Abstract
Motivation
DNA sequencing archives have grown to enormous scales in recent years, and thousands of human genomes have already been sequenced. The size of these data sets has made searching the raw read data infeasible without high-performance data-query technology. Additionally, it is challenging to search a repository of short-read data using relational logic and to apply that logic across samples from multiple whole-genome sequencing samples.Results
We have built a compact, efficiently-indexed database that contains the raw read data for over 250 human genomes, encompassing trillions of bases of DNA, and that allows users to search these data in real-time. The Terabase Search Engine enables retrieval from this database of all the reads for any genomic location in a matter of seconds. Users can search using a range of positions or a specific sequence that is aligned to the genome on the fly.Availability and implementation
Public access to the Terabase Search Engine database is available at http://tse.idies.jhu.edu.Supplementary information
Supplementary data are available at Bioinformatics online.Free full text

The Terabase Search Engine: a large-scale relational database of short-read sequences
Associated Data
Abstract
Motivation
DNA sequencing archives have grown to enormous scales in recent years, and thousands of human genomes have already been sequenced. The size of these data sets has made searching the raw read data infeasible without high-performance data-query technology. Additionally, it is challenging to search a repository of short-read data using relational logic and to apply that logic across samples from multiple whole-genome sequencing samples.
Results
We have built a compact, efficiently-indexed database that contains the raw read data for over 250 human genomes, encompassing trillions of bases of DNA, and that allows users to search these data in real-time. The Terabase Search Engine enables retrieval from this database of all the reads for any genomic location in a matter of seconds. Users can search using a range of positions or a specific sequence that is aligned to the genome on the fly.
Availability and implementation
Public access to the Terabase Search Engine database is available at http://tse.idies.jhu.edu.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The 1000 Genomes Project, which began in 2008, had within a few years created the largest public archive of human genome data ever seen (The 1000 Genomes Project Consortium, 2015). The project website now contains sequence data from many thousands of individuals, ranging from sparse sampling of the exome to deep sequencing of the entire genome. Since its inception, other large projects have been announced and some, notably the Simons Genome Diversity Project (Mallick et al., 2016), have also released large numbers of deeply sequenced genomes. The websites that provide access to data from these and similar projects have provided interfaces that let users explore the single-nucleotide variations (Shringarpure and Bustamente, 2015) and download large raw data files, but the data sets are too large to permit real-time searching.
Searching sequence databases has been a mainstay of genomics research for three decades. The BLAST program (Altschul et al., 1997) and other alignment methods provide a means to inquire whether any of millions of sequences match a query sequence. However, no system has previously been available that would allow a user to compare a sequence to the raw read data from hundreds or thousands of deeply sequenced human genomes. Instead, data producers have aligned reads from human sequence projects to the reference genome [currently GRCh38 (Schneider et al., 2017)], using programs such as Bowtie2 (Langmead and Salzberg, 2012) or BWA (Li and Durbin, 2009), and from these alignments have produced summaries that contain only the locations where a target genome differs from the reference. These summaries, in the form of variant call format (VCF) files, are far smaller than the raw data, and can readily be shared or displayed in graphical genome browsers.
Nonetheless, some scientific questions can only be answered with access to the raw human read data. For example, if one is looking for a sizeable insertion in an individual genome, a VCF file will not provide an answer because the novel DNA in the individual would simply fail to align. Or if one wants to look for extra copies of a tandem repeating sequence, such as those that affect the severity of cystic fibrosis (Guo et al., 2011), the raw reads provide much more accurate information than a simple variant file. Motivated by these and other examples, we have designed a database that allows real-time searching and retrieval of very large repositories of human reads.
Implementing a large repository of short-read sequences poses challenges in both data storage and data access. The amount of space required for storage can be minimized by converting raw sequencer or read-aligner output into compressed formats more suitable for large-scale archival. When large numbers of reads all derive from the same species (e.g. human), one can use the genome itself as part of the storage format, and save storage space by encoding the differences for each read (Hsi-Yang Fritz et al., 2011), a strategy used in the current CRAM format.
