Abstract
Free full text

Engineered CRISPR-Cas9 nuclease with expanded targeting space
Abstract
The RNA-guided endonuclease Cas9 cleaves its target DNA and is a powerful genome-editing tool. However, the widely used Streptococcus pyogenes Cas9 enzyme (SpCas9) requires an NGG protospacer adjacent motif (PAM) for target recognition, thereby restricting the targetable genomic loci. Here, we report a rationally engineered SpCas9 variant (SpCas9-NG) that can recognize relaxed NG PAMs. The crystal structure revealed that the loss of the base-specific interaction with the third nucleobase is compensated by newly introduced non–base-specific interactions, thereby enabling the NG PAM recognition. We showed that SpCas9-NG induces indels at endogenous target sites bearing NG PAMs in human cells. Furthermore, we found that the fusion of SpCas9-NG and the activation-induced cytidine deaminase (AID) mediates the C-to-T conversion at target sites with NG PAMs in human cells.
The CRISPR RNA-guided endonuclease Cas9 cleaves double-stranded DNA targets complementary to the RNA guide (1) (fig. S1) and has been harnessed for genome editing in eukaryotic cells (2). However, the widely used Cas9 from S. pyogenes (SpCas9) strictly recognizes an NGG sequence (where N is any nucleobase) as the protospacer adjacent motif (PAM) (3), thereby restricting the targetable genomic loci. Structure-guided directed evolution approaches to address this limitation yielded several SpCas9 variants with altered PAM specificities, such as the SpCas9 VQR and VRER variants, which recognize the NGA and NGCG PAMs, respectively (4). In addition, Cas9 and Cas12a (also known as Cpf1) enzymes with distinct PAM specificities, such as Staphylococcus aureus Cas9 (SaCas9) (5), Acidaminococcus sp. Cas12a (AsCas12a), and Lachnospiraceae bacterium Cas12a (LbCas12a) (6), have extended the targeting range in CRISPR-Cas-mediated genome editing.
To expand the targeting range of CRISPR-Cas9, we sought to engineer a SpCas9 variant with relaxed preferences for the third nucleobase of the PAM. Previous studies revealed that the second and third nucleobases in the NGG PAM are recognized by Arg1333 and Arg1335 of SpCas9, respectively (7) (fig. S2). We thus hypothesized that the PAM constraint can be reduced by eliminating the base-specific interaction between Arg1335 and the third G, and compensating for the loss of this base-specific interaction by introducing non-base-specific interactions with the PAM duplex. We first measured the in vitro cleavage activities of purified wild-type SpCas9 and the R1335A (Arg1335 → Ala) mutant toward a target plasmid with the TGG PAM and confirmed that, whereas wild-type SpCas9 efficiently cleaves the TGG target, R1335A has almost no activity (fig. S3, A to C).
We next examined whether the R1335A activity is restored by the substitution of residues surrounding the PAM duplex, and found that the replacements of Leu1111, Gly1218, Ala1322, and Thr1337 with Arg partially restored the activity of the R1335A mutant (fig. S3, A to C). Furthermore, the R1335A/L1111R/G1218R/A1322R/T1337R variant (referred to as ARRRR) efficiently cleaved the TGG target (fig. S3, A to C). However, the cleavage kinetics of ARRRR was slower than that of wildtype SpCas9 (fig. S3, D and E). In the previously reported VQR (D1135V/R1335Q/T1337R) and VRER (D1135V/G1218R/R1335E/T1337R) variants, the D1135V mutation provides interactions with the sugar-phosphate backbone of the PAM duplex (8,9). In addition, molecular modeling suggested that the E1219F mutation forms hydrophobic interactions with the ribose moiety of the second G, and that the R1335V mutation stabilizes Arg1333 and Phe1219 (E1219F) (fig. S3F). Indeed, the addition of the D1135V, E1219F, and R1335V mutations enhanced the cleavage activity (fig. S3, D and E). We designated the R1335V/L1111R/D1135V/G1218R/E1219F/A1322R/T1337R variant as VRVRFRR.
