Abstract
Free full text
An atlas of genetic associations in UK Biobank
Abstract
Genome-wide association studies have revealed many loci contributing to the variation of complex traits, yet the majority of loci that contribute to the heritability of complex traits remain elusive. Large study populations with sufficient statistical power are required to detect the small effect sizes of the yet unidentified genetic variants. However, the analysis of huge cohorts, like UK Biobank, is challenging. Here we present an atlas of genetic associations for 118 non-binary and 660 binary traits of 452,264 UK Biobank participants of white descent. Results are compiled in a publicly accessible database that allows querying genome-wide association results for 9,113,133 genetic variants, as well as downloading whole GWAS summary statistics for over 30 million imputed genetic variants (>23 billion phenotype-genotype pairs). Our atlas of associations (GeneATLAS, http://geneatlas.roslin.ed.ac.uk) will help researchers to query UK Biobank results in an easy and uniform way without the need to incur in high computational costs.
Introduction
Most human traits are complex and influenced by the combined effect of large numbers of small genetic and environmental effects1. Genome-wide association studies (GWAS) have identified many genetic variants influencing many complex traits. The largest genetic effects were discovered with modest sample sizes, with researchers subsequently joining efforts to increase the size of the study cohorts, thus allowing them to identify much smaller genetic effects. The UK Biobank2, a large prospective epidemiological study comprising approximately 500,000 deeply phenotyped individuals from the United Kingdom, has been genotyped using an array that comprises 847,441 genetic polymorphisms, with a view to identifying new genetic variants in a uniformly genotyped and phenotyped cohort of unprecedented size, both in terms of the number of samples and number of traits.
The unprecedented size of this cohort has raised a number of analytical challenges3. First, storing, managing and analysing the circa 90 million genetic variants for around half a million individuals is, in itself, a substantial endeavour. Second, the collection of samples at this scale has brought up an analytical challenge, as the cohort is structured by familial relationships and ethnicity. For instance, many relatives were unintentionally collected in the cohort, and removing them from the analyses as traditionally done in GWAS would entail a substantial loss of statistical power. Third, although recent developments have reduced the computational costs4, fitting a Linear Mixed Model (LMM), the standard analytical technique to perform GWAS when there is population or familial structure, at this scale and for this number of traits, entails a computational burden which may be beyond the means of many research labs.
The objective of the current study was to perform GWAS for 778 traits in UK Biobank, adjusting for the effect of relatedness to minimise the loss of statistical power whilst reducing false positives due to familial and population structure, in individuals of white ancestry and to make a searchable atlas of genetic associations in UK Biobank for the benefit of the research community.
Results
Data overview
In July 2017, the UK Biobank released genotyped data from circa 490,000 individuals of largely white descent genotyped for 805,426 genetic variants. We performed GWASs for 660 binary traits and 118 non-binary traits, the latter including continuous traits and traits with multiple ordered categories (Supplementary Table 1). For each of these traits we fitted LMMs to test for association with 623,944 genotyped and 30,798,054 imputed genetic polymorphisms imputed using the Haplotype Reference Consortium5 as reference panel, as well as 310 imputed HLA alleles. All successfully tested polymorphisms are shown in the database (GeneATLAS, http://geneatlas.roslin.ed.ac.uk) or associated downloadable files to allow individual researchers to apply their own quality control thresholds. The summary results presented here are based on the quality controlled imputed polymorphisms (9,113,133 variants after filtering) of 452,264 individuals (Methods).
The phenotypes selected comprise a mix of baseline measurements (e.g. height), self-reported traits at recruitment (e.g. self-reported depression), and Hospital Episode Statistics (i.e. data collected during hospital admissions) as well as cancer diagnoses from the appropriate UK Cancer Registry. Since UK Biobank is a recently stablished prospective cohort, we allowed for potential differences in statistical power among binary and non-binary traits by splitting the presentation of the data into non-binary and binary traits.
To demonstrate the power of using large datasets (so called, Big Data), we first explored how the analysis of increasingly large sample sizes enable new discoveries, and reduce bias when estimating the effect sizes of GWAS hits (Fig. 1 and Supplementary Note). Our results show that the number of GWAS hits increased linearly with the sample size with no sign of saturation, thus suggesting that increasing the size of cohorts like UK Biobank would continue to yield new discoveries. We also observed that the estimated allelic effects of GWAS hits obtained from decreasing sample sizes were generally larger, which is in agreement with a Winner’s Curse effect6 (Fig. 1).
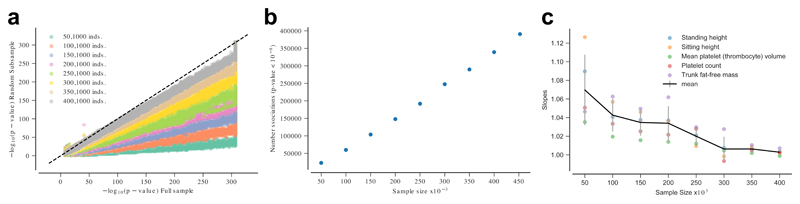
(a) Comparison between the p-values (two-sided t-test) obtained using the whole cohort (452,264 individuals) and random subsamples of increasing sizes. The plot shows only the results for the genetic variants associated with a p-value < 10-8 in the whole cohort. (b) Total number of detected associated variants (two-sided t-test) at a threshold of p-value < 10-8 as a function of the sample size. (c) Slope of the effect sizes of the GWAS hits obtained in random subsamples of increasing size vs the same effect sizes estimated in the whole cohort. Slopes larger than one indicate an inflation on the effect estimates in the smaller sample. The black line joints the mean at each sample size shown. Error bars indicate the standard deviation.