As with any compressed data, however, optimal compression requires additional computation for both compression and decompression. The first step is to align every read to the reference genome. This is a computationally-intensive operation, but alignment information adds significant value to raw sequencer reads for almost all searches of the read data. In addition to permitting more efficient storage of the read, associating each read with a reference-sequence locus makes it possible to search the archive for reads that map within a defined region of reference-sequence locations. One can also use the locations to create a natural index ordering of the reads in the archive, based on where each read maps to the reference.
2 Materials and methods
The Terabase Search Engine (TSE) was designed and implemented as a repository of whole-genome sequencing (WGS) samples whose reads have been aligned to an appropriate reference genome. All of the information in the original sequencer output resides in the TSE database. This information is stored in indexed tables managed by a general-purpose commercial relational-database management system and distributed across multiple database-server instances in order to support the concurrent execution of queries and to scale efficiently as new data is accumulated on additional server computers.
All reads were aligned to the human reference genome, GRCh38, as a pre-processing step in order to load the reads into the database. Alignments were performed using the Arioc GPU-based aligner (Wilton et al., 2015), using computers with a minimum of 128 GB of system memory and multiple NVidia GPU devices. Most of the alignments were performed on two computers equipped with dual Intel Xeon 12-core (24-hyperthread) CPUs running at 2.1 GHz and three NVidia K40 GPUs. The remaining alignments were carried out in a GPU cluster where each node had two Intel Broadwell 12-core CPUs at 2.6 GHz and two NVidia K80 GPUs.
Five computers with dual Intel Xeon 6-core CPUs at 2.1 GHz were configured as database-server instances, each running Microsoft SQL Server 2016 under Microsoft Windows Server 2012 R2. Each machine was configured with 128 GB of system memory and with a single NVidia GTX750i GPU. These computers were also provisioned with 8TB of SSD drives as well as a 1TB NVMe device; these high-bandwidth disk devices were used for ‘transient’ data operations (intermediate tables for data loading, sorting and indexing) as well as for high-usage, randomly-accessed database indexes.
2.1 Whole-genome sequencing data
The TSE data comprises 266 human WGS samples from public repositories and published studies. Of these WGS samples, 247 were obtained from the Simons Foundation Human Genome Diversity Project (Mallick et al., 2016), 17 from the 1000 Genomes Project and two from other sources (Ajay et al., 2011; Illumina Corporation, 2012).
The minimal criteria for inclusion of WGS samples in the TSE relational database were:
paired-end reads generated by Illumina sequencing technology
reads at least 100 bp long
at least 30× total coverage of each genome.
These minimum requirements ensured that reads from different WGS samples were sequenced using similar experimental technique and that search queries could be constructed in a consistent manner without the need to account for variations between samples in read length or the presence of paired-end mates.
2.2 Software implementation
Transformation of raw sequencer read data into a format that can be accessed in the TSE relational database was a four-step process:
Read-alignment
Loading data into database tables
Indexing
Data validation and backup.
Each of these steps was implemented in a pair of parameterized XML-scripted workflows that automate the entire process. The amount of time required to load one full WGS sample into the database depends primarily on the size of the sample. For the WGS samples described above, the end-to-end elapsed time to execute the workflows was about 12 h.
2.3 Read-alignment
We parameterized the read-aligner to report at most two valid mappings for each sequencer read, because two mappings suffice to provide evidence for the computation of mapping quality. We also specified Smith–Waterman alignment-score parameters that permitted the aligner to optimize mappings for reads with localized structural differences from the reference (local alignments, +2 −
− 6 −5
6 −5 −
− 3, minimum score 2N/2). These scoring parameters were designed to match the parameters used by default in Bowtie 2.
3, minimum score 2N/2). These scoring parameters were designed to match the parameters used by default in Bowtie 2.
