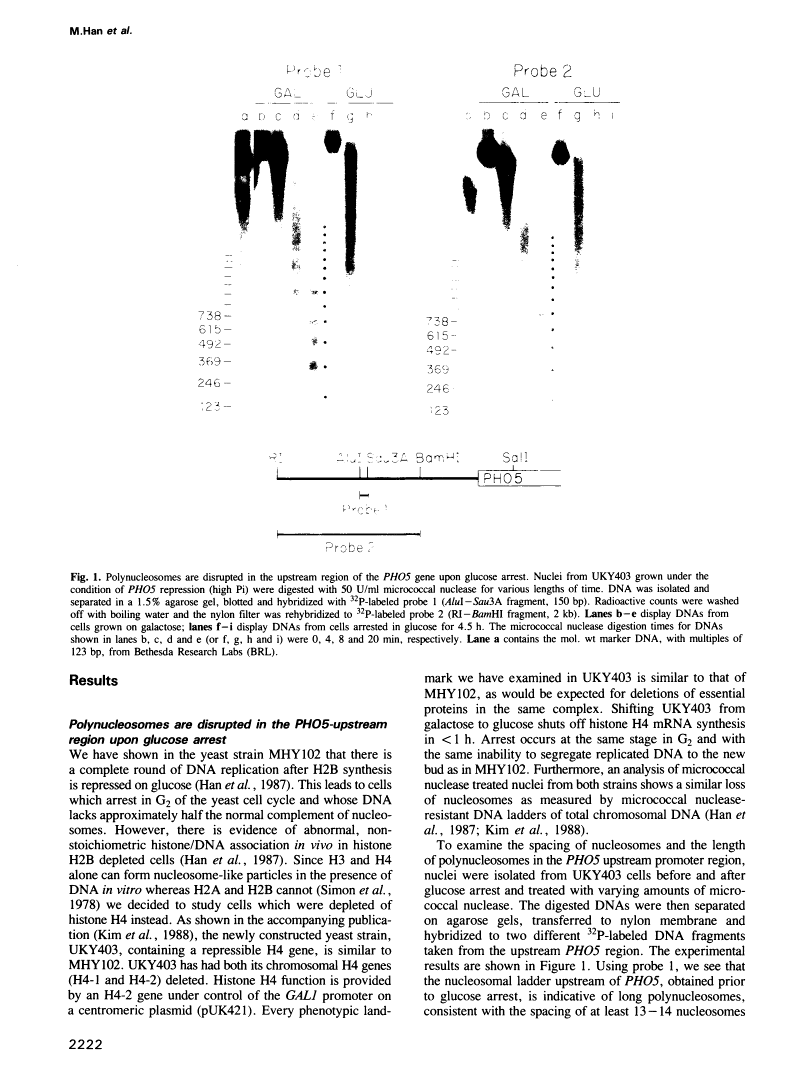
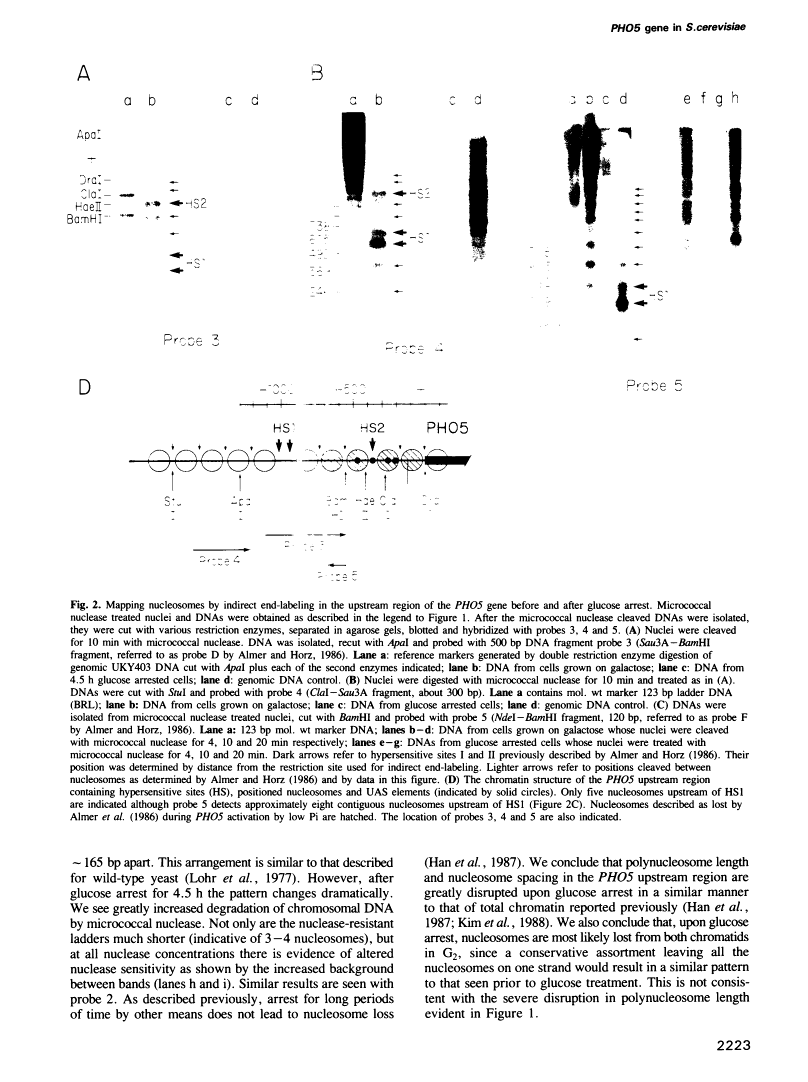
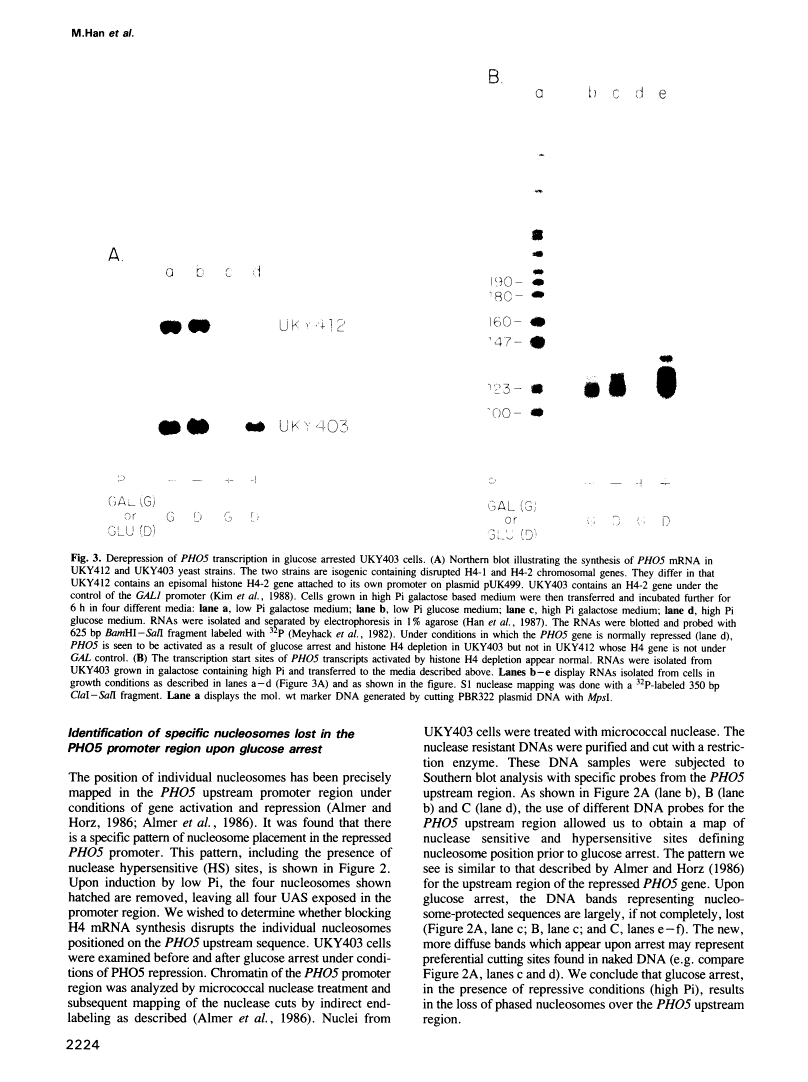
Abstract
Free full text

Depletion of histone H4 and nucleosomes activates the PHO5 gene in Saccharomyces cerevisiae.