Distribution of GWAS hits among non-binary trait
Just below 5 million of the circa 1 billion tests performed across 118 non-binary traits were significant at a conventional genome wide threshold (P<10-8) (Supplementary Table 2), and 3,117,904 were significant after Bonferroni correction (P<0.05/9,113,133*118). The significant associations where distributed across 74,471 leading polymorphisms mapping to 38,651 independent loci (Methods, Fig. 2, Supplementary Table 3). A substantial proportion of these associations (13.0%) were within the HLA region (Supplementary Table 2).
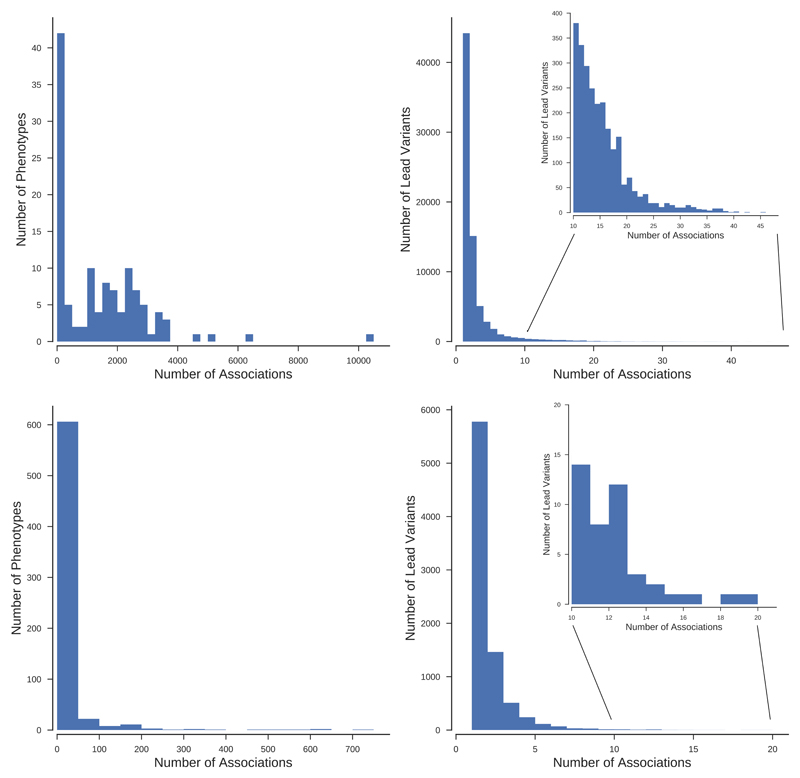
The panels show results for each phenotype (left) and independent lead variant (right) for non-binary (top) and binary (bottom) phenotypes.
About 9.5% of the tested polymorphisms reached genome-wide significant thresholds (P<10-8) for at least one of the 118 tested traits, whilst 82% of the tested polymorphisms were associated with at least one of these 118 traits at a significance level of 10-2 (Supplementary Table 4). There were 20,393 genetic variants each associated with more than 30 of the tested non-binary traits (Figs. 2 and and3,3, Supplementary Fig. 1). A cluster of nine variants in a 9kb region including the genotyped intronic variant rs1421085 within the FTO gene had the largest number of genome-wide significant associations outside the HLA region, all nine variants being found to be associated with 58 traits (Fig. 3 and Supplementary Fig. 1). The genotyped variant rs1421085 at the FTO locus also had the largest average significance across non-binary traits (P<10-74) (Supplementary Fig. 2), which was largely contributed by the associations to anthropometric traits such as BMI and Weight which showed some of the strongest associations (P<10-300). The HLA region contained 362 genetic variants which were significantly (P<10-8) associated with 50 or more of the non-binary traits compared to only 128 such variants in the remaining autosomal variants. About 36% of the analyzed imputed HLA alleles were significant (P<10-8) for at least one trait (Supplementary Fig. 3). Six traits ('Standing height', 'Sitting height', 'Platelet count', 'Mean platelet (thrombocyte) volume', ’Trunk predicted mass’, ‘Trunk fat-free mass’) had over 100,000 significant associations (P<10-8) each distributed across 25,352 different independent lead genetic variants (Methods). Over 94% of the non-binary traits had more than 100 genome-wide significant hits distributed in 74,442 different leading genetic variants.
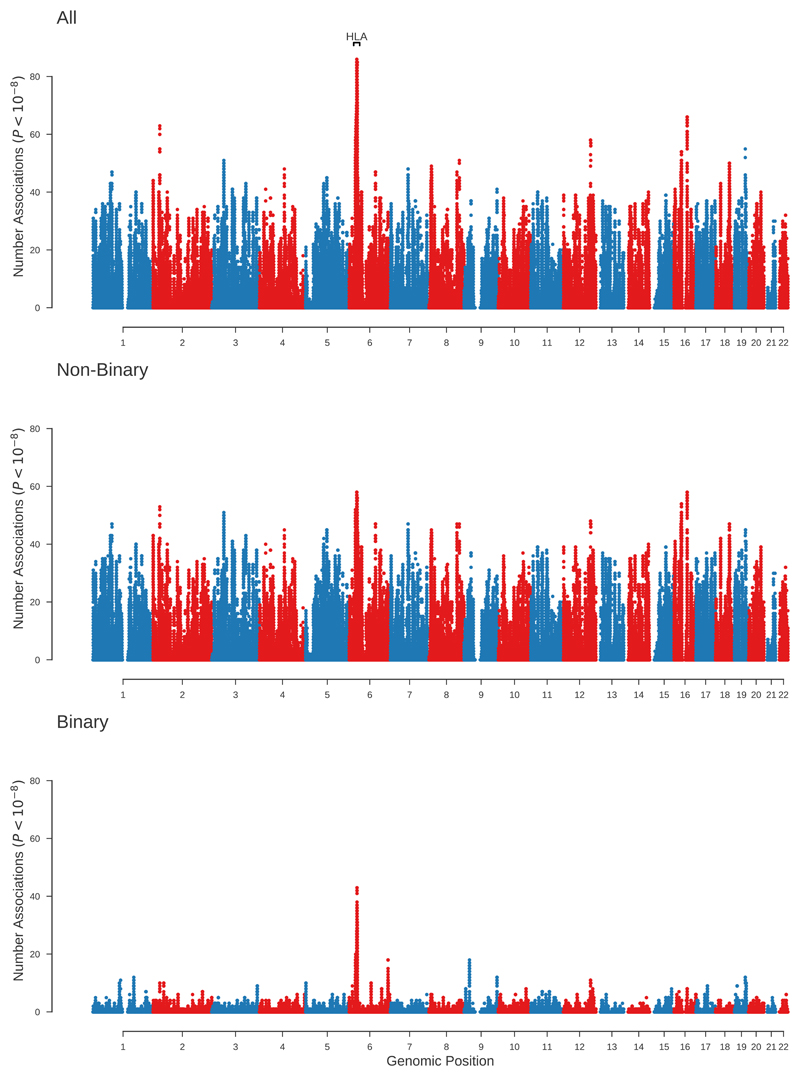
The panels show the number of significant associations at each tested genetic variant for all traits, non-binary and binary phenotypes. The HLA region (±10Mb) is indicated.
