Abstract
Free full text

An Emerging Issue in Oncogenic Virology: the Role of Beta Human Papillomavirus Types in the Development of Cutaneous Squamous Cell Carcinoma
ABSTRACT
Evidence suggests that beta human papillomaviruses (HPVs), together with ultraviolet radiation, contribute to the development of cutaneous squamous cell carcinoma. Beta HPVs appear to be not the main drivers of carcinogenesis but rather facilitators of the accumulation of ultraviolet-induced DNA mutations. Beta HPVs are promoters of skin carcinogenesis, although they are dispensable for the maintenance of the malignant phenotype. Therefore, beta HPV represents a target for skin cancer prevention, especially in high-risk populations.
INTRODUCTION
The human papillomavirus (HPV) family comprises approximately 200 types that are able to infect the mucosal and cutaneous epithelia (1). They are subdivided into genera and species in the HPV phylogenetic tree according to the DNA sequence of the late gene L1 (2). The genus Alphapapillomavirus includes the mucosal high-risk (HR) HPV types that have been clearly associated with the development of cervical and anal cancers and a subset of other genital tract cancers, such as vulvar, vaginal, and penile carcinomas, as well as a subset of head and neck cancers (3). More than 40 years of research have elucidated many of the HR HPV mechanisms involved in cancer development. The products of two early genes, E6 and E7, are major oncoproteins able to deregulate many key cellular events and greatly facilitate the malignant transformation of the infected cells. Classic examples of processes targeted by HR HPV E6 and/or E7 oncoproteins are the cell cycle, DNA repair, apoptosis, senescence, and the immune response (4). In the context of the viral life cycle, these activities of E6 and E7 are essential to guarantee viral DNA replication and progeny production. As a side effect, they facilitate the accumulation of chromosomal abnormalities, leading to cancer development. Despite the accumulation of this DNA damage, constant expression of the viral oncogenes is required for the maintenance of the transformed phenotype. Indeed, experiments in in vitro models have shown that silencing of E6 and E7 expression in cervical cancer-derived cell lines, such as CaSki or HeLa, severely affects cell viability (5,–8). Because of the biological properties of the HR HPV E6 and E7 in directly targeting several cellular proteins/pathways, HPV-positive cancer cells do not usually accumulate many mutations compared with cancers associated with other environmental factors. For instance, HPV-negative oropharyngeal cancers have more DNA mutations, which are often linked to tobacco use and/or alcohol consumption, compared with HPV-positive oropharyngeal cancers (9).
years of research have elucidated many of the HR HPV mechanisms involved in cancer development. The products of two early genes, E6 and E7, are major oncoproteins able to deregulate many key cellular events and greatly facilitate the malignant transformation of the infected cells. Classic examples of processes targeted by HR HPV E6 and/or E7 oncoproteins are the cell cycle, DNA repair, apoptosis, senescence, and the immune response (4). In the context of the viral life cycle, these activities of E6 and E7 are essential to guarantee viral DNA replication and progeny production. As a side effect, they facilitate the accumulation of chromosomal abnormalities, leading to cancer development. Despite the accumulation of this DNA damage, constant expression of the viral oncogenes is required for the maintenance of the transformed phenotype. Indeed, experiments in in vitro models have shown that silencing of E6 and E7 expression in cervical cancer-derived cell lines, such as CaSki or HeLa, severely affects cell viability (5,–8). Because of the biological properties of the HR HPV E6 and E7 in directly targeting several cellular proteins/pathways, HPV-positive cancer cells do not usually accumulate many mutations compared with cancers associated with other environmental factors. For instance, HPV-negative oropharyngeal cancers have more DNA mutations, which are often linked to tobacco use and/or alcohol consumption, compared with HPV-positive oropharyngeal cancers (9).
Similar to the HR HPV types, cutaneous beta HPV types have also been implicated in carcinogenesis, although the model of carcinogenesis is quite different. Epidemiological and biological studies support the model of synergistic cooperation between cutaneous beta HPV types and UV radiation in the development of cutaneous squamous cell carcinoma (cSCC). Importantly, beta HPV infection appears to play a role in an initial phase of skin carcinogenesis, but it is not essential for the viability of the tumor cells once they have become malignant.
