Abstract
Free full text

Adipsin, the adipocyte serine protease: gene structure and control of expression by tumor necrosis factor.
Abstract
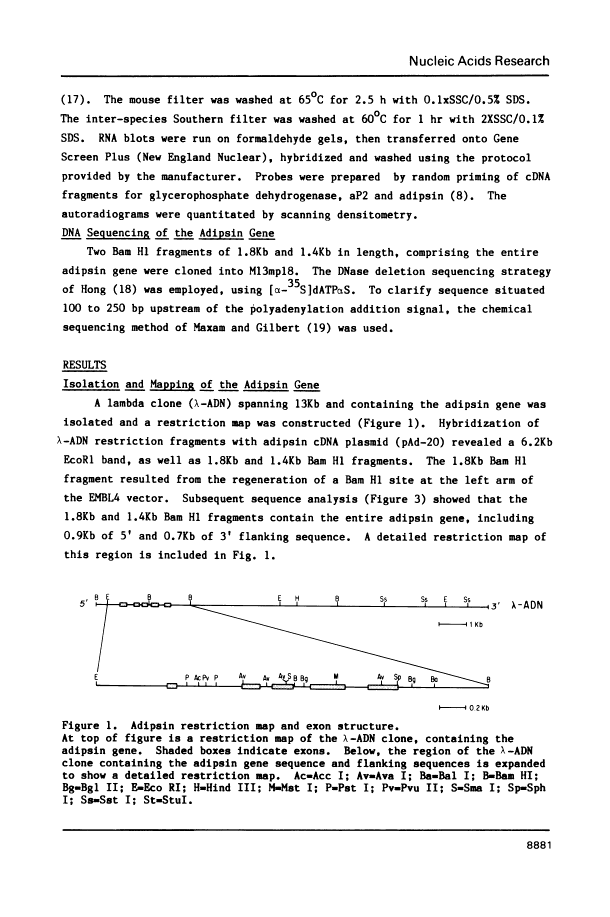
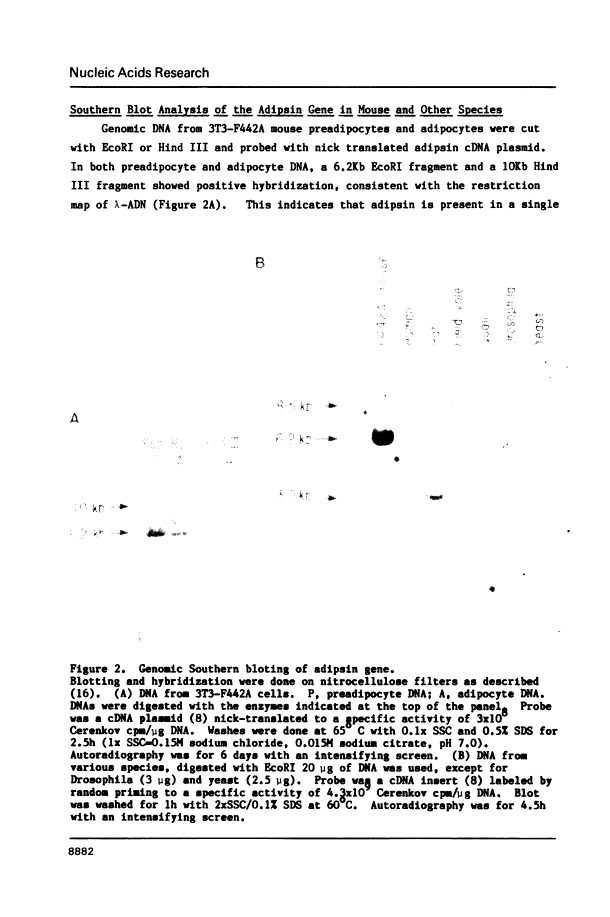
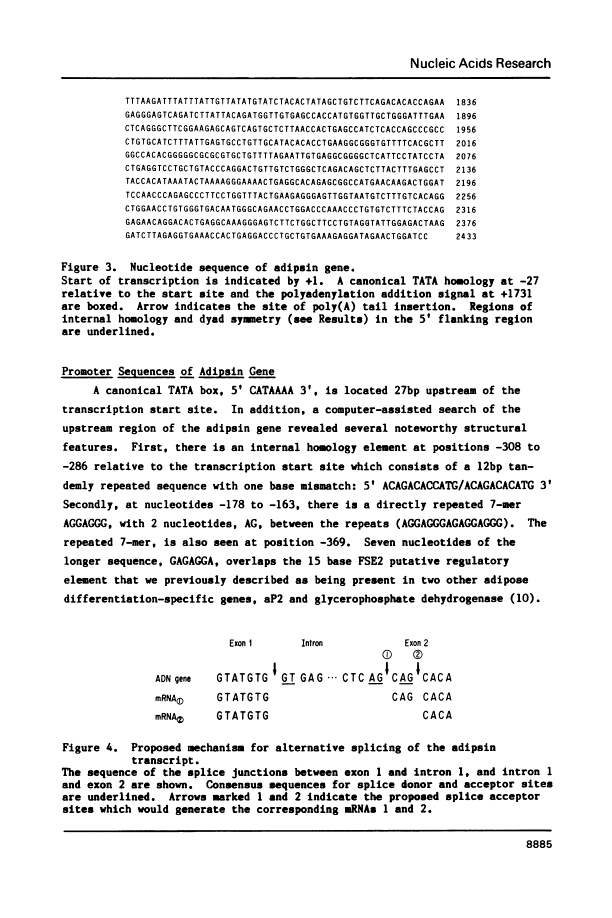
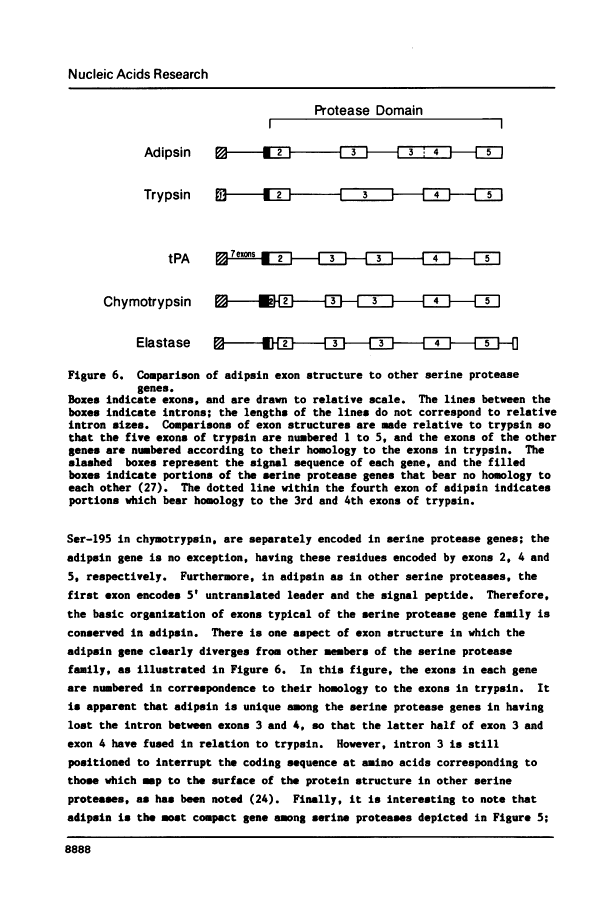
We have isolated, mapped and sequenced adipsin, the adipocyte differentiation-dependent serine protease gene. This gene, which is present in a single form in the mouse, spans 1.7 kilobases and contains five exons. While the basic exon structure characteristic of serine protease genes is conserved in adipsin, there is also a fusion of two exons that are separate in other serine proteases. The sequence data also suggests a mechanism of alternative splicing which appears to account for the generation of two adipsin mRNA species differing by only three nucleotides and encoding two different signal peptides. To investigate the control of adipsin expression we have examined the effects of tumor necrosis factor (TNF) on adipocytes. The level of adipsin RNA is dramatically decreased by hormone treatment, but the change occurs more slowly than for other fat cell mRNAs, such as glycerophosphate dehydrogenase. These results show that adipsin is a novel serine protease gene whose expression is regulated by a macrophage-derived factor which modulates expression of other adipocyte-specific RNAs.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.2M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Rubin CS, Lai E, Rosen OM. Acquisition of increased hormone sensitivity during in vitro adipocyte development. J Biol Chem. 1977 May 25;252(10):3554–3557. [Abstract] [Google Scholar]
- Reed BC, Kaufmann SH, Mackall JC, Student AK, Lane MD. Alterations in insulin binding accompanying differentiation of 3T3-L1 preadipocytes. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4876–4880. [Europe PMC free article] [Abstract] [Google Scholar]
- Rubin CS, Hirsch A, Fung C, Rosen OM. Development of hormone receptors and hormonal responsiveness in vitro. Insulin receptors and insulin sensitivity in the preadipocyte and adipocyte forms of 3T3-L1 cells. J Biol Chem. 1978 Oct 25;253(20):7570–7578. [Abstract] [Google Scholar]
- Karlsson FA, Grunfeld C, Kahn CR, Roth J. Regulation of insulin receptors and insulin responsiveness in 3T3-L1 fatty fibroblasts. Endocrinology. 1979 May;104(5):1383–1392. [Abstract] [Google Scholar]
