Notch signaling pathways, which are usually amplified by treatment with a pathway agonist.24, 26, 60 Although these pathways did not feature prominently in ToppGene enrichment analysis, all three were upregulated in Hallmark gene-list enrichment analyses of the primary dataset before the exclusion of highly variable genes (Figures S4D–S4F). In a murine Ift140 HoxB7-Cre knockout model, transcriptional changes in these signaling pathways were not observed until postnatal days 15–20.61 The present study, however, demonstrates pathway changes consistent with the existing literature within a model that at a transcriptional level most closely resembles first-trimester human fetal kidney.6
Gene enrichment analysis of downregulated DGE reflected disturbances to apical and basolateral plasma membranes, the apical junction complex, and transporter activities associated with these cellular membranes (Figure 4D). Additional downregulated GO outputs of interest included “negative regulation of epithelial cell proliferation” (GO:0050680) and “axonemal dynein complex assembly” (GO:0070286) (Figures S4C and S4D). RNA-seq of selected differentially expressed genes from these GO terms was validated by qPCR (Figure S5).
Spheroid Culture Is Impaired in IFT140−/− Epithelial Cells Sorted from Kidney Organoids
The differential expression profiling highlighted downregulation of genes expressed in the apical part of the cell and the cell junction (Figures 5A and 5B). The functional validation of apicobasal polarity defects by 3D epithelial spheroid culture has previously been applied in the study of ciliopathies.33 Accordingly, MACS-sorted EPCAM+ cells from both PR and GC organoids were subjected to spheroid culture and stained with markers of apical junctions (ZO-1 and CDH1) and apicobasal polarity (acetylated tubulin and β-catenin) (Figure 5C). Consistently fewer PR EPCAM+ cells than GC EPCAM+ cells developed spheroids with polarized epithelium (PR = 34.0% [25.4–43.7], GC = 53.0% [43.2–62.5]; p = 0.01; Figure 5D). This confirms the utility of spheroid culture as an assay for evaluating cellular polarization by using proband-derived renal epithelium and indicates an underlying defect in cell-cell interactions potentially present during human development. Assessment of primary cilia per nucleus was technically challenging in whole 3D organoids, but manual counting in cultured cysts was possible. Cilia per nucleus were lower in PR cysts than in GC cysts and were also more variable (PR = 83.7% [74.7–89.3], GC = 93.8% [91.1–96.7]; p < 0.01; Figure S6).
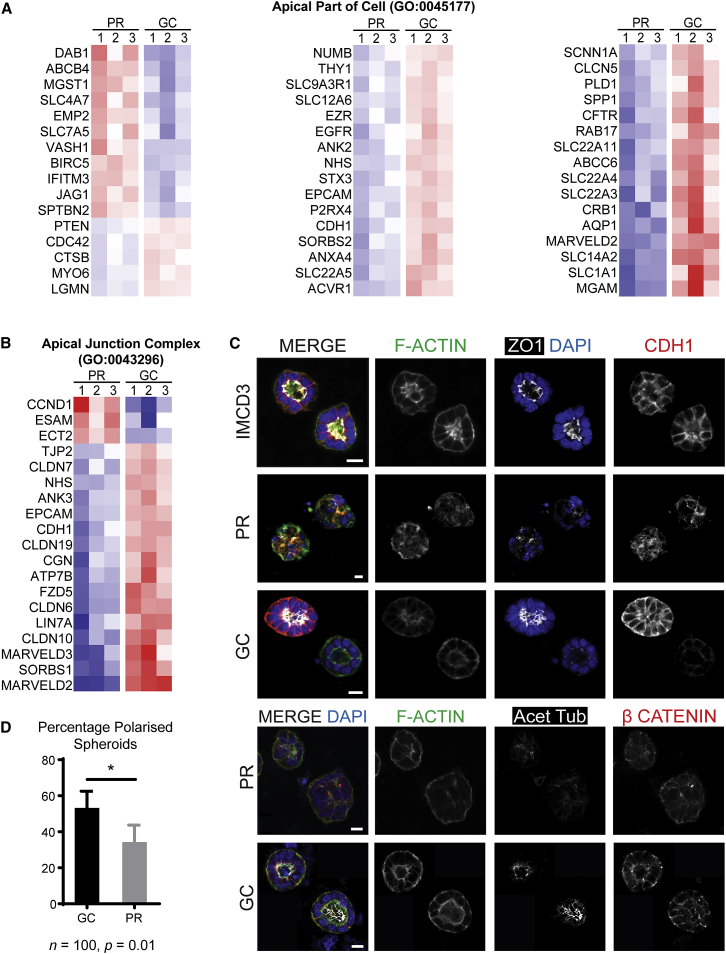
Spheroid Culture Demonstrates Functional Assay of Disturbed Apicobasal Polarity in Proband Organoid Epithelium
(A and B) Heatmaps demonstrating significant downregulation of components of (A) the apical part of the cell and (B) the apical cell junction in the PR line (red, expression above the mean; blue, expression below the mean).