Beta HPV types and epidermodysplasia verruciformis.
The first cutaneous beta HPV types, HPV5 and HPV8, were isolated from the skin of patients with epidermodysplasia verruciformis (EV), an autosomal recessive inherited skin disorder characterized by the development of wart-like lesions in several parts of the body that frequently progress to cSCC in UV-exposed areas (10). More than 50 beta HPV types have been isolated and fully characterized so far, although additional beta HPV types may exist, because partial genome sequences of novel putative beta HPV types have been reported (11, 12). Beta HPV types are subdivided into five different species, beta-1, beta-2, beta-3, beta-4, and beta-5 (1). Beta-1 and beta-2 HPVs are the most prevalent types in the skin, whereas the other species are rarely detected in the skin and comprise very few of the HPV types, i.e., beta-3 (n =
= 4), beta-4 (n
4), beta-4 (n =
= 1), and beta-5 (n
1), and beta-5 (n =
= 2) (13).
2) (13).
Epidemiology of beta HPV infection.
Similar to EV patients, organ transplant recipients (OTRs) are at high risk of beta HPV infection as a result of immunosuppression (14,–16). For example, of more than 600 organ transplant patients enrolled in two multicentered European cohort studies, approximately 50% had DNA in their eyebrow hairs (EBH) corresponding to five or more beta HPV types soon after organ transplantation (17). Beta HPV DNA is also frequently present in the skin of immunocompetent individuals, with prevalence estimates ranging from 39% to 91% (18,–20). The variation in beta HPV prevalence observed across study populations may be due to differences in the number of beta HPV types tested, the distributions of other skin cancer risk factors, and, importantly, the types of specimens analyzed. For example, a recent cross-sectional analysis of baseline results from the VIRUSCAN cohort study demonstrated that the overall prevalence of at least one beta HPV type (46 types measured) was higher in skin swabs (92%) than in EBH follicles (73%) from the same individuals. Interestingly, there was a strong correlation across the two sites with respect to the prevalence and intensity of type-specific infections, as well as the number of HPV types for which an individual was positive (21).
Beta HPV infection is present in the skin of infants and young children (16, 22) and appears to be acquired early in life, most likely through direct contact with the skin of the parents (23). Beta HPV infection can persist for 6 months or longer in plucked EBH from healthy adults (24, 25), and based on a single study that measured persistence at two anatomical sites, beta HPV has been shown to persist longer in skin swabs (11.3
months or longer in plucked EBH from healthy adults (24, 25), and based on a single study that measured persistence at two anatomical sites, beta HPV has been shown to persist longer in skin swabs (11.3 months) than in EBH (8.6
months) than in EBH (8.6 months) (18). Antibodies against the major capsid protein L1 for the most commonly detected beta HPV types in the skin are present in the blood of a large proportion of adults (26,–28). Risk factors for beta HPV infection and/or persistence include older age, history of a blistering sunburn (18, 29), and history of organ transplantation (26). Similarly, beta HPV seroreactivity is associated with skin phenotypic factors, such as poor tanning ability and cutaneous sensitivity to sunlight exposure (30, 31).
months) (18). Antibodies against the major capsid protein L1 for the most commonly detected beta HPV types in the skin are present in the blood of a large proportion of adults (26,–28). Risk factors for beta HPV infection and/or persistence include older age, history of a blistering sunburn (18, 29), and history of organ transplantation (26). Similarly, beta HPV seroreactivity is associated with skin phenotypic factors, such as poor tanning ability and cutaneous sensitivity to sunlight exposure (30, 31).