Considering the criteria for inclusion of genetic polymorphisms on the genotyping array (Supplementary Table 5), the HLA polymorphisms were the most enriched for associations with at least one non-binary trait (88% had a P<10-8), followed by the Cardiometabolic, Autoimmune/Inflammatory and ApoE criteria, whilst the lowest enrichment was for two low frequency variants categories (“Genome-wide coverage for low frequency variants” and “Rare, possibly disease causing, mutations”). Less than 8 in 100 of these polymorphisms were associated with any non-binary trait (Supplementary Table 5).
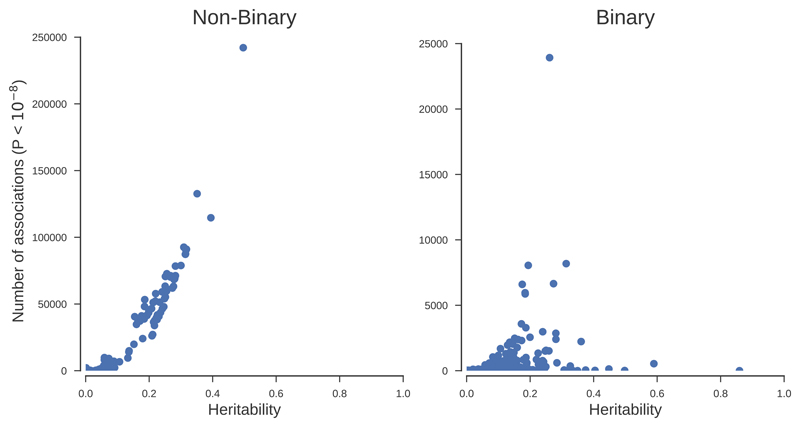
We found a significant correlation (r=0.93, P<10-51) between the number of hits and the SNP heritability of the traits, suggesting that the number of loci affecting a trait might be proportional to the heritability of the trait (Fig. 4, Supplementary Fig. 4). Consistent with this model and variation in the distribution of linkage disequilibrium across the genome, the correlation of the SNP heritability with the number of identified independent lead variants was similarly high (r=0.88, P<10-38). The number of hits (P<10-8) per chromosome was highly correlated (r=0.86) with the length of the chromosome covered by the genotyped SNPs (Supplementary Fig. 5, Supplementary Table 6). Although this correlation could arise under a polygenic model where the length of the chromosome is correlated with the number of possible variants affecting the traits, the simplest explanation is that it arises as a consequence of the correlation of chromosomal length and number of tested variants per chromosome. Comparing the fit of two nested models to explain the number of hits per chromosome as a function of number of tested genetic variants and length of the chromosome or just the number of genetic variants was consistent with the number of GWAS hits per chromosome correlating with the length of the chromosome rather than the number of tested variants (Methods).
HLA and surrounding 10Mb region were excluded for non-binary and binary phenotypes respectively.
Standing height was the trait with the largest number of hits (Fig. 5) with 261,908 significantly associated variants distributed across 10,374 independent lead variants. We estimated that the leading polymorphisms across the 118 traits studied are distributed among 38,651 independent loci, therefore 27% of these independent loci contribute to the variation of height, as expected by a highly polygenic trait7. We also computed the proportion of tested genetic variants associated with at least one disease (P < 10-8) that are also associated with height and BMI at different thresholds (Supplementary Table 7). At a threshold of 10-8, ~28% and ~7% of the genetic variants associated for height and BMI, respectively, were also associated with at least one disease. This is important for the interpretation of Mendelian Randomisation studies as it is likely that one of the critical assumptions to demonstrate causality, that is, that there is no pleiotropy between the exposure and the outcome, may be broken for many exposure-outcome pairs.