The read-aligner assigned a unique 64-bit integer identifier through which the read’s provenance can be determined (Supplementary Fig. S1). The 64-bit value was constructed as a set of bit fields that identify the WGS sample that originally contained the read, a mate-pair specifier and a flag that indicated whether the read mapping was the highest-scoring ‘primary’ mapping or a ‘secondary’ mapping with the second-highest alignment score.
We also configured the read-aligner to emit results in binary-formatted files that could be bulk loaded directly into SQL database tables. We limited the amount of data in each SQL bulk-format file to 10 MB so that the alignment results for each WGS sample were separated into a set of 50 or more files. This made it possible to decrease the time required to transfer data into the database by loading multiple discrete subsets of the data concurrently into database tables.
2.4 Data loading
For each aligned WGS sample, we used the native binary ‘bulk load’ mechanism of the database-server to transfer each file of alignment results into a corresponding database table. Because each such data transfer is a single-threaded, serial operation, we carried out multiple bulk load operations concurrently on disjoint subsets (partitions) of the alignment results. The tables were then combined into a single unified table. The contents of this table were validated, compared with previously-loaded data to eliminate duplication, indexed and finally backed up to compressed archival storage.
To conserve storage space, we stored each read’s sequence using lossless 3-bit (ACGTN) binary run-length encoding of the differences between the reference genome and the actual sequence data (Supplementary Appendix A1). For the base quality scores associated with each read sequence we used dynamic 3-bit (8-value) binning (Supplementary Appendix A2); this level of quantization provides a significant reduction in storage space while delivering sufficient fidelity for downstream applications such as variant calling, which suffer no loss in accuracy with quantized quality values (Yu et al., 2015).
To store mates without valid mappings, we used a run-length encoded 3-bit (ACGTN) representation of the entire read sequence. For these reads, we also computed a 64-bit integer signature value that is used for locality-sensitive hashing and similarity searching (Zola, 2014).
2.5 Indexing
We created SQL indexes that best served the types of queries we expected to be most common. In particular, read mappings were indexed and stored according to the reference-genome location at which they mapped. The intention was to provide optimal performance for queries for a set of reads with mappings associated with a short contiguous region of the reference genome, as in the following model:
select…from mapping_table where POS between…and…
This indexing strategy does not preclude other kinds of queries, but queries are best optimized for speed when they contain a predicate that uses the POS-based indexes.
2.6 Data validation
We relied on low-level data-integrity validations internal to the database-server to detect data-transmission errors and other potential sources of data corruption. In addition, we analyzed the distribution of mapping positions (mapping widths, number of mappings per chromosome, proportion of reads without valid mappings) to prevent systematic errors in read-alignment and data loading.
2.7 Searching the database
Once data resides in the database, it is searched either by writing SQL queries or through a set of pre-compiled SQL stored procedures. The stored procedures support queries through a website that provides basic visualization of read-mapping distributions and allows for extraction of read-alignments from the database in SAM format.
The TSE also supports searches using a given query sequence. The implementation uses Bowtie 2 to identify reference-sequence regions where the query sequence has valid mappings. The TSE then returns all of the reads in the TSE database that have valid mappings in those regions.
For reads that do not have valid mappings in the reference genome, the TSE supports a mechanism for returning unmapped reads whose sequences are similar to a given query sequence (Wilton, 2017). This implementation computes a Jaccard similarity index between the query sequence and each unmapped read sequence, using the pre-computed 64-bit signature values associated with each unmapped read. It then computes Smith–Waterman alignments between the query sequence and all unmapped read sequences with a sufficiently high Jaccard similarity index and returns those reads associated with high alignment scores.
2.8 Mapping footprint
Because the TSE database contains paired-end reads, it is useful to devise queries that relate the mapping of each mate in a pair to a region of interest in the reference genome. Individually, the mapping for each mate may either intersect some part of the region (that is, at least one mapped base lies within the region) or be completely covered by the region (all mapped bases lie within the region). When the mappings for both mates are considered together, there are several additional possible relationships between the ‘mapping footprint’ and the region.