Epidemiological studies of the influence of UV radiation exposure on cutaneous HPV infection have been limited by measurement errors associated with self-reported past sun exposures and/or the use of skin phenotypes as proxies for sun-related exposure or damage. Baseline results from the VIRUSCAN cohort study (D. E. Rollison, unpublished data) demonstrate significant associations between UV radiation exposure, as measured using spectrophotometer readings of skin pigmentation, and beta HPV seropositivity, with those in the highest tertile of UV radiation exposure having a significant 93% increase in the odds of beta HPV seropositivity compared with those in the lowest tertile (odds ratio, 1.93; 95% confidence interval, 1.19 to 3.16; P for trend = 0.02). One possible explanation for these findings is the influence of UV radiation on the immune system. UV radiation exposure has been shown to correlate with subpopulations of circulating regulatory T cells (32), which could, in turn, influence beta HPV acquisition, replication, and/or persistence. Additional studies of immune function and beta HPV infection are needed, including studies conducted in immunocompetent individuals, in order to identify those at highest risk of beta HPV infection.
Independent studies provided evidence for the presence of beta HPV types in different anatomical sites other than skin. Beta HPV DNAs have been detected in the oral mucosal epithelium, eyebrow hairs, and penile and external genital lesions (reviewed in reference 33). However, there is no evidence for beta HPV involvement in pathological conditions at the anatomical sites listed above.
Epidemiological studies of beta HPV and cutaneous SCC.
As described above, the first lines of evidence for a link between beta HPV types and cSCC originated from studies of EV patients. The fact that impairment of the immune system in OTRs increases the risk of beta HPV infection and cSCC development further corroborates this association (34). A large proportion of OTRs develop cSCC within 15 years of the initiation of the immunosuppression treatment (34), correlating with multiple high-viral-load beta HPV infections in EBH (35). Most recently, extended follow-up of two multicentered European OTR study populations demonstrated that presence of DNA corresponding to five or more beta HPV types and higher viral loads in EBH soon after organ transplantation were associated with significant 70 to 80% increased risks of subsequent cSCC after more than 10
years of the initiation of the immunosuppression treatment (34), correlating with multiple high-viral-load beta HPV infections in EBH (35). Most recently, extended follow-up of two multicentered European OTR study populations demonstrated that presence of DNA corresponding to five or more beta HPV types and higher viral loads in EBH soon after organ transplantation were associated with significant 70 to 80% increased risks of subsequent cSCC after more than 10 years of follow-up (17).
years of follow-up (17).
Given that the beta HPV types are also abundantly present in the skin of the general population, epidemiological studies aiming to evaluate the role of cutaneous HPV infection in skin carcinogenesis in immunocompetent individuals have been difficult to design and have incorporated a variety of biomarkers for the measurement of past or present infection with beta HPV (seroreactivity, viral DNA in EBH, viral DNA in skin swabs, and viral DNA in tumors), further complicating the comparison of results across studies. Nevertheless, many observational studies of immunocompetent individuals have consistently demonstrated that markers of beta HPV infections are weakly, but significantly, associated with a history of cSCC. Indeed, compared with the general population, cSCC patients are more frequently positive for viral DNA corresponding to at least one beta HPV type in the skin and/or EBH, as well as for antibodies against the major capsid protein L1 (30, 36,–41). Notably, studies that also incorporated basal cell carcinoma cases observed no associations between beta HPV seropositivity and basal cell carcinoma (27, 28, 42), indicating that the observed associations are specific to cSCC.
Previous studies have included different numbers of beta HPV types in the multiplex assays used to measure HPV DNA in cutaneous tissues or circulating antibodies to HPV capsids, with more recent studies incorporating more types as novel types continue to be identified. Epidemiological studies of cutaneous HPV have conventionally defined exposure as positivity for “at least one” beta HPV type, although this approach limits the comparison of results across studies. HPV type-specific analyses are important to determine whether there are individual beta HPV types that confer a majority of the HPV-associated cSCC risk, because identification of these types would be required for vaccine development. Although not all studies have measured the same beta HPV types and/or reported type-specific associations, a recent meta-analysis of 14 case-control studies in immunocompetent individuals demonstrated that types 5, 8, 15, 17, 20, 24, 36, and 38 were statistically significantly associated with an increased risk of cSCC, with adjusted pooled odds ratios (95% confidence intervals) of 1.4 (1.18 to 1.66), 1.39 (1.16 to 1.66), 1.25 (1.04 to 1.50), 1.34 (1.19 to 1.52), 1.38 (1.21 to 1.59), 1.26 (1.09 to 1.44), 1.23 (1.01 to 1.50), and 1.37 (1.13 to 1.67), respectively (43). As more beta HPV types are discovered and incorporated into epidemiological studies of SCC risk, the probability of false discovery resulting from multiple-comparison testing increases; therefore, future studies need to incorporate larger sample sizes to detect differences after correction for false discovery. Individual HPV types consistently demonstrated to be associated with SCC risk can inform future mechanistic studies of type-specific transforming properties.