- Spiegelman BM, Green H. Control of specific protein biosynthesis during the adipose conversion of 3T3 cells. J Biol Chem. 1980 Sep 25;255(18):8811–8818. [Abstract] [Google Scholar]
- Spiegelman BM, Frank M, Green H. Molecular cloning of mRNA from 3T3 adipocytes. Regulation of mRNA content for glycerophosphate dehydrogenase and other differentiation-dependent proteins during adipocyte development. J Biol Chem. 1983 Aug 25;258(16):10083–10089. [Abstract] [Google Scholar]
- Cook KS, Groves DL, Min HY, Spiegelman BM. A developmentally regulated mRNA from 3T3 adipocytes encodes a novel serine protease homologue. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6480–6484. [Europe PMC free article] [Abstract] [Google Scholar]
- Hunt CR, Ro JH, Dobson DE, Min HY, Spiegelman BM. Adipocyte P2 gene: developmental expression and homology of 5'-flanking sequences among fat cell-specific genes. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3786–3790. [Europe PMC free article] [Abstract] [Google Scholar]
- Zezulak KM, Green H. Specificity of gene expression in adipocytes. Mol Cell Biol. 1985 Feb;5(2):419–421. [Europe PMC free article] [Abstract] [Google Scholar]
- Cook KS, Hunt CR, Spiegelman BM. Developmentally regulated mRNAs in 3T3-adipocytes: analysis of transcriptional control. J Cell Biol. 1985 Feb;100(2):514–520. [Europe PMC free article] [Abstract] [Google Scholar]
- Djian P, Phillips M, Green H. The activation of specific gene transcription in the adipose conversion of 3T3 cells. J Cell Physiol. 1985 Sep;124(3):554–556. [Abstract] [Google Scholar]
- Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. [Abstract] [Google Scholar]
- Frischauf AM, Lehrach H, Poustka A, Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. [Abstract] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. [Abstract] [Google Scholar]
- Hong GF. A systemic DNA sequencing strategy. J Mol Biol. 1982 Jul 5;158(3):539–549. [Abstract] [Google Scholar]
- Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. [Abstract] [Google Scholar]
- Breathnach R, Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. [Abstract] [Google Scholar]
- Mount SM. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. [Europe PMC free article] [Abstract] [Google Scholar]
- Beutler B, Cerami A. Cachectin and tumour necrosis factor as two sides of the same biological coin. Nature. 1986 Apr 17;320(6063):584–588. [Abstract] [Google Scholar]