We next measured the cleavage activities of VRVRFRR toward the target plasmid with TGN PAMs. Relative to wild-type SpCas9, VRVRFRR slowly but more efficiently cleaved the TGA, TGT, and TGC targets (Fig. 1, A to C, and fig. S4). Although VRVRFRR is less active than wild-type SpCas9 and SaCas9, its cleavage activity was comparable to those of AsCas12a and LbCas12a (5, 6) (fig. S5). Using an in vitro PAM identification assay (10), we confirmed that whereas wildtype SpCas9 is specific to NGG PAMs, VRVRFRR preferentially recognizes NG PAMs (Fig. 1D and fig. S6). Although the PAM identification assay revealed that VRVRFRR slightly recognizes NAN PAMs (Fig. 1D), in vitro cleavage experiments demonstrated that VRVRFRR is less active toward TAN PAMs than toward TGN PAMs (fig. S7). Thus, we concluded that VRVRFRR recognizes a relaxed PAM, and we refer to this variant as SpCas9-NG, as it has increased activity on NGH (H = A, T, or C) PAMs, albeit with reduced relative activity on NGC.
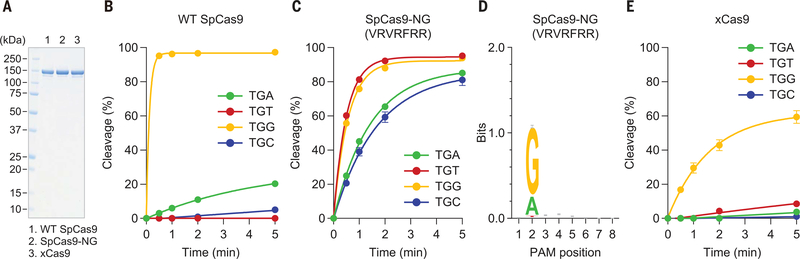
(A) SDS-polyacrylamide gel electrophoresis analysis of wild-type SpCas9, SpCas9-NG, and xCas9. (B, C, and E) In vitro DNA cleavage activities of wild-type SpCas9 (B), SpCas9-NG (C), and xCas9 (E) toward the TGN PAM targets. Data are means ± SD (n = 3). (D) PAM preference of SpCas9-NG.
We compared SpCas9-NG with the xCas9 enzyme (a SpCas9 variant with the A262T/R324L/S409I/E480K/E543D/M694I/E1219V mutations), which was engineered via directed evolution and recognizes NG PAMs (11) (fig. S8). We measured their in vitro cleavage activities toward the target plasmid with the TGN PAMs. Under our assay conditions (50 nM Cas9), xCas9 showed almost no activity toward the TGA, TGT, and TGC targets, and it cleaved the TGG target with lower efficiency than did SpCas9-NG (Fig. 1, A to C and E, and fig. S9A). At a higher concentration (200 nM Cas9), xCas9 cleaved the TGA, TGT, and TGC targets (fig. S9, D and G); nonetheless, the cleavage kinetics of xCas9 was slower than that of SpCas9-NG (fig. S9, C, D, F, and G), and the cleavage activity of xCas9 toward the TGA, TGT, and TGC targets was comparable to or lower than that of wild-type SpCas9 toward the TGA target (fig. S9, B, D, E, and G). These results demonstrate that SpCas9-NG outperforms xCas9 in recognizing NGH PAMs in vitro.
To clarify the NG PAM recognition mechanism, we determined the crystal structure of SpCas9- NG, in complex with a single guide RNA and its target DNA containing the TGG PAM, at 2.7 Å resolution (fig. S10A and table S1). The second G in the PAM (dG2*) forms bidentate hydrogen bonds with Arg1333 (Fig. 2A), which is stabilized by Val1335 (R1335V). The third G (dG3*) forms a hydrogen bond with Arg1337 (T1337R) (Fig. 2A), consistent with the preference of SpCas9-NG for the third G (Fig. 1C). Arg1111 (L1111R), Val1135 (D1135V), and Phe1219 (E1219F) interact with the sugar-phosphate backbone of the PAM duplex, whereas Arg1218 (G1218R) does not directly interact with the DNA backbone (Fig. 2B). A structural comparison with the SpCas9 R-loop complex (12) suggested that Arg1322 (A1322R) interacts with the nontarget DNA strand (fig. S10B).
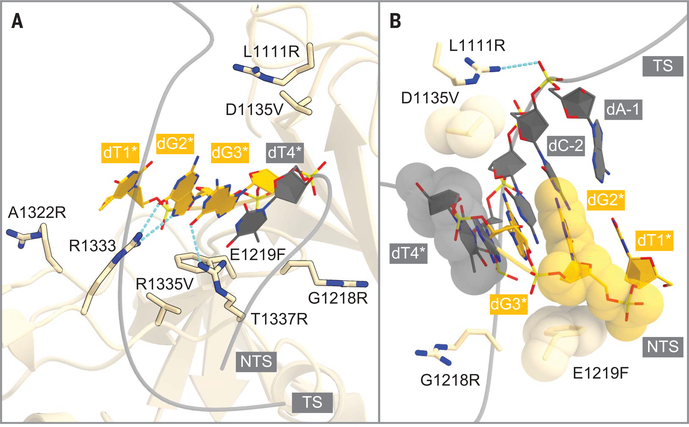
(A) Recognition of the PAM duplex. Arg1333 and the substituted residues are shown as stick models. (B) Non-base-specific interactions between the PAM duplex and the substituted residues. TS, target strand; NTS, nontarget strand.