In contrast with the cancers associated with mucosal HR HPV types, in which each malignant cell contains at least one copy of viral DNA, much less than one copy of viral DNA per cancer cell has been detected in cSCC (44). These findings indicate that viral DNA is not homogeneously present in all cancer cells. In addition, quantitative PCR revealed that the viral load in cSCC is lower than that detected in the premalignant skin lesions actinic keratosis (45). In addition, a very few studies have evaluated the presence of beta HPV transcripts in skin lesions. One study showed that E6 and E7 genes of beta 8, 9, and 15 are weakly expressed in actinic keratosis and SCC (46). In contrast, no beta HPV transcripts were found in SCC by high-throughput next-generation sequencing techniques (44, 47).
Taken together, these data indicate that the beta HPV types may play a role at an early stage of skin carcinogenesis and are no longer required for the viability of the malignant cell after development of the skin tumor. This scenario is compatible with the involvement of additional carcinogens in skin cancer development. UV radiation has been established as a key risk factor for the development of nonmelanoma skin cancer (48,–50). Thus, it is possible that beta HPV types, in order to guarantee the completion of their life cycle even in the presence of cellular stresses, deregulate fundamental pathways of host cells, facilitating the accumulation of UV-induced DNA damage. Some epidemiological evidence supports this model of synergy between cutaneous beta HPV types and UV radiation exposure. For example, a U.S. population-based case-control study demonstrated greater risks of SCC associated with a tendency to burn when exposed to the sun and 10 or more lifetime painful sunburns among those who were beta HPV seropositive compared with those who were beta HPV seronegative (27). A significant interaction was also observed between beta HPV seropositivity and fair skin phenotype in relation to SCC risk among study participants in the Netherlands and Australia (40, 51). Importantly, no prospective studies have evaluated the interaction between UV exposure and beta HPV infection in relation to SCC risk, nor has the interaction been investigated using DNA-based markers of beta HPV infection. Results from the ongoing VIRUSCAN cohort study should provide important information on the temporality of sun exposure, HPV infection, and SCC risk (21, 52).
Experimental evidence is also mounting in support of a synergistic model of skin carcinogenesis involving UV radiation exposure and beta HPV infection. Although only a small number of the beta HPV types have been studied for their biological properties so far, many independent investigations provide clear evidence for the transforming activities of E6 and E7 from some beta HPV types, as described below.
Biological properties of beta HPV E6 and E7 oncoproteins.
Because of the findings for mucosal HR HPV that highlighted the transforming activities of E6 and E7, many biological studies on beta HPV types have focused on E6 and E7 proteins. Findings using in vitro experimental models, including established cell lines and primary human keratinocytes, provide clear lines of evidence for the transforming activities of the two viral proteins from some beta HPV types (10, 33, 53, 54). Similar to the mucosal HR HPV types, beta HPV types are able to alter the networks regulated by the tumor suppressor gene products retinoblastoma (pRb) and p53, with consequent loss of control of the cell cycle, apoptosis, and DNA repair (33). It appears that beta HPV types use different mechanisms in targeting p53. For instance, HPV38 E7 induces accumulation of a specific form of p53 that activates the expression of its antagonist ΔNp73α, which in turn hampers the proapoptotic functions of p53 (55, 56). In contrast, HPV49, via E6, induces p53 degradation via the proteasome pathway, as shown for the mucosal HR HPV types (57). In agreement with these findings, two studies have shown that E6 and E7 from a few beta HPV types (e.g., beta-2 HPV38 and beta-3 HPV49) efficiently immortalize primary human keratinocytes, which are the natural host of HPVs (57, 58).