- Torti FM, Dieckmann B, Beutler B, Cerami A, Ringold GM. A macrophage factor inhibits adipocyte gene expression: an in vitro model of cachexia. Science. 1985 Aug 30;229(4716):867–869. [Abstract] [Google Scholar]
- Phillips M, Djian P, Green H. The nucleotide sequence of three genes participating in the adipose differentiation of 3T3 cells. J Biol Chem. 1986 Aug 15;261(23):10821–10827. [Abstract] [Google Scholar]
- Craik CS, Choo QL, Swift GH, Quinto C, MacDonald RJ, Rutter WJ. Structure of two related rat pancreatic trypsin genes. J Biol Chem. 1984 Nov 25;259(22):14255–14264. [Abstract] [Google Scholar]
- Swift GH, Craik CS, Stary SJ, Quinto C, Lahaie RG, Rutter WJ, MacDonald RJ. Structure of the two related elastase genes expressed in the rat pancreas. J Biol Chem. 1984 Nov 25;259(22):14271–14278. [Abstract] [Google Scholar]
- Rogers J. Exon shuffling and intron insertion in serine protease genes. Nature. 1985 Jun 6;315(6019):458–459. [Abstract] [Google Scholar]
- Mason AJ, Evans BA, Cox DR, Shine J, Richards RI. Structure of mouse kallikrein gene family suggests a role in specific processing of biologically active peptides. Nature. 1983 May 26;303(5915):300–307. [Abstract] [Google Scholar]
- Evans BA, Richards RI. Genes for the alpha and gamma subunits of mouse nerve growth factor are contiguous. EMBO J. 1985 Jan;4(1):133–138. [Europe PMC free article] [Abstract] [Google Scholar]
- van Leeuwen BH, Evans BA, Tregear GW, Richards RI. Mouse glandular kallikrein genes. Identification, structure, and expression of the renal kallikrein gene. J Biol Chem. 1986 Apr 25;261(12):5529–5535. [Abstract] [Google Scholar]
- Bell GI, Quinto C, Quiroga M, Valenzuela P, Craik CS, Rutter WJ. Isolation and sequence of a rat chymotrypsin B gene. J Biol Chem. 1984 Nov 25;259(22):14265–14270. [Abstract] [Google Scholar]
- Nagamine Y, Pearson D, Altus MS, Reich E. cDNA and gene nucleotide sequence of porcine plasminogen activator. Nucleic Acids Res. 1984 Dec 21;12(24):9525–9541. [Europe PMC free article] [Abstract] [Google Scholar]
- Fransen L, Müller R, Marmenout A, Tavernier J, Van der Heyden J, Kawashima E, Chollet A, Tizard R, Van Heuverswyn H, Van Vliet A, et al. Molecular cloning of mouse tumour necrosis factor cDNA and its eukaryotic expression. Nucleic Acids Res. 1985 Jun 25;13(12):4417–4429. [Europe PMC free article] [Abstract] [Google Scholar]