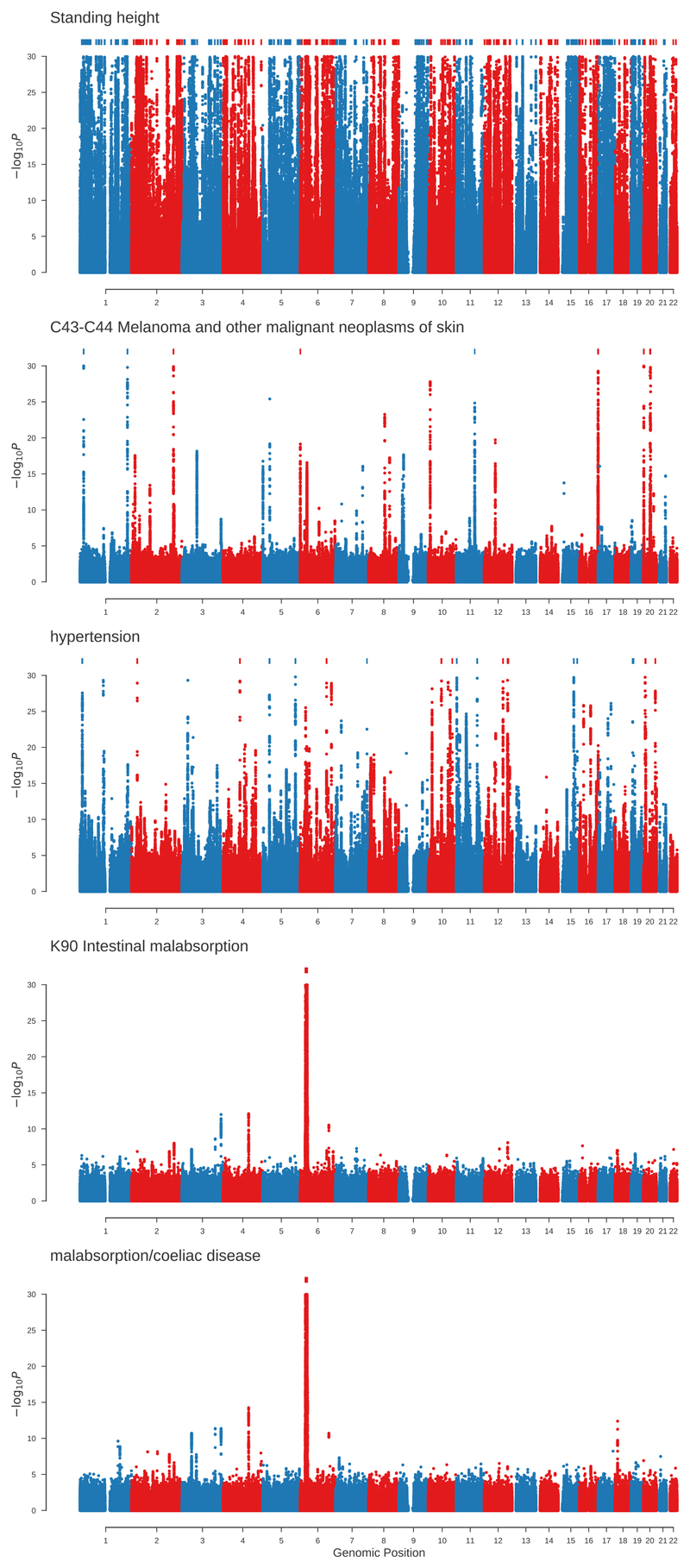
Manhattan plots for the phenotypes with the largest number of genome wide significant associations (two-sided t-test, P < 10-8) within each of these categories: non-binary phenotypes, cancer registry, self-reported non-cancer illness, clinically defined disease from hospital episode statistics and matching self-reported disease to the clinically defined disease from hospital episode statistics. From top to bottom: non-binary phenotypes (Standing height), cancer registry (Melanoma and other malignant neoplasms of skin), self-reported non-cancer illness (hypertension), clinically defined malabsorption, and self-reported malabsorption. Genetic variants with P < 10-30 are indicated by marks along the top of each plot.
Distribution of GWAS hits among binary traits
The binary trait with the largest number of cases was self-reported hypertension, with an average across binary traits of 6,593 cases (Supplementary Table 1). Of the 660 binary phenotypes 86 were specific to one sex (Supplementary Table 1). Individuals of the unaffected sex were excluded from the analysis for these phenotypes (Methods). Consistent with the reduced statistical power to detect association with binary phenotypes (mainly diseases) compared to non-binary traits we detected 393,023 associations at a P<10-8 (Supplementary Table 2), 61% of those were within the HLA region. Similarly, almost half (i.e. 48%) of the analyzed imputed HLA alleles were significant (P<10-8) for at least one binary trait (Supplementary Fig. 3). Approximately 1 in 15,000 of the genotype-phenotype pairs was genome-wide significant (P<10-8) for binary traits, whilst approximately 1 in 200 genotype-phenotype pairs were significant (P<10-8) for non-binary traits. Among the tested genetic variants, one in ~80 was associated with at least one binary trait, whilst one in ~10 was associated with one non-binary trait. Only genetic variants within the HLA region were associated with more than 20 binary traits each (Figs. 3, Supplementary Fig. 1 and 6).
We found a positive correlation (r=0.64, P<10-76 in the observed scale, r=0.56, P<10-53 in the liability scale) between the heritability of the binary trait and the number of genome-wide significant variants, albeit of smaller magnitude to that found for the non-binary traits (Fig. 4). Some of these traits were obvious outliers as they had large heritabilities but few significantly associated variants. The three largest heritabilities for binary traits were for three autoimmune diseases (ankylosing spondylitis, coeliac disease and seropositive rheumatoid arthritis) but few significant variants were found outside the HLA region for these traits. For instance, 5,704 out of 5,706 genome-wide significant associations for ankylosing spondylitis were within the HLA region.
Among the categories for inclusion of genetic variants in the genotyping array there was a substantial enrichment for HLA (79%), ApoE (48%), and Cancer common variants (40%). The categories with the lowest enrichment were genome-wide coverage for low frequency variants (0.15%) and tags for Neanderthal ancestry (0.8%) (Supplementary Table 5).