Importantly, beta HPV types target pathways that are known to be altered in skin carcinogenesis, such as the Notch signaling pathway (59). Members of the Notch (1,–4) family play a crucial role in tissue development and homeostasis by coordinating differentiation and cellular proliferation. In particular, Notch1 is a negative regulator of proliferation by activating the expression of the cell cycle inhibitor p21Cip1/Waf1 (60). Studies in mouse models have shown that a loss of Notch1 in the skin increases the susceptibility to SCC induced by the two-stage chemical carcinogenesis protocol (61, 62). Whole-genome sequencing studies revealed that Notch1 is one of the most prominent mutated genes in human cSCC (63). Beta HPV E6 oncoprotein deregulates the Notch signaling pathway via direct interaction with Mastermind-like transcriptional coactivator 1 (MAML1) (64,–68). The beta HPV E6-MAML1 interaction results in the inhibition of Notch1-regulated gene expression, with consequent alteration of cellular proliferation and differentiation (69).
In agreement with the above-mentioned skin carcinogenesis model of cooperation between UV radiation and beta HPV types, beta HPV E6 oncoproteins are able to prevent the activation of UV-induced apoptosis by promoting the degradation of BAK via the proteasome pathway (70,–72). This cellular protein belongs to the BCL2 family and exerts a proapoptotic function, being released from the mitochondria upon UV-induced cellular stress. Thus, it is plausible to predict that the E6-mediated BAK degradation, by allowing the survival of UV-damaged cells, facilitates the accumulation of DNA mutations. The high susceptibility of beta HPV-infected cells to UV-induced DNA mutations is also supported by additional viral properties to deregulate the apoptosis and the DNA repair machinery. Indeed, E6 from beta-1 HPV5 and HPV8 binds to and promotes degradation of the p53 transcriptional coactivator p300 (73), with consequent inhibition of calcium-mediated differentiation as well as a decrease in the efficiency of repair of UV-induced DNA damage (74, 75). In contrast, beta-2 HPV38 E6 weakly interacts with p300 and does not induce its degradation (73,–75). However, it appears that even for HPV38 E6, the interaction with p300 is relevant for cellular transformation (76). An HPV38 E6 mutant unable to associate with p300 loses the capacity to prevent p53-dependent apoptosis. In addition, this HPV38 E6 mutant is not able, together with E7, to immortalize primary human keratinocytes (76). Alterations of integrin network are linked to skin carcinogenesis. Accordingly, studies in monolayer and organotypic cultures showed that beta HPV8 is able to alter the integrin network, promoting invasion of human keratinocytes into the dermis (77,–79).
Several mouse models have further confirmed the transforming properties of beta HPV proteins and their cooperation with UV radiation in promoting cSCC. Transgenic (Tg) mice expressing E6 and E7 from different beta HPV types in the skin under the control of a keratinocyte-specific promoter have increased susceptibility to UV-induced carcinogenesis (80,–82). Long-term UV exposure of Tg mice expressing beta-2 HPV38 E6/E7 genes in the basal layer of the epidermis under the control of the cytokeratin K14 promoter (K14) resulted in a high incidence of cSCC, whereas the same UV radiation doses did not cause the development of any type of skin lesions in wild-type animals (82). The fact that K14 HPV38 E6/E7 Tg mice did not develop any skin disease during their life span in the absence of UV radiation provides additional evidence for the synergism between beta HPV oncoproteins and UV radiation in promoting cSCC. A more recent study showed that the high susceptibility of K14 HPV38 E6/E7 Tg mice to UV-mediated skin carcinogenesis tightly correlates with the accumulation of DNA mutations. Whole-exome sequencing of DNA extracted from cSCC of long-term UV-irradiated K14 HPV38 E6/E7 Tg mice revealed that the vast majority of the somatic mutations detected in SCCs were mutations that are also prevalent in the UV-induced mutational signature. Importantly, the pattern of mutated genes in the skin lesions of UV-irradiated K14 HPV38 E6/E7 closely resembled that detected in human cSCC, with the highest mutation rates in p53 and Notch genes. Because both of the mutated genes encode proteins targeted by the beta HPV types, it is plausible to hypothesize that after accumulation of DNA mutations, the expression of the viral genes becomes dispensable. Using a conditional expression system based on Cre-Lox recombination, the same study showed that beta HPV E6 and E7 act only at an initial stage of carcinogenesis by potentiating the deleterious effects of UV radiation, but they do not contribute to the maintenance of cSCC (83). This scenario fully supports the concept that beta HPV types act with a hit-and-run mechanism, being facilitators of the accumulation of UV-induced DNA mutations and, consequently, of SCC development. Most importantly, this provides an explanation for the low HPV DNA prevalence in human skin lesions.