To assess the activity of SpCas9-NG in mammalian cells, we next measured the formation of indels (insertions or deletions) induced by wildtype SpCas9, SpCas9-NG, and xCas9 at 69 endogenous target sites with NGN PAMs in human embryonic kidney (HEK) 293FT cells (table S2). As expected, wild-type SpCas9 induced indels predominantly at the NGG sites, with some recognition of NGA sites (Fig. 3, A and B). Wild-type SpCas9 achieved a >20% indel rate at NGG (17 of 17 sites) and NGA sites (3 of 19 sites), but not NGT (0 of 18 sites) or NGC sites (0 of 15 sites) (Fig. 3, A and B). In contrast, SpCas9-NG edited NGA, NGT, and NGG sites, with lower activity at NGC (Fig. 3, A and B), consistent with the in vitro cleavage preference for the third D (D = A, T, or G) PAM nucleotide (Fig. 1C). SpCas9-NG achieved a >20% indel rate at 13 NGG, 13 NGA, 15 NGT, and 5 NGC sites (Fig. 3, A and B). Relative to SpCas9-NG, xCas9 had lower editing efficiency at all of the NGH sites (Fig. 3, A and B), consistent with our in vitro cleavage data (Fig. 1E). xCas9 induced a >20% indel rate at 15 NGG sites, five NGA sites, and four NGT sites, but none at NGC sites (Fig. 3, A and B).
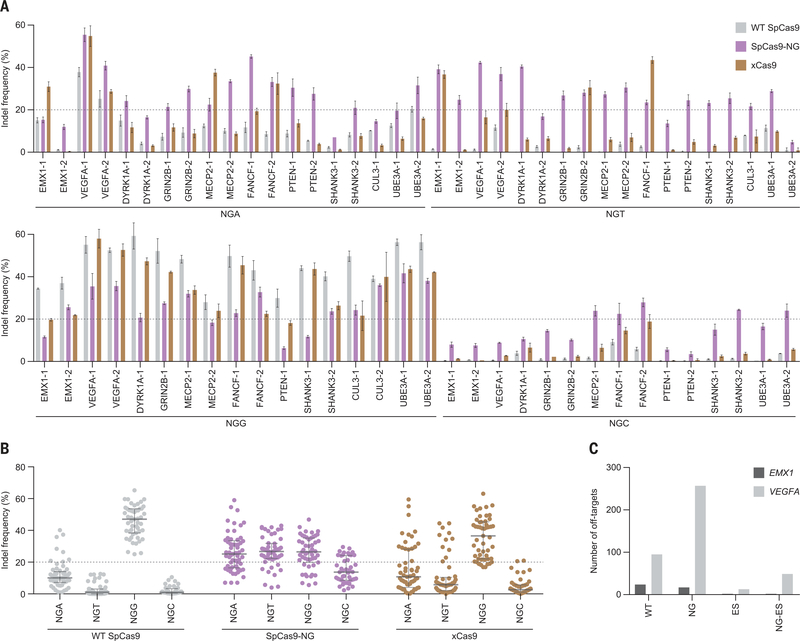
(A) Indel formation efficiencies of wild-type SpCas9, SpCas9-NG, and xCas9 at the 69 endogenous target sites in HEK293FT cells. Data are means ± SD (n = 3). (B) Summary of the editing efficiencies of SpCas9, SpCas9-NG, and xCas9. Medians and first and third quartiles are shown. In (A) and (B), 20% indel frequency is indicated by dashed lines. (C) Specificities of wild-type SpCas9, SpCas9-NG, and the enhanced-specificity versions of SpCas9 (ES) and SpCas9-NG (NG-ES). The off-target cleavages were evaluated by GUIDE-seq.