We show three examples of Manhattan plots for binary traits (Fig. 5). The first example shows where there are associations with skin cancer (i.e melanoma and other malignant neoplasms of the skin). There are 4795 variants associated (P<10-8) with skin cancer distributed among 172 independent lead variants (Supplementary Table 3). We found associations in genetic variants in or around known susceptibility genes (e.g. MC1R, IRF4, TERT, TYR) for melanoma8, but also genes like FOXP1 (rs13316357, P=1.5x10-15) associated with basal cell carcinoma9. The other two examples show the similarity between the results of one of the self-reported and clinically defined traits available in UK Biobank. The Manhattan plots for self-reported and clinically defined coeliac disease are very similar but not identical, which suggests that generally there will be benefit in analyzing both clinically and self-reported traits.
Heritability Estimates
Heritability estimates inform about the contribution of genetics to the observed phenotypic variation. The heritability of many of the 778 traits analysed here has never been reported, but even if they have been reported it is useful to know how much phenotypic variation is captured by genetic variants in a cohort of the size and interest of UK Biobank. The majority (78%) of the traits analyzed had a significant SNP-heritability (P<0.05; Fig. 6), with the largest SNP-heritability being for ankylosing spondylitis, which was 0.86 on the liability scale. The mean and median heritability among those estimates that were significant were 0.12 and 0.08, respectively. Mean heritabilities were significantly different for binary and non-binary traits (h2Non-binary=0.17; h2Binary=0.10; P=4x10-12). A total of thirty-six traits, all binary, had a heritability estimate close to zero (h2Liability < 10-4). Only seven of those thirty-six traits had no genome-wide significant hits (P<10-8), with nine having more than ten significant hits, self-reported gastritis having the largest number of hits with 41. This scenario could arise for monogenic and oligogenic traits for which the model assumptions do not hold or because of false positives. The Manhattan plots for the traits that had the largest numbers of hits seem more consistent with these hits being false positives or perhaps lack of power to detect heritability than with the violation of the model assumptions (Supplementary Fig. 7).
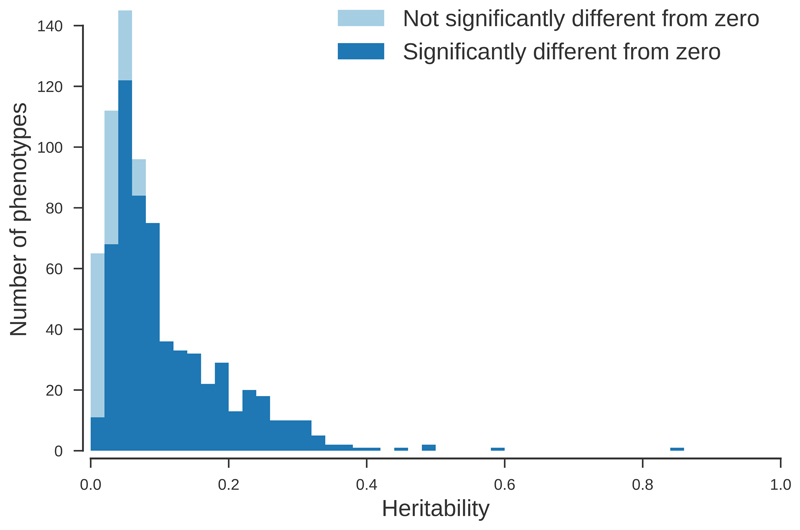
Colours indicate the fraction of phenotypes with heritability significantly (P < 0.05, Chi-squared test, see Online Methods for details) different from zero in each bin.
Estimates of genetic and environmental correlations show that for 15% of the pairs of non-binary traits the genetic and environmental correlation changes sign (Supplementary Fig. 8, GeneATLAS web page). Across all pairs of non-binary traits for which the genetic and environmental correlation had the same sign the absolute value of the genetic correlation was smaller in 31% of the cases. Overall, taking into account the size of observed heritabilities, this suggests that the phenotypic covariance of many of these traits is likely driven by the environment and not genetics (average (covg/cove)=0.24, among traits where covg and cove have the same sign).
Phenotypic prediction from genetic markers
We computed genomic predictions (that is, models of phenotypic prediction based on genetic markers) for all 692 non-gender dependent traits using Genomic Best Linear Predictions (GBLUP)10 (Methods). GBLUP estimates polygenic risk scores assuming that all fitted variants have an effect. It has been argued that this method has several advantages to traditional polygenetic risk scores from GWAS hits10,11. Some of the traits for which we developed GBLUP models did indeed reach large prediction accuracies (Fig. 7), which was further increased when we used additional covariates such as gender or sex. The largest prediction accuracy for a non-binary trait was for height which was 0.59, whilst the largest discriminative ability for a binary trait was 0.82 for self-reported malabsorption/coeliac disease. We observed a large correlation between the prediction accuracy and the trait heritability (Fig. 7 and Supplementary Table 8). Furthermore, we previously developed a model that predicted the benefit of having increasingly large training datasets for prediction of complex traits in UK Biobank11,12. Our current accuracy of prediction for anthropomorphic traits is very similar to the ones we previously predicted we would achieve with this training set11 (Supplementary Fig. 9).