(C) Immunofluorescent images from spheroid culture of EPCAM+ MACS-sorted epithelial cells demonstrate examples of poorly polarized structures (PR), well-polarized structures (GC), and reference images from IMCD3 cells. Scale bars, 10 μm.
(D) PR EPCAM+ cells were less able to establish polarized spheroids than GC controls (PR = 34.0% [25.4–43.7], GC = 53.0% [43.2–62.5]; p = 0.01; error bars represent 95% CI). The asterisk signifies adjusted p < 0.01.
Discussion
This study presents the prospective application of kidney organoids regenerated from the iPSCs of an individual with inherited kidney disease for functional genomic disease modeling. We demonstrate the utility of an isogenic, gene-corrected control iPSC clone for the functional validation of an isolated, proband-specific genetic variant. The demonstration of such a clear restoration of renal ciliary morphology by gene correction of a single mutated allele of an autosomal-recessive genotype supports the likely efficacy of this approach for validation of novel VUS in future studies. Previous reports of iPSC-based renal disease modeling have relied on the generation of mutations by CRISPR/Cas9-induced non-homologous end joining in wild-type human cell lines.62, 63 Proband iPSC-derived kidney organoids model not only the proband-specific alleles for any candidate variant but also the genomic background of that individual, allowing for contributions to phenotype from gene modifiers or allowing multigene disorders to be dissected. Importantly, if a modeled candidate VUS proves ultimately non-disease associated, data derived from the study of the proband-derived organoids continue to express the bona fide VUS, potentially permitting a retrospective diagnosis.
Although the functional genomic study of human ciliopathy has previously relied on animal models, immortalized-cell-based models, or proband-derived fibroblast monolayers, an increasing body of evidence advocates that differences in ciliary biology occur between species64, 65, 66, 67, 68, 69 and between cell types in the same individual.70 This emphasizes that modeling a human renal disease in a human renal tissue model is essential for the future application to affected individuals in a clinical setting. Additionally, the generation of transgenic models of allelic insufficiency by knockdown with morpholino oligomers, mRNA silencing, or gene knockout fails to recapitulate the specific allelic dysfunction in the affected individual. This is especially pertinent for a gene such as IFT140, which is frequently characterized by a compound-heterozygous genotype that creates a spectrum of gene dysfunction and variable phenotypes as a result of different allelic permutations.45, 47 Indeed, the c.634G>A allele present in this proband has been previously studied by expression of a FLAG-tagged variant transcript in immortalized RPE cells, suggested mislocalization of this variant IFT140 protein.45 Our results, however, suggest the lack of a stable transcript from the c.634G>A allele and a resulting 50% reduction in mRNA and protein, consistent with probable mRNA transcript decay. The ability to examine the specific transcriptional consequence of a single-base-pair mutation against an isogenic control is a unique advantage of this disease-modeling approach.