The TSE supports queries that specify any of these mapping footprints (Fig. 1). It also allows arbitrary combinations, so that (for example) all pairs with a mapping where both mates ‘touch’ a region may be found by aggregating ‘cover-cover’ and ‘intersect-intersect’ mapping footprints.
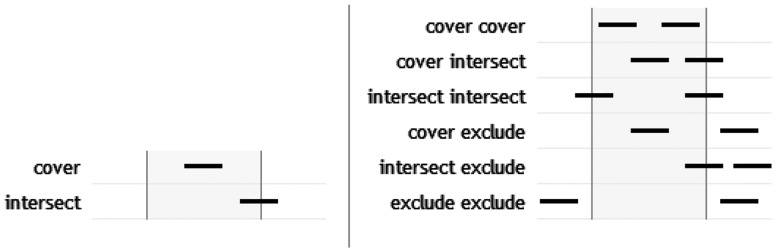
Mapping footprints. The mapping footprint of a read describes the relationship between the region covered by a read mapping and an arbitrary region on the reference genome. For an unpaired mate, the mapping footprint may be covered by the reference region (that is, every mapped base in the read lies within the region) or intersected (at least one mapped base lies within the region and at least one mapped base lies outside it). For paired-end mappings, the mapping footprint is described by the relationship of the reference region to the mappings of both mates
3 Results
The current TSE database contains WGS samples from 266 individuals (164 male, 102 female). In total, the genomes contain 354.8 billion reads (length 94–102), of which 340.4 billion (95.9%) have valid mappings to the human genome. Of the 14.4 billion (4.1%) unmapped reads, there are 6.6 billion distinct read sequences; i.e. many of them are duplicates.
On average, each WGS sample contained about 662 million paired-end reads (1.32 billion mates, 44× coverage), of which 94.5% had proper (concordant) mappings. For mapped reads, the average alignment score was 193 and the average mapping quality was 50 (Supplementary Figs S2 and S3).
3.1 Data loading and storage
The data for each WGS sample was obtained from a pair of FASTQ-formatted files downloaded from their repositories of origin. Read-alignment throughput for individual WGS samples ranged from 25 000 to 95 000 alignments/second (roughly 4–8 h elapsed), depending on the sample and upon available GPU hardware. Data loading, indexing and validation required an additional 3–7 h per WGS sample.
The TSE data for all of the WGS samples in the database (including indexes and metadata) occupies 50.5 TB of disk space, distributed across five database-server instances.
3.2 Query performance
For queries that define a contiguous region of interest in the human reference genome, the TSE database can retrieve ~10 000 reads per second. Depending upon the actual coverage of the region of interest, all reads mapped entirely within a region 1000 bases wide can be obtained within about 10 s.
Queries that involve similarity searching of unmapped reads require ~30 s to return both the locations at which a specified query sequence maps to the human reference genome (on-the-fly alignment) as well as a set of unmapped reads whose sequence is similar to the query sequence.
4 Discussion
The TSE implementation demonstrates that it is feasible to transform large volumes of raw DNA sequences into a format that can be efficiently queried using relational-database operations. The work of loading data into a relational-database storage format yields the ability to compose complex relational queries. As a tool for the execution of relational operations, a SQL database provides both higher speed and greater flexibility than the use of flat file formats such as SAM, BAM and VCF, which are the dominant formats in the field of genomics today. A relational database-server implementation can use optimizations such as multithreading and asynchronous file input/output to accelerate queries. It can also analyze the relational logic implicit in complex SQL queries so as to choose optimal strategies for sorting, merging, and index usage.