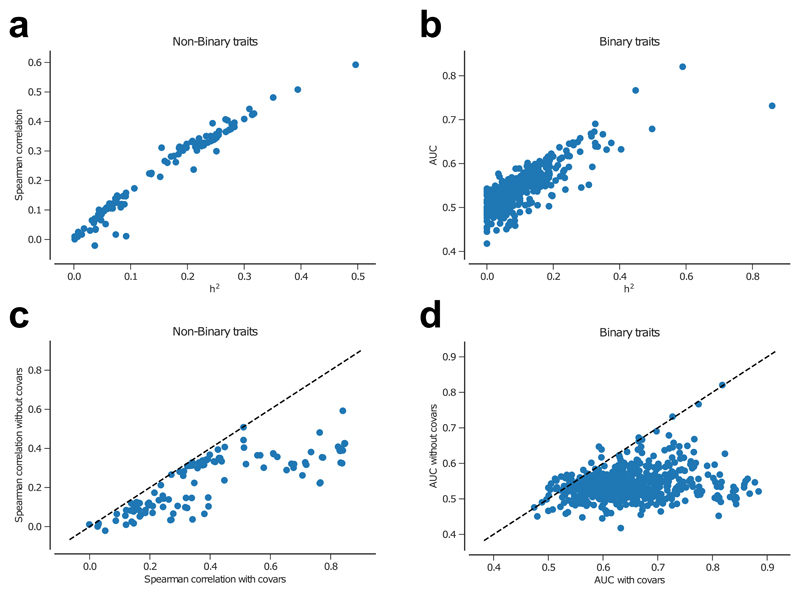
Accuracy of phenotypic prediction as a function of the estimated SNP-heritability for (a) non-binary traits and (b) binary traits when no covariates were used for prediction. Comparison between prediction accuracy when covariates are included or not included for (c) non-binary traits and (d) binary traits.
Discusion
We used circa 452,000 related and unrelated UK Biobank participants of white ethnicity to build the largest atlas of genetic associations to date. Summary statistics for 778 traits will be available to the research community to help them gain further insight into the genetic architecture of complex traits. Unlike other currently available databases, like the GWAS catalog (which contains ~39,366 unique SNP-trait associations), our database includes significant and non-significant associations, thus providing an unbiased view of phenotype-genotype associations across a large number of traits within a single cohort. In addition, the database contains 182,266 independent genotype-phenotype associations, genetic and environmental correlations, and estimates of SNP heritability to allow researchers to perform their own filters on what a meaningful association or heritability is. We hope this database will be useful to those working on complex traits genetics, but also to those that have not got the expertise or capabilities to perform analyses at this scale.
Online Methods
Phenotypes
In total we analysed 778 phenotypes in UK Biobank participants of white ethnicity. These included 657 binary phenotypes generated from self-reported disease status (UK Biobank field 20002), ICD10 codes from hospitalization events (UK Biobank fields 41202 and 41204), and ICD10 codes from cancer registries (UK Biobank fields 40006), as well as a further 3 binary and 118 non-binary (comprising continuous and ordered integral measures) phenotypes from across the UK Biobank. Amongst the 660 binary phenotypes 86 exhibited either a complete lack of cases in one sex or a strong imbalance in prevalence in the two sexes, i.e, the ratio between the smaller and larger prevalence was <0.02. Of these 86 phenotypes 72 where specific to women. We only included individuals of the appropriate sex, i.e., the sex with higher prevalence, in the analysis of these sex specific phenotypes. A description of each phenotype, its category and the relevant UK Biobank fields can be found in Supplementary Table 1 and Gene ATLAS website. The non-binary phenotypes were not scale transformed, so the units of the effect sizes are in the units reported in the UK Biobank database. The phenotypes for individuals with negative coding were replaced with the corresponding value (Supplementary Table 9). We also ordered the keys for the ordinal phenotypes with unordered keys in the UK Biobank database (Supplementary Table 10). The individuals with a phenotype departing 10 standard deviations from their gender mean were set as missing for traits with a value type defined as “Integer” or “Continuous” by UK Biobank. The exceptions to this were Number of self-reported cancers (134-0.0), Number of self-reported non-cancer illnesses (135-0.0), Nucleated red blood cell percentage (30230-0.0), Nucleated red blood cell count (30170-0.0), and Frequency of solarium/sunlamp use (2277-0.0) which were left as reported by UK Biobank. Some of the traits analysed have some redundancy that has been left for completeness. That is, some of these traits were measured in different ways during the study (e.g. weight) or are analysed as self-reported traits and clinical traits (e.g. malabsorption). For disease traits all individuals reporting a disease code were coded as cases with all other individuals considered controls. Only non-disease phenotypes with missing data rate < 5% were selected for analysis. For these phenotypes missing values were imputed to the age and sex specific mean in the study cohort.
Analysis Checks
Extensive validation steps were performed to ensure the reliability of the data (Supplementary Material). These steps included, for instance, a comparison of effect sizes with previous results from GWAS published in GWAS Catalog (Supplementary Figs. 10-18), the investigation of how the polygenicity of the traits drive inflation factors in GWAS (Supplementary Fig. 19), and comparisons with repeated analyses where the non-binary phenotypes containing at least 500 different values were transformed using a rank-based normal transformation (Supplementary Note, Supplementary Table 11, and Supplementary Fig. 20). The results are in good agreement. Since the statistical power may be different in some cases, the results are available at the GeneATLAS web. Furthermore, the comparison between our heritability estimations with previously published heritabilities showed a good agreement (Supplementary Fig. 21 and Supplementary Table 12) when comparing ten traits. In addition, we computed the Q-Q plots (Supplementary Fig. 22, and summary plots in GeneATLAS website). We also checked whether there were any areas depleted of associations, that is, that showed few significant associations (Supplementary Fig. 23 and 24). Finally, we compared the coherence of the effect size directions estimated with the whole cohort and subsets of it of different sizes (Supplementary Table 13).
Genotypes
The genotypes of the UK Biobank participants were assayed using either of two genotyping arrays, the Affymetrix UK BiLEVE Axiom or Affymetrix UK Biobank Axiom array. These arrays were augmented by imputation of ~90 million genetic variants from the Haplotype Reference Consortium5, the thousand genomes13 and the UK 10K13 projects. Full details regarding these data have been published elsewhere14.
We excluded individuals who were identified by the UK Biobank as outliers based on either genotyping missingness rate or heterogeneity, whose sex inferred from the genotypes did not match their self-reported sex and who were not of white ancestry (based on both, self-reported ethnicity and those from whom one of the two first genomic principal components did not fall within 5 standard deviations from the mean). Finally, we removed individuals with a missingness >5% across variants which passed our quality control procedure and those that have a missing phenotype for 40 or more traits. The resulting study cohort comprised 452,264 individuals.