Using GUIDE-seq (genome-wide, unbiased identification of double-stranded breaks enabled by sequencing) (13), we examined the specificity of wild-type SpCas9 and SpCas9-NG in human cells at two previously characterized target sites (EMX1 and VEGFA). We found that wild-type SpCas9 and SpCas9-NG have comparable numbers of off-target sites for the EMX1 target, and that the K848A/K1003A/R1060A mutations in the high-fidelity SpCas9 variant [eSpCas9(1.1)] (14) substantially reduced the off-target cleavage by SpCas9-NG (Fig. 3C and fig. S11). These results show that SpCas9-NG has cleavage specificity comparable to that of wild-type SpCas9, and that its specificity can be enhanced by the high-fidelity mutations. As expected, SpCas9-NG had a different off-target profile with off-target sites harboring NGH PAMs (fig. S11), further confirming the relaxed PAM recognition.
The nuclease-inactive version of SpCas9 can be applied to numerous technologies such as base editing (15,16). We examined whether the SpCas9-NG D10A nickase fused to the activation-induced cytidine deaminase (nSpCas9-NG-AID, referred to as Target-AID-NG) mediates C-to-T conversion at 32 endogenous target sites with NG PAMs in human cells (table S3). nSpCas9-AID (Target- AID) efficiently induced C-to-T conversion at the NGG sites, with lower activity at NGA sites and no activity at NGT or NGC sites (Fig. 4A and figs. S12A and S13). In contrast, Target- AID-NG showed base-editing activity toward all PAMs assessed, albeit with lower efficiency at NGC sites (Fig. 4A and figs. S12A and S13), consistent with the indel data (Fig. 3A). In addition, Target-AID-NG showed higher base-editing efficiencies than did the xCas9 D10A nickase fused to the APOBEC1 cytidine deaminase (xCas9-BE4) at most of the tested poly-C–containing target sites (Fig. 4B and figs. S12B and S13). These results demonstrate that the catalytically inactive version of SpCas9-NG can serve as a useful RNA-guided DNA targeting platform.
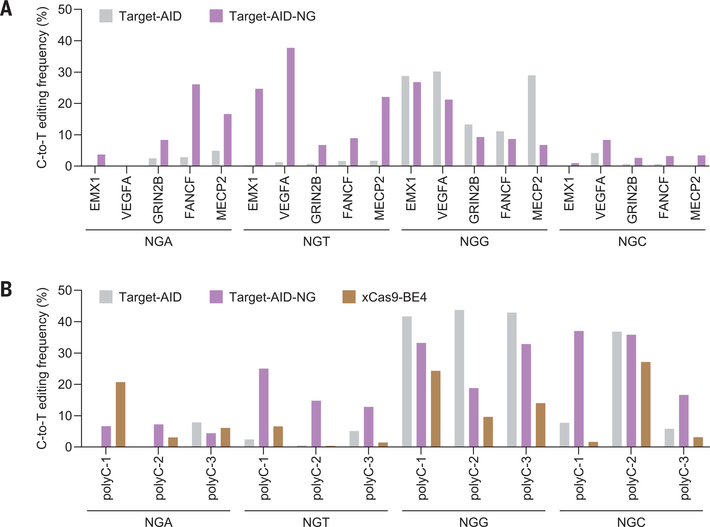
(A and B) C-to-T conversion efficiencies at the 20 endogenous target sites (Target-AID and Target-AID-NG) (A) and at the 12 poly-C-containing target sites (Target-AID, Target-AID-NG, and xCas9-BE4) (B) in HEK293Tcells. The experiments were performed at least twice, and similar results were obtained.
In this study, we engineered an SpCas9-NG variant with an increased targeting range (fig. S14). Nonetheless, the cleavage activity of SpCas9-NG is lower than that of wild-type SpCas9 at NGG sites, and SpCas9-NG is less active at NGC sites relative to NGD sites. Thus, it will be important to improve the activity of SpCas9-NG by further molecular engineering. Overall, the rationally designed SpCas9-NG and its high-fidelity variants can serve as useful genome-editing tools with increased versatility across genomes.
ACKNOWLEDGMENTS
We thank A. Kurabayashi for assistance with sample preparation, T. Yamano for Cas12a preparation, B. P. Kleinstiver and J. K. Joung for assistance with GUIDE-seq, and the beamline scientists at BL41XU at SPring-8 for assistance with data collection.