Abstract
We have previously constructed a yeast strain (UKY403) whose sole histone H4 gene is under control of the GAL1 promoter. This yeast arrests in G2 upon glucose treatment as a result of histone H4 depletion. The yeast PHO5 gene contains phase nucleosomes covering promoter (UAS) sequences in the PHO5 repressed state and it has been suggested that nucleosomes prevent the binding of positively acting factors to these UAS sequences. Using UKY403 we examined the length of polynucleosomes and nucleosome phasing in the PHO5 upstream region by the use of micrococcal nuclease and indirect end-labeling. It was found that glucose arrest led to a severe disruption in PHO5 chromatin structure and that most nucleosomes had their position altered or were lost from the PHO5 promoter region. Cell undergoing nucleosome depletion synthesized large quantities of accurate PHO5 transcripts even under repressive, high inorganic phosphate conditions. Histone H4 depletion did not appear to affect the repression or activation of another inducible yeast gene, CUP1. Arrest with landmarks in early G1 (in the cell division cycle mutant cdc28) or in various stages of G2 (in cdc15, cdc17 and cdc20) does not activate PHO5; nor does arrest due to chromosome topology changes (in top2 or the top1top2 topoisomerase mutants). cdc14, which has its arrest landmark at a similar point in the cell cycle as cdc15, does derepress PHO5. However, since it also leads to derepression of CUP1 it is probably functioning through an independent mechanism. Therefore, our data suggest that nucleosomes regulate PHO5 transcription.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.7M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Click on the image to see a larger version.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Simon RH, Camerini-Otero RD, Felsenfeld G. An octamer of histones H3 and H4 forms a compact complex with DNA of nucleosome size. Nucleic Acids Res. 1978 Dec;5(12):4805–4818. [Europe PMC free article] [Abstract] [Google Scholar]
- Smith MM, Andrésson OS. DNA sequences of yeast H3 and H4 histone genes from two non-allelic gene sets encode identical H3 and H4 proteins. J Mol Biol. 1983 Sep 25;169(3):663–690. [Abstract] [Google Scholar]
- Struhl K. Promoter elements, regulatory elements, and chromatin structure of the yeast his3 gene. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):901–910. [Abstract] [Google Scholar]
- Szent-Györgyi C, Finkelstein DB, Garrard WT. Sharp boundaries demarcate the chromatin structure of a yeast heat-shock gene. J Mol Biol. 1987 Jan 5;193(1):71–80. [Abstract] [Google Scholar]
- Toh-e A, Inouye S, Oshima Y. Structure and function of the PHO82-pho4 locus controlling the synthesis of repressible acid phosphatase of Saccharomyces cerevisiae. J Bacteriol. 1981 Jan;145(1):221–232. [Europe PMC free article] [Abstract] [Google Scholar]
- Weintraub H, Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. [Abstract] [Google Scholar]
- Workman JL, Roeder RG. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell. 1987 Nov 20;51(4):613–622. [Abstract] [Google Scholar]
- Wu C, Gilbert W. Tissue-specific exposure of chromatin structure at the 5' terminus of the rat preproinsulin II gene. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1577–1580. [Europe PMC free article] [Abstract] [Google Scholar]
- Almer A, Hörz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 1986 Oct;5(10):2681–2687. [Europe PMC free article] [Abstract] [Google Scholar]