The beta HPV Tg animal models are complemented by mouse models that can be naturally infected by the corresponding PV. Mus musculus papillomavirus 1 (MmuPV1) is able to infect immunodeficient animals as well as immunocompetent standard laboratory mouse strains (84). Experiments with this animal model clearly showed a link between UV radiation, MmuPV1, immunosuppression, and the development of skin lesions (84, 85). Notably, studies using in vitro experimental models showed that E6 oncoprotein from beta-1 HPV8 and MmuPV1 display several functional similarities in targeting cellular pathways (54, 69). In particular, both proteins are able to inhibit transforming growth factor beta (TGF-β) and Notch signaling by binding to the SMAD2/SMAD3 and MAML1 transcription factors, respectively (69).
Other important findings on the role of an infectious agent in UV-mediated skin carcinogenesis were provided by experiments in the African multimammate mouse Mastomys coucha, formerly taxonomically classified as Mastomys natalensis. This mouse model is naturally infected by Mastomys natalensis papillomavirus (MnPV) in the skin and promotes the formation of skin lesions without integrating the host genome. A recent study has shown that long-term UV radiation induces a higher number of cSCCs in infected animals than in virus-free controls. In addition, some of the cSCCs in infected animals were poorly differentiated and nonkeratinizing, containing small amounts of or even lacking viral DNA (86). These findings provide additional support for the hit-and-run mechanism of HPV in UV-mediated skin carcinogenesis.
CONCLUSIONS
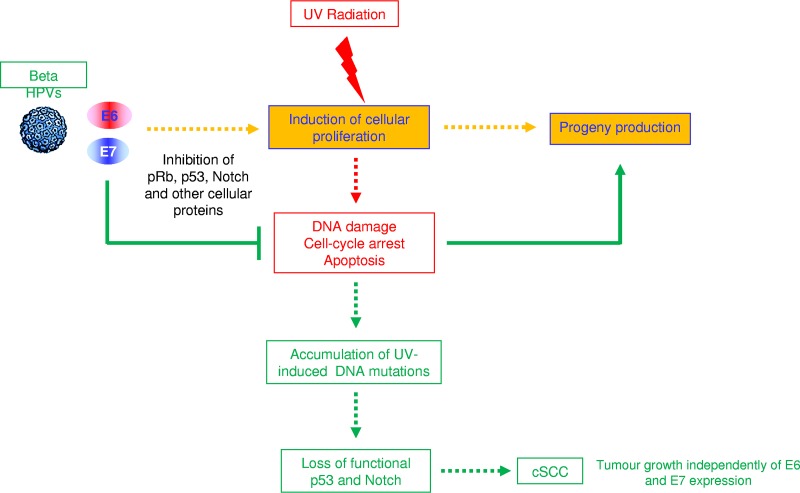
Emerging lines of evidence support models of beta HPV-associated carcinogenesis that differ from classic mucosal HR HPV driving carcinogenesis. In order to complete the life cycle, beta HPV types have developed mechanisms to promote cellular proliferation, even in the presence of constant stress, such as UV radiation. Consequently, these viruses facilitate the accumulation of DNA damage induced by UV radiation (Fig. 1). Once UV promotes mutations of genes that encode proteins involved in beta HPV-targeted cellular pathways (e.g., p53 and Notch), the expression of the viral oncogene becomes irrelevant for the growth of the cancer cells. In agreement with this model, biological and epidemiological findings support the concept that the beta HPV types are required at an early stage of skin carcinogenesis. These new models of HPV-driven carcinogenesis present opportunities for the development of novel strategies to decrease the incidence of cSCC, in particular in high-risk populations, such as OTRs. Taking into consideration all the findings available for the prophylactic vaccine against mucosal HR HPV types, the development of a beta HPV vaccine is highly feasible. These novel strategies could include vaccines based on the major capsid protein L1 as well as on the minor capsid protein L2 (for a review, see reference 53). An enhanced understanding of the synergy between beta HPV infection and UV radiation in cSCC development can also provide the foundation for future studies aiming to identify novel interactions between HPV infections and other carcinogenic risk factors.