Funding: Supported by JST (JPMJPR13L8) and JSPS (26291010 and 15H01463) (H.N.); JSPS (16J06287) (S.I.); JSPS (17H01394), AMED (JP18gm5010001), Takeda Science Foundation, NICHD (P01HD087157 and R01HD088412), and the Bill & Melinda Gates Foundation (OPP1160866) (M.I.); NEDO (Genome Editing Program), AMED (17g6110007h0002), JST (10814) and JSPS (18H02428) (N.Y.); NIMH (5DP1-MH100706 and 1R01-MH110049), NIDDK (5R01DK097768-03), and the New York Stem Cell, Simons, Paul G. Allen Family, and Vallee foundations (F.Z.); AMED (16am0301002h0003) and CSTI, SIP (O.N.). F.Z. is a New York Stem Cell Foundation Robertson Investigator.
Footnotes
Competing interests: H.N. is a scientific adviser for EdiGENE. F.Z. is a co-founder and scientific adviser for Editas Medicine, Pairwise Plants, Beam Therapeutics, and Arbor Biotechnologies. F.Z. serves as a director for Beam Therapeutics and Arbor Biotechnologies. O.N. is a co-founder, board member, and scientific adviser for EdiGENE. H.N., H.H., and O.N. have filed a patent application related to this work.
Data and materials availability: The atomic coordinates of SpCas9-NG have been deposited in the Protein Data Bank with PDB code 6AI6. All data are available in the manuscript or the supplementary material.
REFERENCES AND NOTES
Full text links
Read article at publisher's site: https://doi.org/10.1126/science.aas9129
Read article for free, from open access legal sources, via Unpaywall:
https://science.sciencemag.org/content/sci/361/6408/1259.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1126/science.aas9129
Article citations
The Evolution of Nucleic Acid-Based Diagnosis Methods from the (pre-)CRISPR to CRISPR era and the Associated Machine/Deep Learning Approaches in Relevant RNA Design.
Methods Mol Biol, 2847:241-300, 01 Jan 2025
Cited by: 0 articles | PMID: 39312149
Comprehensive evaluation and prediction of editing outcomes for near-PAMless adenine and cytosine base editors.
Commun Biol, 7(1):1389, 25 Oct 2024
Cited by: 0 articles | PMID: 39455714 | PMCID: PMC11511846
Amphiphilic shuttle peptide delivers base editor ribonucleoprotein to correct the CFTR R553X mutation in well-differentiated airway epithelial cells.
Nucleic Acids Res, 52(19):11911-11925, 01 Oct 2024
Cited by: 0 articles | PMID: 39315713 | PMCID: PMC11514481
Base editing screens define the genetic landscape of cancer drug resistance mechanisms.
Nat Genet, 56(11):2479-2492, 18 Oct 2024
Cited by: 0 articles | PMID: 39424923 | PMCID: PMC11549056
Use of paired Cas9-NG nickase and truncated sgRNAs for single-nucleotide microbial genome editing.
Front Genome Ed, 6:1471720, 26 Sep 2024
Cited by: 0 articles | PMID: 39391173 | PMCID: PMC11464485
Go to all (506) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe
-
(1 citation)
PDBe - 6AI6View structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Structural Basis for the Altered PAM Specificities of Engineered CRISPR-Cas9.
Mol Cell, 61(6):886-894, 01 Mar 2016
Cited by: 80 articles | PMID: 26990991
Structural Plasticity of PAM Recognition by Engineered Variants of the RNA-Guided Endonuclease Cas9.
Mol Cell, 61(6):895-902, 01 Mar 2016
Cited by: 99 articles | PMID: 26990992 | PMCID: PMC5065715
Expanding the scope of CRISPR/Cas9-mediated genome editing in plants using an xCas9 and Cas9-NG hybrid.
J Integr Plant Biol, 62(4):398-402, 09 Jan 2020
Cited by: 17 articles | PMID: 31702097
Cas9, Cpf1 and C2c1/2/3-What's next?
Bioengineered, 8(3):265-273, 31 Jan 2017
Cited by: 49 articles | PMID: 28140746 | PMCID: PMC5470521
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Bill & Melinda Gates Foundation - Grand Challenges Explorations Initiative (1)
Grant ID: OPP1160866
Japan Agency for Medical Research and Development
NHGRI NIH HHS (2)
Grant ID: R01 HG009761
Grant ID: RM1 HG006193
NHLBI NIH HHS (1)
Grant ID: DP1 HL141201
NICHD NIH HHS (2)
Grant ID: P01 HD087157
Grant ID: R01 HD088412
NIDDK NIH HHS (1)
Grant ID: R01 DK097768
NIMH NIH HHS (2)
Grant ID: DP1 MH100706
Grant ID: R01 MH110049