Transcriptional profiling of epithelial cells sorted from PR organoids and GC controls yielded results consistent with the evolving literature on IFT140 dysfunction. Integral components of dynein motor assembly were found to be downregulated in PR organoid epithelium. This observation supports a growing body of evidence for a novel role of IFT140 (and other IFT-A proteins) in maintaining retrograde IFT activity by ciliary import and assembly of dynein motor components rather than just linking ciliary proteins to the dynein motor for axonemal descent.71
The apparent success of our approach is likely to be critically associated with using proband and isogenic corrected lines to reduce genetic variation between test and control. However, acknowledging the variability in gene expression within a regenerative disease-modeling platform is essential for the accurate interpretation of disease-associated transcriptional change. Here, we have applied a unique approach that is based upon an extensive prior evaluation of the source of transcriptional variability between kidney organoids.50 It is possible that a portion of those genes removed from the analysis did represent genes of relevance to the disease state rather than simple experimental variation. To investigate this further, an isolated gene enrichment analysis of the excluded 570 genes returned GO terms associated with plasma membrane transporter activity and kidney or urogenital system development. Such terms are related to the relative maturation of nephrons within organoids, which we have shown does vary but could also be attributed to IFT140 variants. Importantly, where a gene was associated with the disease state in our analyses, we investigated changes in all genes for linked GO terms in order to more comprehensively evaluate pathway changes.
Over 40 genes have been identified as related to NPHP, and all are linked to the primary cilium-centrosome complex.10 Within this group, a variety of proposed disease mechanisms exist for NPHP gene products, but no unifying pathogenetic mechanism is known. Cell junction, apicobasal polarity, and cytoskeleton terms featured strongly in our DGE analysis. Although these pathways have not previously been described in IFT140 dysfunction, they have been implicated in NPHP pathobiology since the discovery of the first NPHP genes.12, 14, 72, 73 A cytoplasmic role for IFT-B protein IFT54 (encoded by TRAF3IP1 [OMIM: 607380]) in the negative regulation of microtubule stabilization by its interaction with MAP4 has recently been described.13 Affected individuals had only mildly disturbed ciliary morphology but an over-stabilized microtubule cytoskeleton, and the authors proposed that defective delivery of protein complexes to cell junctions and cell membranes leads to impaired polarity, differentiation, and degeneration of the renal tubular epithelium.13 In another study, nphp4 (OMIM: 607215) knockdown in Xenopus epidermis disrupted the subapical actin cytoskeleton, impairing communication with ciliary basal bodies and motile cilia function.14 NPHP1 and INVS also interact with β-tubulin.72 In the demonstration of disease mechanisms for IFT140, which are well established for other NPHP genes, we demonstrate how the cumulative examination of further NPHP genotypes via organoid modeling might better illustrate the common and/or distinguishing pathways mediating NPHP disease. Additionally, the ToppGene Human Phenotype output of “vitreoretinal degeneration” closely resembled the proband’s non-renal phenotype, validating the fidelity of the disease model and differential expression analysis.
Existing research on renal ciliopathy suggests epithelial dysfunction as the cause of disease; hence, the epithelial component of the organoids was purified and examined.10 As a multicellular 3D disease model, any segment of the nephron or non-epithelial cell type could be isolated from differentiated tissue by this method depending on the disease in question and the availability of an antibody for a specifying cell-surface marker. The ability to fractionate different cell types from the organoids could theoretically illustrate the differential contribution of each cell type to the disease pathobiology. For example, the interstitial expansion seen in vivo in NPHP raises the possibility of stromal contribution to disease pathobiology.74 Although a stromal element of the organoid exists,6 it is yet to be sufficiently characterized as renal stroma and as such was not studied in this work.