ACKNOWLEDGMENTS
We apologize to those authors whose important contributions to HPV research could not be cited or adequately discussed due to space limitations. We thank the IARC Director, Christopher P. Wild, and all members of the Infections and Cancer Biology Group for their constant support.
The work performed in the groups is partially supported by grants from the European Commission, HPV-AHEAD (grant FP7-HEALTH-2011-282562), INSERM (grant ENV201610), the Fondation ARC pour la Recherche sur le Cancer (grant JA 20151203192), Deutsche Krebshilfe (grant 110259), and the National Cancer Institute at the National Institutes of Health (grant 1R01-CA17758).
REFERENCES
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.01003-18
Read article for free, from open access legal sources, via Unpaywall:
https://jvi.asm.org/content/jvi/93/7/e01003-18.full.pdf
Citations & impact
Impact metrics
Article citations
Commensal HPVs Have Evolved to Be More Immunogenic Compared with High-Risk α-HPVs.
Vaccines (Basel), 12(7):749, 07 Jul 2024
Cited by: 0 articles | PMID: 39066387 | PMCID: PMC11281416
Vaccination with a Human Papillomavirus L2 Multimer Provides Broad Protection against 17 Human Papillomavirus Types in the Mouse Cervicovaginal Challenge Model.
Vaccines (Basel), 12(6):689, 20 Jun 2024
Cited by: 0 articles | PMID: 38932417 | PMCID: PMC11209485
Cutaneous Squamous Cell Carcinoma: An Updated Review.
Cancers (Basel), 16(10):1800, 08 May 2024
Cited by: 2 articles | PMID: 38791879 | PMCID: PMC11119634
Review Free full text in Europe PMC
Molecular mechanisms of human papilloma virus related skin cancers: A review.
Medicine (Baltimore), 103(21):e38202, 01 May 2024
Cited by: 0 articles | PMID: 38787972 | PMCID: PMC11124606
Review Free full text in Europe PMC
Advancements in elucidating the pathogenesis of actinic keratosis: present state and future prospects.
Front Med (Lausanne), 11:1330491, 19 Mar 2024
Cited by: 1 article | PMID: 38566927 | PMCID: PMC10985158
Review Free full text in Europe PMC
Go to all (65) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Immunity to commensal papillomaviruses protects against skin cancer.
Nature, 575(7783):519-522, 30 Oct 2019
Cited by: 53 articles | PMID: 31666702 | PMCID: PMC6872936
HPV and skin carcinogenesis.
Papillomavirus Res, 7:129-131, 03 Apr 2019
Cited by: 34 articles | PMID: 30953864 | PMCID: PMC6460321
Review Free full text in Europe PMC
Human Papillomaviruses and Skin Cancer.
Adv Exp Med Biol, 1268:195-209, 01 Jan 2020
Cited by: 11 articles | PMID: 32918220
Review
The interplay of UV and cutaneous papillomavirus infection in skin cancer development.
PLoS Pathog, 13(11):e1006723, 30 Nov 2017
Cited by: 31 articles | PMID: 29190285 | PMCID: PMC5708609
Funding
Funders who supported this work.
NCI NIH HHS (1)
Grant ID: R01 CA177586
World Health Organization (1)
WHO generic grant number for open-access policy
World Organization
Grant ID: 001
 c
c