4.1 SQL queries
Although the TSE database provides access to WGS samples for only 266 individuals, the amount of data represented in the database would make it unwieldy if a large proportion of the data had to be accessed in order to generate query results. Even computationally simple queries (for example, generating a histogram of values from one mapping-table column) can take several hours to process because all of the data in the table must be read in order to produce an aggregate result.
Our strategy for using the database efficiently is therefore to create queries such that only an efficiently-defined subset of data rows is used, and then to further manipulate the data to produce desired results. Because WGS samples were loaded separately into the TSE database and indexed primarily by reference-sequence mapping location, high-performance queries of the data filter their results first by sample and/or by mapping region. A subset of the database that has been filtered in this way may still contain millions of data rows, but result sets of this size can be manipulated in seconds on currently available hardware.
Even queries that cannot immediately be defined in terms of a known genomic region can be managed in this way. In particular, it is possible to extract reads from the database based on their similarity to a given query sequence by first aligning the query sequence to the human reference genome. The result of this procedure is to identify one or more regions where the query sequence has a valid mapping; these regions can then be used for efficient queries of the TSE read-mapping data.
4.2 Interactive access
In addition to direct SQL queries of the data, it is useful to visualize the distribution of read mappings within a specified region of the reference genome. For this reason, we developed a simple web-browser-based coverage-visualization tool that displays the reference-sequence positions at which reads in each of the WGS samples are mapped to the reference genome (Fig. 2). High-variability regions with lower coverage are visually apparent in this display, yet it is possible to examine the individual reads within each WGS sample within the same display context. This same tool can be used to save the reads in individual samples (or, if desired, all of the available samples) for subsequent download in SAM format.
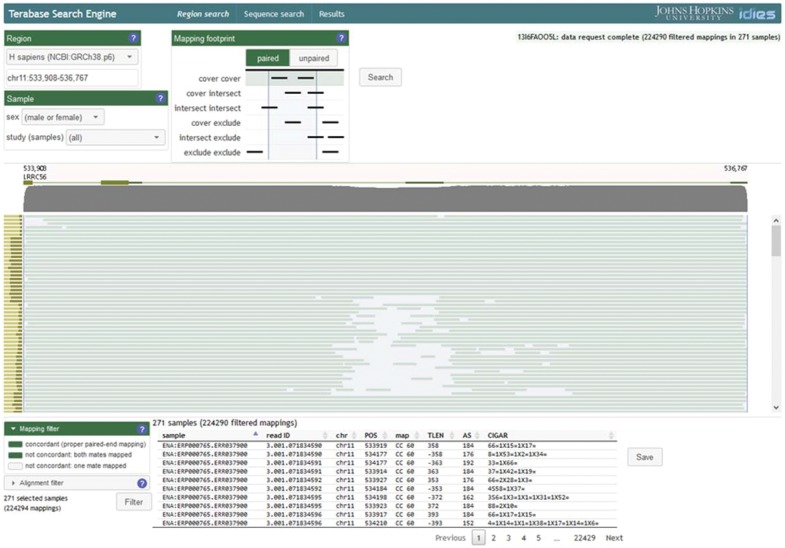
TSE web user interface. Search criteria (‘Region’, ‘Sample’, ‘Mapping footprint’) are specified prior to searching the database to visualize per-sample coverage. The result of such a search is a coverage map in which each horizontal bar represents coverage for one WGS sample. The vertical (left) histogram represents the total number of reads for each WGS sample. The horizontal (top) histogram indicates total coverage for each reference-genome location. Individual reads may be visualized and saved for subsequent processing by selecting specific WGS samples and by applying ‘mapping filter’ criteria
The TSE visual interface emphasizes the relational nature of the underlying implementation. Instead of providing a ‘service’ with a set of pre-defined query types, the TSE web pages are designed to facilitate the parameterization of relational operations on the raw sequencer reads in the database. In other words, a query such as ‘does a particular variant exist at a specified genome locus?’ is executed through the TSE as a request for mapped reads at or near a particular locus; the additional work of characterizing the variants at that locus is left to the user.