- Pennica D, Hayflick JS, Bringman TS, Palladino MA, Goeddel DV. Cloning and expression in Escherichia coli of the cDNA for murine tumor necrosis factor. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6060–6064. [Europe PMC free article] [Abstract] [Google Scholar]
- Dreyfuss G, Adam SA, Choi YD. Physical change in cytoplasmic messenger ribonucleoproteins in cells treated with inhibitors of mRNA transcription. Mol Cell Biol. 1984 Mar;4(3):415–423. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/14.22.8879
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc311917?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/nar/14.22.8879
Article citations
Adipsin promotes bone marrow adiposity by priming mesenchymal stem cells.
Elife, 10:e69209, 22 Jun 2021
Cited by: 32 articles | PMID: 34155972 | PMCID: PMC8219379
Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat Chem Biol, 12(12):1105-1110, 24 Oct 2016
Cited by: 37 articles | PMID: 27775713
Small cell lung cancer: Recruitment of macrophages by circulating tumor cells.
Oncoimmunology, 5(3):e1093277, 29 Oct 2015
Cited by: 51 articles | PMID: 27141354 | PMCID: PMC4839345
Revisiting the adipocyte: a model for integration of cytokine signaling in the regulation of energy metabolism.
Am J Physiol Endocrinol Metab, 309(8):E691-714, 01 Sep 2015
Cited by: 137 articles | PMID: 26330344
Review
FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis.
Cell Res, 24(12):1403-1419, 21 Nov 2014
Cited by: 655 articles | PMID: 25412662 | PMCID: PMC4260349
Go to all (46) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Adipsin: a circulating serine protease homolog secreted by adipose tissue and sciatic nerve.
Science, 237(4813):402-405, 01 Jul 1987
Cited by: 206 articles | PMID: 3299705
Adipocyte P2 gene: developmental expression and homology of 5'-flanking sequences among fat cell-specific genes.
Proc Natl Acad Sci U S A, 83(11):3786-3790, 01 Jun 1986
Cited by: 181 articles | PMID: 3520554 | PMCID: PMC323608
Differentiation dependent biphasic regulation of adipsin gene expression by insulin and insulin-like growth factor-1 in 3T3-F442A adipocytes.
Endocrinology, 127(6):2898-2906, 01 Dec 1990
Cited by: 11 articles | PMID: 2249632
Regulation of gene expression in the adipocyte: implications for obesity and proto-oncogene function.
Trends Genet, 4(7):203-207, 01 Jul 1988
Cited by: 19 articles | PMID: 3070869
Review
Funding
Funders who supported this work.
NIADDK NIH HHS (2)
Grant ID: AM31405
Grant ID: AM34605