In this article, the study of a previously validated genotype directed the assays performed (e.g., ciliary number, length, and morphology). In any future study of a previously unvalidated VUS, such guidance might not be available, and the most appropriate assay might not be obvious. This raises the prospect of false negatives and false positives. The use of isogenic lines and normalization for highly variable genes will be important for the removal of false positives. With respect to false negatives in the case of NPHP, a number of cell-line-based functional genomic studies in this field have added pathway agonists (e.g., hedgehog modifiers and forskolin) to amplify readouts of dysfunctional molecular pathways.56, 57, 60 Although this might also be applicable in organoids, it is not known whether such treatments would alter patterning rather than reveal a relevant disease response. Indeed, even when using cell lines to model NPHP, it is not known whether this “stress” pushes the model beyond the boundary of what remains relevant to the original disease or affected individual. Hence, this is not a unique challenge to organoids. What we do demonstrate here is the application of an unbiased quantitative approach to the evaluation of cilia morphology. This approach could be applied to other NPHP models, and further development could integrate machine learning to support a fully automated high-throughput assay for the purposes of proband-specific compound screening without the need to dissociate the regenerated tissue.
The application of kidney organoids to the in vitro modeling of other types of inherited kidney disease raises specific challenges with respect to the disease studied. The transcriptional profile of the organoid derived by our protocol most closely resembles first-trimester human fetal kidney.6 This suggests that diseases with antenatal or infantile disease onset (e.g., congenital nephrotic syndrome [OMIM: 256300] and autosomal-recessive polycystic kidney disease [OMIM: 606702]) could be more readily studied than those with adult onset (e.g., ADPKD). Without a patent vasculature, urinary drainage system, or urine production, modeling of congenital anomalies of the kidney and urinary tract (or glomerulopathies, such as congenital nephrotic syndrome and Alport syndrome [OMIM: 301050 and 203780]) is less feasible. Prenatal, and hence low, expression of electrolyte channels and transporters could also limit the modeling of some tubulopathies. However, improving the degree of cellular maturation of the organoid model by refining culture conditions and introducing flow75, 76, 77 will most likely expand the repertoire of kidney diseases amenable to disease modeling with iPSC-derived models.
This study demonstrates that kidney organoids derived from proband iPSCs are capable of expressing an inherited developmental renal disease phenotype. Together with CRISPR/Cas9-mediated gene correction, kidney organoids represent a novel functional genomic platform for the validation of novel genomic variants. This study also demonstrates the study of human renal disease via a proband-derived, 3D, in vitro model. The fidelity of transcriptional profiling for the identification of not only established IFT140 disease mechanisms but also those implicated for other NPHP genes heralds the ability to refine the common cellular pathways in NPHP through the future study of other known and novel genotypes. This study was enabled via coordinated clinical and laboratory research arms able to provide a comprehensive clinical and research genomic service for the rapid identification of novel VUS in a proband-specific disease model. The future applications of kidney organoids as a disease model could include high-throughput personalized therapeutic screening, which could feed back to the affected individual within such a multidisciplinary clinical and experimental team approach.
Acknowledgments
We thank Pei Xuan Er and Irene Ghobrial for assistance with iPSC derivation and maintenance, Alison Graham for iPSC pluripotency flow cytometry, and Andrew Lonsdale for assistance with bioinformatic graphical outputs. This study was funded by the National Health and Medical Research Council of Australia (NHMRC) (GNT1098654), Kidney Health Australia, the Royal Brisbane and Women's Hospital Foundation, the Royal Children’s Hospital Foundation, and the Murdoch Children’s Research Institute. The Murdoch Childrens Research Institute is supported by the Victorian Government’s Operational Infrastructure Support Program. T.A.F. is a recipient of the NHMRC Postgraduate Scholarship (GNT1114409) and RACP Jacquot Award. M.H.L. is an NHMRC Senior Principal Research Fellow and has consulted for and received research funding from Organovo Inc.
Footnotes
Supplemental Data include six figures and seven tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.03.014
Web Resources
Broad Institute Hallmark Gene Sets, http://software.broadinstitute.org/gsea/msigdb
Gene Expression Omnibus, https://www.ncbi.nlm.nih.gov/geo/
gnomAD, http://gnomad.broadinstitute.org
Human Splicing Finder, http://www.umd.be/HSF3/index.html
OMIM, http://www.omim.org
STRING, https://string-db.org/
UCSC Genome Browser, https://genome.ucsc.edu/
Supplemental Data
References
Articles from American Journal of Human Genetics are provided here courtesy of American Society of Human Genetics
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.ajhg.2018.03.014
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S0002929718301034/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Advancements in Research on Genetic Kidney Diseases Using Human-Induced Pluripotent Stem Cell-Derived Kidney Organoids.