4.3 TSE query examples
The benefit of an interactive design based on relational operations is that a TSE user can craft queries that identify sequencer reads related to any genomic region, with full access to the alignment results associated with each read. Typical queries might include any of the following:
prevalence of a SNP
identifying an inversion
prevalence of an uncatalogued deletion
variations in the length of a tandem repeat
prevalence of a polymorphic L1 (LINE-1) insertion.
The Supplementary Material contains details and screen snapshots of each of these potential use cases (Supplementary Appendixes A3–A7) as well as visualizations of the query results exported from the TSE and imported into the Integrative Genome Viewer (Thorvaldsdóttir et al., 2013).
5 Conclusions
As the above examples suggest, the fact that the TSE implements search queries as relational operations makes it possible to identify sets of raw sequencer reads that meet a wide variety of complex search conditions. The TSE’s visual interface is, in effect, a simplified representation of a set of parameterized relational-database queries. It is possible, of course, to query the TSE in a straightforward manner—for example, to retrieve all reads whose mappings lie entirely within a specified region of the genome—but the approach used in the TSE also supports the rapid retrieval of more precisely-defined results from a large repository of sequencer reads.
In terms of performance, these examples each execute in fewer than 15 s when executed against a TSE database that contains about 355 billion reads representing 266 individual WGS samples. This level of performance is typical for TSE queries where the primary criterion for selecting reads is their mapping location within a fairly small (up to 1000 bp) region of the reference genome. Other relational queries are feasible as well, but query patterns that involve scanning the entire database of reads (for example, a request for the average alignment score across all mapped reads) will be limited by disk hardware speeds and may execute in minutes or hours, depending on the complexity of the query.
The TSE database schema was designed to support 1000–10 000 human WGS samples, although we were unable to find anywhere near that number of public-domain samples with which to initialize the database. Because queries execute concurrently on independent database-server instances, database performance scales efficiently as new server instances are added to accommodate additional data.
With these performance constraints in mind, the TSE implementation demonstrates that one may quickly and interactively achieve tangible results for queries against hundreds of billions of raw reads. This performance is made possible by leveraging the query-optimization capabilities of a database-server and using database tables and indexes whose layout facilitates the optimization of meaningful data queries. In this way, the TSE embodies a general-purpose tool that makes it possible to provide real-time access to sequencing reads from hundreds or thousands of WGS projects.
Acknowledgements
The authors thank Alyza M. Skaist for assistance with development and testing of the search examples.
Funding
This work was supported in part by NIH grants R01-HG007196 and R01-HL129239.
Conflict of Interest: none declared.
References
- Ajay S.S. et al. (2011) Accurate and comprehensive sequencing of personal genomes. Genome Res., 21, 1498–1505. [Europe PMC free article] [Abstract] [Google Scholar]
- Altschul S.F. et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. PMCID: PMC146917 [Europe PMC free article] [Abstract] [Google Scholar]
- Guo X. et al. (2011) Mucin variable number tandem repeat polymorphisms and severity of cystic fibrosis lung disease: significant association with MUC5AC. PLoS One, 6, e25452.. [Europe PMC free article] [Abstract] [Google Scholar]
- Hsi-Yang Fritz M. et al. (2011) Efficient storage of high throughput DNA sequencing data using reference-based compression. Genome Res., 21, 734–740. [Europe PMC free article] [Abstract] [Google Scholar]
- Illumina Corporation (2012) Whole Human Genome Sequencing of an African Male Individual (HapMap: NA18507) Using the Illumina HiSeq 2500 and Paired 100 Base Reads. ENA accession number PRJEB2892.