From the genotyped data we only retained bi-allelic autosomal variants which were assayed by both genotyping arrays employed by UK Biobank. We furthermore excluded variants which had failed UK Biobank quality control procedures in any of the genotyping batches. Additionally, for imputed and genotyped variants, we excluded variants with P < 10-50 for departure from Hardy-Weinberg, computed on a subset of 344,057 unrelated (Kinship coefficient < 0.0442) individuals in the White-British subset of the study cohort, and with a missingness rate > 2% in the study cohort. Although we analysed all imputed variants and all genotyped variants with MAF > 10-4 (all results available on the GeneATLAS website), only imputed variants with MAF>10-3 in the study cohort and imputation score larger than 0.9 were used for the summary results presented here. This cut-off corresponds to less than 905 occurrences of the minor allele in the study cohort. We also filtered the HLA imputed alleles that were present in fewer than 10 individuals.
GWAS Analysis
To test each genetic variant whilst taking into account population structure in UK Biobank (e.g. presence of related individuals or local structure), we used a Linear Mixed Model. Specifically, the model takes the form
where y is the vector of phenotypes, X, is the matrix of fixed effects, and β the effect size of these effects. We included as fixed effects sex, array batch, UK Biobank Assessment Center, age, age2, and the leading 20 genomic principal components as computed by UK Biobank. g is the polygenic effect that captures the population structure, fitted as a random effect. It follows the distribution
Fitting one instance of such a LMM model is computationally very demanding. Following a naïve approach, the required computational time increasing with the cube of the sample size, ~O(N3), and the memory requirements with the square of the sample size, ~O(N2). Consequently, fitting a single model on a cohort of the size of UK Biobank is challenging, and fitting millions of these models, one for each analysed genetic variant and phenotype is not feasible with standard computational and statistical approaches. To address this problem, we took advantage of three different tools. First, we used a large supercomputer, and DISSECT3 to speed up the calculations (e.g. computing the GRM eigen-decomposition required 5,040 processor cores working together for ~10h, and using ~5TB of memory). Second, we computed the full eigen decomposition of the GRM, A = ΛΣΛT, where Λ is the matrix of eigenvectors, and Σ is a diagonal matrix containing the eigenvalues. This allowed us to transform all the other model matrices, y, X, and ϵ to the new space where the GRM is diagonal. Although the eigen-decomposition is a computationally intensive process, once diagonalized, the computational time of fitting a model is reduced considerably to ~O(N), thus enabling us to perform several tests using Mixed Linear Models on a cohort of hundreds of thousands of individuals. Finally we performed over 23 billion tests using a two-step approximation that optimizes the computational resources15. The first step of the approximation fits a LMM that adjusts by the relevant fix (e.g. age, sex, etc.) and random effects (genetic effects) to each trait, the second step uses the residuals of LMM to test (two-tailed t-test on effect sizes) all available genetic markers for significance in a linear model. We corrected for the polygenic effect using a Leave-One-Chromosome-Out (LOCO) approach16.
HLA Region
We defined the HLA region as the region of chromosome 6 spanning base pairs 28,866,528 to 33,775,446. Throughout all analyses we included 10Mb either side of the above HLA region to account for LD with variants outside this region.
The imputed HLA alleles were tested using the same GWAS model described above, where the independent variable is the best guess allele reported dosage from the HLA imputed values (UK Biobank field 22182). We tested the alleles using two models. A model where the number of copies of each HLA allele for each locus was tested independently as a fixed effect, and a second model where the number of copies of all alleles in a given locus were tested together as fixed effects in the same model (i.e. an omnibus test)17.
Estimation of Genetic Parameters
In order to estimate heritabilities and genetic correlations we fitted LMMs for each trait with a GRM containing all common (MAF > 5%) autosomal genetic variants which passed QC. The heritability was estimated as
Lead variants and Independent Loci
We clustered GWAS results into independent lead variants using the --clump option of the PLINK 1.9 software20,21. Specifically, for each trait individually, we clustered GWAS results by selecting genome wide significant variants as lead variants and assigning to them unassigned variants within 10Mb, that have P<10-2 and a r2 > 0.3 with the lead variant. To compute the total number of independent loci across all traits, we performed the same clustering on the lead variants across all traits, choosing the lowest p-value for variants which were lead variants in different traits.
Relation of number of associations and chromosome length
We regressed the number of significant associations (P<10-8) across traits for each chromosome on the covered length of the chromosome, i.e., distance in base pairs of the first and last tested genetic variants, and the number of genetic variants tested on the chromosome. For chromosome 6 we excluded the HLA region and variants contained therein from the statistics. We compared the full model to one with either the chromosomal length or number of tested genetic variants removed using the likelihood ratio test. The full model was not significantly better than the model containing only chromosomal length (P=0.08) but was significantly better than the model containing only the number of genetic variants (P=0.004). Both reduced models were significant when compared to a null model containing only an intercept.
Phenotypic prediction
The effect of all common genetic variants (MAF>0.05) were estimated together as a random effect using the model,
where μ is the mean term and ei the residual for individual i. L is the number of fixed effects, xil being the value for the fixed effect l at individual i and βl the estimated effect of the fixed effect l. We fitted the same covariates as in the GWAS analyses. M is the number of markers and zij is the standardised genotype of individual i at marker j. The vector of effects of random common genetic variants a is distributed as
The prediction of the phenotype
where sij is the number of copies of the reference allele at marker j of individual i, M is the number of markers used for the prediction, and aj the effect of marker j.