Cells, 13(14):1190, 13 Jul 2024
Cited by: 0 articles | PMID: 39056771 | PMCID: PMC11274677
Review Free full text in Europe PMC
Patient-derived and gene-edited pluripotent stem cells lacking NPHP1 recapitulate juvenile nephronophthisis in abnormalities of primary cilia and renal cyst formation.
Front Cell Dev Biol, 12:1370723, 26 Jun 2024
Cited by: 0 articles | PMID: 38989059 | PMCID: PMC11233770
The reprotoxic adverse side effects of neurogenic and neuroprotective drugs: current use of human organoid modeling as a potential alternative to preclinical models.
Front Pharmacol, 15:1412188, 14 Jun 2024
Cited by: 1 article | PMID: 38948466
Review
Kidney Organoid Modeling of WT1 Mutations Reveals Key Regulatory Paths Underlying Podocyte Development.
Adv Sci (Weinh), 11(29):e2308556, 29 May 2024
Cited by: 1 article | PMID: 38810140 | PMCID: PMC11304319
Advancements in therapeutic development: kidney organoids and organs on a chip.
Kidney Int, 105(4):702-708, 29 Jan 2024
Cited by: 1 article | PMID: 38296026
Review
Go to all (102) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Diseases (Showing 19 of 19)
- (2 citations) OMIM - 256300
- (1 citation) OMIM - 600105
- (1 citation) OMIM - 601678
- (1 citation) OMIM - 266900
- (1 citation) OMIM - 610805
- (1 citation) OMIM - 617781
- (1 citation) OMIM - 220100
- (1 citation) OMIM - 116952
- (1 citation) OMIM - 614620
- (1 citation) OMIM - 607380
- (1 citation) OMIM - 173900
- (1 citation) OMIM - 613835
- (1 citation) OMIM - 203780
- (1 citation) OMIM - 604210
- (1 citation) OMIM - 301050
- (1 citation) OMIM - 143400
- (1 citation) OMIM - 607215
- (1 citation) OMIM - 606702
- (1 citation) OMIM - 604766
Show less
GEO - Gene Expression Omnibus
- (2 citations) GEO - GSE107230
Quick GO (2)
- (1 citation) GO - GO0050680
- (1 citation) GO - GO0070286
RefSeq - NCBI Reference Sequence Database
- (2 citations) RefSeq - NM_014714.3
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Gene Correction Reverses Ciliopathy and Photoreceptor Loss in iPSC-Derived Retinal Organoids from Retinitis Pigmentosa Patients.
Stem Cell Reports, 10(4):1267-1281, 08 Mar 2018
Cited by: 120 articles | PMID: 29526738 | PMCID: PMC5998840
Partial uniparental isodisomy of chromosome 16 unmasks a deleterious biallelic mutation in IFT140 that causes Mainzer-Saldino syndrome.
Hum Genomics, 11(1):16, 19 Jul 2017
Cited by: 12 articles | PMID: 28724397 | PMCID: PMC5517791
Generation of infant- and pediatric-derived urinary induced pluripotent stem cells competent to form kidney organoids.
Pediatr Res, 87(4):647-655, 19 Oct 2019
Cited by: 23 articles | PMID: 31629364
Nephronophthisis-associated ciliopathies.
J Am Soc Nephrol, 18(6):1855-1871, 18 May 2007
Cited by: 232 articles | PMID: 17513324
Review
Funding
Funders who supported this work.
Kidney Health Australia
Murdoch Children’s Research Institute
NHMRC
NHMRC Postgraduate Scholarship (1)
Grant ID: GNT1114409
National Health and Medical Research Council of Australia (1)
Grant ID: GNT1098654