- Almer A, Rudolph H, Hinnen A, Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986 Oct;5(10):2689–2696. [Europe PMC free article] [Abstract] [Google Scholar]
- Benezra R, Cantor CR, Axel R. Nucleosomes are phased along the mouse beta-major globin gene in erythroid and nonerythroid cells. Cell. 1986 Mar 14;44(5):697–704. [Abstract] [Google Scholar]
- Becker P, Renkawitz R, Schütz G. Tissue-specific DNaseI hypersensitive sites in the 5'-flanking sequences of the tryptophan oxygenase and the tyrosine aminotransferase genes. EMBO J. 1984 Sep;3(9):2015–2020. [Europe PMC free article] [Abstract] [Google Scholar]
- Berk AJ, Sharp PA. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. [Abstract] [Google Scholar]
- Kramer RA, Andersen N. Isolation of yeast genes with mRNA levels controlled by phosphate concentration. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6541–6545. [Europe PMC free article] [Abstract] [Google Scholar]
- Brill SJ, DiNardo S, Voelkel-Meiman K, Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature. 326(6111):414–416. [Abstract] [Google Scholar]
- Cusick ME, DePamphilis ML, Wassarman PM. Dispersive segregation of nucleosomes during replication of simian virus 40 chromosomes. J Mol Biol. 1984 Sep 15;178(2):249–271. [Abstract] [Google Scholar]
- Drlica K. Biology of bacterial deoxyribonucleic acid topoisomerases. Microbiol Rev. 1984 Dec;48(4):273–289. [Europe PMC free article] [Abstract] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. [Abstract] [Google Scholar]
- Garel A, Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. [Europe PMC free article] [Abstract] [Google Scholar]
- Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. [Abstract] [Google Scholar]
- Han M, Chang M, Kim UJ, Grunstein M. Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell. 1987 Feb 27;48(4):589–597. [Abstract] [Google Scholar]
- Holm C, Goto T, Wang JC, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985 Jun;41(2):553–563. [Abstract] [Google Scholar]
- Kim UJ, Han M, Kayne P, Grunstein M. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. EMBO J. 1988 Jul;7(7):2211–2219. [Europe PMC free article] [Abstract] [Google Scholar]
- Koren R, LeVitre J, Bostian KA. Isolation of the positive-acting regulatory gene PHO4 from Saccharomyces cerevisiae. Gene. 1986;41(2-3):271–280. [Abstract] [Google Scholar]
- Labarca C, Paigen K. A simple, rapid, and sensitive DNA assay procedure. Anal Biochem. 1980 Mar 1;102(2):344–352. [Abstract] [Google Scholar]
- Lemire JM, Willcocks T, Halvorson HO, Bostian KA. Regulation of repressible acid phosphatase gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Aug;5(8):2131–2141. [Europe PMC free article] [Abstract] [Google Scholar]
- Lohr D. Organization of the GAL1-GAL10 intergenic control region chromatin. Nucleic Acids Res. 1984 Nov 26;12(22):8457–8474. [Europe PMC free article] [Abstract] [Google Scholar]
- Lohr D, Hereford L. Yeast chromatin is uniformly digested by DNase-I. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4285–4288. [Europe PMC free article] [Abstract] [Google Scholar]
- Lohr D, Corden J, Tatchell K, Kovacic RT, Van Holde KE. Comparative subunit structure of HeLa, yeast, and chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1977 Jan;74(1):79–83. [Europe PMC free article] [Abstract] [Google Scholar]