- Langmead B., Salzberg S.L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods, 9, 357–359. [Europe PMC free article] [Abstract] [Google Scholar]
- Li H., Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25, 1754–1760. [Europe PMC free article] [Abstract] [Google Scholar]
- Mallick S. et al. (2016) The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature, 538, 201–206. [Europe PMC free article] [Abstract] [Google Scholar]
- Schneider V.A. et al. (2017) Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res., 27, 849–864. [Europe PMC free article] [Abstract] [Google Scholar]
- Shringarpure S.S., Bustamante C.D. (2015) Privacy risks from genomic data-sharing beacons. Am. J. Hum. Genet., 97, 631–646. [Europe PMC free article] [Abstract] [Google Scholar]
- The 1000 Genomes Project Consortium. (2015) A global reference for human genetic variation. Nature, 526, 68–74. [Europe PMC free article] [Abstract] [Google Scholar]
- Thorvaldsdóttir H. et al. (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform., 14, 178–192. [Europe PMC free article] [Abstract] [Google Scholar]
- Wilton R. (2017) GPU-accelerated similarity searching in a Database of short DNA sequences In: Presented at the NVidia GPU Technology Conference, San Jose, California, 2017. [Google Scholar]
- Wilton R. et al. (2015) Arioc: high-throughput read alignment with GPU-accelerated exploration of the seed-and-extend search space. PeerJ., 3, e808.. [Europe PMC free article] [Abstract] [Google Scholar]
- Yu Y.W. et al. (2015) Quality score compression improves genotyping accuracy. Nat. Biotechnol., 33, 240–243. [Europe PMC free article] [Abstract] [Google Scholar]
- Zola J. (2014) Constructing similarity graphs from large-scale biological sequence collections. In Proceedings of the Thirteenth IEEE International Workshop on High Performance Computational Biology (HiCOMB), pp. 500–507.
Articles from Bioinformatics are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/bioinformatics/bty657
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/bioinformatics/article-pdf/35/4/665/27838499/bty657.pdf
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/bioinformatics/bty657
Article citations
Short-read aligner performance in germline variant identification.
Bioinformatics, 39(8):btad480, 01 Aug 2023
Cited by: 1 article | PMID: 37527006 | PMCID: PMC10421969
Review Free full text in Europe PMC
Data-Rich Spatial Profiling of Cancer Tissue: Astronomy Informs Pathology.
Clin Cancer Res, 28(16):3417-3424, 01 Aug 2022
Cited by: 1 article | PMID: 35522154 | PMCID: PMC9378428
Bio-Strings: A Relational Database Data-Type for Dealing with Large Biosequences.
BioTech (Basel), 11(3):31, 30 Jul 2022
Cited by: 9 articles | PMID: 35997339 | PMCID: PMC9472027
PIDS: A User-Friendly Plant DNA Fingerprint Database Management System.
Genes (Basel), 11(4):E373, 30 Mar 2020
Cited by: 0 articles | PMID: 32235513 | PMCID: PMC7230844
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
VarSome: the human genomic variant search engine.
Bioinformatics, 35(11):1978-1980, 01 Jun 2019
Cited by: 867 articles | PMID: 30376034 | PMCID: PMC6546127
GIGGLE: a search engine for large-scale integrated genome analysis.
Nat Methods, 15(2):123-126, 08 Jan 2018
Cited by: 98 articles | PMID: 29309061 | PMCID: PMC5872823
GEMINI: a computationally-efficient search engine for large gene expression datasets.
BMC Bioinformatics, 17:102, 24 Feb 2016
Cited by: 4 articles | PMID: 26911289 | PMCID: PMC4765211
A space and time-efficient index for the compacted colored de Bruijn graph.
Bioinformatics, 34(13):i169-i177, 01 Jul 2018
Cited by: 33 articles | PMID: 29949982 | PMCID: PMC6022659
Funding
Funders who supported this work.
NHGRI NIH HHS (1)
Grant ID: R01 HG007196
NHLBI NIH HHS (1)
Grant ID: R01 HL129239
NIH (2)
Grant ID: R01-HG007196
Grant ID: R01-HL129239