We used 407,669 genetically confirmed white British to train the models and 44,595 whites of non-British descent to validate the models. We restricted this analysis to the 692 non-gender specific phenotypes. Prediction accuracies for non-binary traits were computed as the Spearman correlation between the predicted and the real phenotype of white participants of non-British descent after correcting by the estimated effect of the used covariates. Prediction accuracies for binary traits were computed as the Area Under the Curve (AUC) of a Receiver Operating Characteristic (ROC) curve using the predicted and the real phenotypes of white individuals of non-British descent.
Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary linked to this article.
Code availability
The source code of DISSECT, the tool 700 used for GWAS and heritability estimations, is freely available at https://www.dissect.ed.ac.uk under GNU Lesser General Public License v3.
Data availability
All summary results from the analyses performed are available at GeneATLAS website, http://geneatlas.roslin.ed.ac.uk/.
Supplementary Material
Supl Tabl 3
Supl Tables
Supl Information
Acknowledgements
This research has been conducted using the UK Biobank Resource under project 788. The work was funded by the Roslin Institute Strategic Programme Grant from the BBSRC (BB/P013732/1) and MRC grant (MR/N003179/1) granted to AT. AT also acknowledge funding from the Medical Research Council and OCX from MRC fellowship MR/R025851/1. Analyses were performed using the ARCHER UK National Supercomputing Service.
Footnotes
Accession Codes
This research has been conducted using the UK Biobank Resource under project 788.
Author Contributions
All authors contributed equally to the design, running of the analyses, and writing of the manuscript.Competing Interest Statement
The authors declare no competing financial interests.
Ethical Compliance
The UK Biobank project was approved by the National Research Ethics Service Committee North West-Haydock (REC reference: 11/NW/0382). An electronic signed consent was obtained from the participants.
URLs
GeneATLAS, http://geneatlas.roslin.ed.ac.uk; UK Biobank, http://www.ukbiobank.ac.uk/; ARCHER UK National Supercomputing Service, http://www.archer.ac.uk; DISSECT, https://www.dissect.ed.ac.uk; GWAS catalog https://www.ebi.ac.uk/gwas/; Affymetrix array https://affymetrix.app.box.com/s/6gc2mcw2s6a7zbb7wijn; PLINK, http://zzz.bwh.harvard.edu/plink/ and http://www.cog-genomics.org/plink/1.9/). BGENIX and BGEN reference implementation, https://bitbucket.org/gavinband/bgen.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/s41588-018-0248-z
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc6707814?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/s41588-018-0248-z
Article citations
Investigating potential drug targets for IgA nephropathy and membranous nephropathy through multi-queue plasma protein analysis: a Mendelian randomization study based on SMR and co-localization analysis.
BioData Min, 17(1):49, 08 Nov 2024
Cited by: 0 articles | PMID: 39516845 | PMCID: PMC11545554
Adverse effects of CXCR2 deficiency in mice reared under non-gnotobiotic conditions.
Sci Rep, 14(1):26159, 30 Oct 2024
Cited by: 0 articles | PMID: 39478033 | PMCID: PMC11525579
Accounting for genetic effect heterogeneity in fine-mapping and improving power to detect gene-environment interactions with SharePro.
Nat Commun, 15(1):9374, 30 Oct 2024
Cited by: 1 article | PMID: 39478020 | PMCID: PMC11526169
A MR-PheWAS and bidirectional Mendelian randomization study: Exploring for causal relationships of pancreatic cancer.
Medicine (Baltimore), 103(41):e40047, 01 Oct 2024
Cited by: 0 articles | PMID: 39465831 | PMCID: PMC11479532
Review Free full text in Europe PMC
The goldmine of GWAS summary statistics: a systematic review of methods and tools.
BioData Min, 17(1):31, 05 Sep 2024
Cited by: 0 articles | PMID: 39238044 | PMCID: PMC11375927
Go to all (322) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
SNPs (2)
- (2 citations) dbSNP - rs1421085
- (1 citation) dbSNP - rs13316357
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Analyses of biomarker traits in diverse UK biobank participants identify associations missed by European-centric analysis strategies.
J Hum Genet, 67(2):87-93, 11 Aug 2021
Cited by: 24 articles | PMID: 34376796 | PMCID: PMC8792153
Transcriptome-wide association study in UK Biobank Europeans identifies associations with blood cell traits.
Hum Mol Genet, 31(14):2333-2347, 01 Jul 2022
Cited by: 8 articles | PMID: 35138379 | PMCID: PMC9307312
Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N=112 117).
Mol Psychiatry, 22(10):1376-1384, 25 Jul 2017
Cited by: 236 articles | PMID: 28937693 | PMCID: PMC5622124
The UK Biobank: A Shining Example of Genome-Wide Association Study Science with the Power to Detect the Murky Complications of Real-World Epidemiology.
Annu Rev Genomics Hum Genet, 23:569-589, 04 May 2022
Cited by: 8 articles | PMID: 35508184
Review
Funding
Funders who supported this work.
Biotechnology and Biological Sciences Research Council (1)
Grant ID: BB/P013732/1
Medical Research Council (7)
Grant ID: HDR-5013
Genomic prediction of anthropomorphic traits using hundreds of thousands of individuals
Dr Albert Tenesa, University of Edinburgh
Grant ID: MR/N003179/1
Vast-scale linear mixed modelling genetic discovery approaches for genome- and exome-wide association analyses to enable therapeutic target validation
Dr Oriol Canela-Xandri, University of Edinburgh
Grant ID: MR/R025851/1
Grant ID: HDR-9004
UK Biobank
Professor Sir Rory Collins, UK Biobank
Grant ID: MC_QA137853
Understanding disease through environment-wide association studies
Dr Albert Tenesa, University of Edinburgh
Grant ID: MR/P015514/1
UK Biobank (core renewal)
Professor Sir Rory Collins, UK Biobank
Grant ID: MC_PC_17228