- Lorch Y, LaPointe JW, Kornberg RD. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987 Apr 24;49(2):203–210. [Abstract] [Google Scholar]
- Nakao J, Miyanohara A, Toh-e A, Matsubara K. Saccharomyces cerevisiae PHO5 promoter region: location and function of the upstream activation site. Mol Cell Biol. 1986 Jul;6(7):2613–2623. [Europe PMC free article] [Abstract] [Google Scholar]
- Pederson DS, Shupe K, Gorovsky MA. Changes in chromatin structure accompany modulation of the rate of transcription of 5S ribosomal genes in Tetrahymena. Nucleic Acids Res. 1984 Nov 26;12(22):8489–8507. [Europe PMC free article] [Abstract] [Google Scholar]
- Proffitt JH. DNase I-hypersensitive sites in the galactose gene cluster of Saccharomyces cerevisiae. Mol Cell Biol. 1985 Jun;5(6):1522–1524. [Europe PMC free article] [Abstract] [Google Scholar]
- Rubin GM. Three forms of the 5.8-S ribosomal RNA species in Saccharomyces cerevisiae. Eur J Biochem. 1974 Jan 3;41(1):197–202. [Abstract] [Google Scholar]
- Rudolph H, Hinnen A. The yeast PHO5 promoter: phosphate-control elements and sequences mediating mRNA start-site selection. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1340–1344. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from The EMBO Journal are provided here courtesy of Nature Publishing Group
Full text links
Read article at publisher's site: https://doi.org/10.1002/j.1460-2075.1988.tb03061.x
Read article for free, from open access legal sources, via Unpaywall:
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/j.1460-2075.1988.tb03061.x
Citations & impact
Impact metrics
Citations of article over time
Article citations
Beyond the tail: the consequence of context in histone post-translational modification and chromatin research.
Biochem J, 481(4):219-244, 01 Feb 2024
Cited by: 3 articles | PMID: 38353483 | PMCID: PMC10903488
Review Free full text in Europe PMC
Nucleosome retention by histone chaperones and remodelers occludes pervasive DNA-protein binding.
Nucleic Acids Res, 51(16):8496-8513, 01 Sep 2023
Cited by: 1 article | PMID: 37493599 | PMCID: PMC10484674
Decidualization of human endometrial stromal cells requires steroid receptor coactivator-3.
Front Reprod Health, 4:1033581, 24 Nov 2022
Cited by: 4 articles | PMID: 36505394 | PMCID: PMC9730893
Measuring the buffering capacity of gene silencing in Saccharomyces cerevisiae.
Proc Natl Acad Sci U S A, 118(49):e2111841118, 01 Dec 2021
Cited by: 6 articles | PMID: 34857629 | PMCID: PMC8670432
Whole-genome methods to define DNA and histone accessibility and long-range interactions in chromatin.
Biochem Soc Trans, 50(1):199-212, 01 Feb 2022
Cited by: 2 articles | PMID: 35166326 | PMCID: PMC9847230
Review Free full text in Europe PMC
Go to all (139) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae.
EMBO J, 7(7):2211-2219, 01 Jul 1988
Cited by: 147 articles | PMID: 3046933 | PMCID: PMC454562
Effects of Sin- versions of histone H4 on yeast chromatin structure and function.
EMBO J, 16(8):2086-2095, 01 Apr 1997
Cited by: 57 articles | PMID: 9155034 | PMCID: PMC1169811
Nucleosome disruption at the yeast PHO5 promoter upon PHO5 induction occurs in the absence of DNA replication.
Cell, 71(5):853-864, 01 Nov 1992
Cited by: 98 articles | PMID: 1423633
Interplay between nucleosomes and transcription factors at the yeast PHO5 promoter.
Semin Cell Biol, 6(4):177-183, 01 Aug 1995
Cited by: 16 articles | PMID: 8562909
Review
Funding
Funders who supported this work.
NIGMS NIH HHS (2)
Grant ID: GM23674
Grant ID: GM31336