Abstract
Free full text

Targeting alterations in the RAF-MEK pathway
Abstract
The MAPK pathway is one of the most commonly mutated oncogenic pathways in cancer. While RAS mutations are the most frequent MAPK alterations, less frequent alterations in downstream components of the pathway, including the RAF and MEK genes, offer promising therapeutic opportunities. In addition to BRAFV600 mutations, for which several approved therapeutic regimens exist, other alterations in RAF and MEK genes may provide more rare, but tractable, targets. However, recent studies have illustrated the complexity of MAPK signaling and highlighted that distinct alterations in these genes may have strikingly different properties. Understanding the unique functional characteristics of specific RAF and MEK alterations, reviewed herein, will be critical for developing effective therapeutic approaches for these targets.
INTRODUCTION
The mitogen-activated protein kinase (MAPK) signaling pathway is critically involved in many important cellular processes. Its dysregulation leads to uncontrolled cellular proliferation, survival, and dedifferentiation. As a consequence, the MAPK pathway is altered or inappropriately activated in a majority of cancers.
Under physiologic conditions, MAPK signaling is triggered through activation of RAS proteins (KRAS, NRAS, and HRAS), a family of small guanine triphosphatases (GTPases) that integrate signals from a variety of upstream sources, most commonly from activated receptor tyrosine kinases (RTKs)(1). These upstream signals lead to activation of guanine nucleotide exchange factors (GEFs), such as son-of-sevenless (SOS), which catalyze the exchange of RAS-bound guanine diphosphate (GDP) for guanine triphosphate (GTP). RAS activity is negatively regulated by GTPase activating proteins (GAPs), such as neurofibromin 1 (NF1) which augment the GTPase activity of RAS to hydrolyze GTP to GDP, thus reverting RAS to its inactive GDP-bound state(2). In its active, GTP-bound state, a conformational change occurs in the Switch I and II regions of RAS, which facilitate interactions with a variety of downstream effectors, including the RAF family of kinases (ARAF, BRAF, and CRAF, the latter of which is encoded by the RAF1 gene)(3-5). Association of RAF proteins with activated RAS through their conserved RAS-binding domains leads to the formation of RAF homodimers (i.e. CRAF-CRAF) or heterodimers (i.e. BRAF-CRAF) with activated RAF kinase activity. For example, prior to binding activated RAS, BRAF is in an autoinhibited conformation in which a short α helix in its activation loop associates and displaces the critical αC helix in the kinase domain in an inactive “out” state. BRAF dimerization and activation loop phosphorylation destabilizes this autoinhibitory interaction to move the BRAF kinase into the helix αC “in” active conformation(6). Once activated, RAF kinases phosphorylate and activate MEK kinases (MEK1 and MEK2, encoded by the MAP2K1 and MAP2K2 genes, respectively), which in turn phosphorylate and activate ERK kinases (ERK1 and ERK2). The activated ERK kinases then phosphorylate a host of critical substrates that regulate key cellular processes.
Given the many important roles of MAPK pathway signaling, activation of the pathway is tightly regulated. There are several levels of negative feedback controls that limit physiologic activation of MAPK signaling. For instance, negative feedback loops from ERK include direct inhibitory phosphorylation of CRAF and BRAF and induction of expression of multiple MAPK phosphatases, such as DUSPs(7). ERK also inhibits the activation of RAS by RTKs by phosphorylating SOS and a variety of RTKs and by inducing the expression of members of the Sprouty family of proteins(8). These feedback signals modulate the output of oncogenic alterations within the pathway, affecting the spectrum of recurrent oncogenic alterations at each level of the pathway with implications for targeted therapy response and drug resistance.
The frequency of genomic alterations in the MAPK pathway decreases in incidence as one moves further downstream in the pathway: across human tumors, RAS mutations occur in 22%, BRAF in 7%, MEK in <1% of cases, and ERK mutations are exceptionally rare. The degree of ERK activation produced by alterations upstream in the pathway (e.g., RAS mutations) is often susceptible to constraint by negative feedback signals, while those further downstream escape negative feedback regulation and can lead to more profound activation of pathway output. In papillary thyroid cancers, where expression of ERK-responsive genes important in iodide transport can be readily assayed, differences in expression of ERK-responsive genes is seen between BRAFV600E mutants (strongly activating) and RAS mutants (less activating)(9).
RAS mutations are by far the most common MAPK alterations observed in human cancer, and RAS signaling and strategies for targeting RAS have been reviewed extensively elsewhere(10). This review will focus on downstream alterations in the MAPK pathway, including alterations in the RAF and MEK genes. Given the unique signaling biology of this pathway, recent studies have suggested that downstream alterations in the MAPK pathway can be broadly characterized into two groups: activators and amplifiers (FIGURE 1). “Activator” alterations lead to constitutive MAPK pathway signaling through ERK activation that is independent of upstream pathway activity. Conversely, “amplifier” alterations are dependent on upstream activity and augment the downstream signal to ERK. Activating alterations strongly activate ERK and are usually mutually exclusive with each other, while amplifying alterations commonly co-occur with other activating mutations upstream in the pathway. Interestingly, the incidence of activating mutations decreases further downstream in the pathway, and the proportion of amplifying alterations increases; activating alterations in downstream components of the pathway, like MEK or ERK, would lead to very high levels of output as they evade feedback signals and may thus have a selective disadvantage. Understanding the unique signaling properties of specific RAF and MEK alterations is key to devising strategies to overcome them. Here, we summarize our current understanding of recurrent alterations in RAF and MEK and their effects on signaling, targeted therapy response, and drug resistance. Based on this mechanistic framework, we outline rational strategies to target these specific alterations.
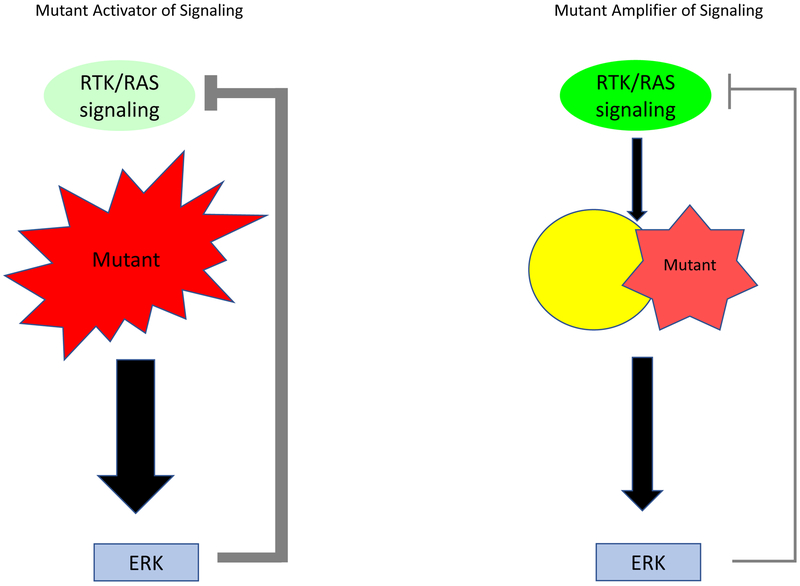
Schema showing the effect of “activator” versus “amplifier” mutants in the MAPK pathway. Activator alterations lead to constitutive MAPK signaling through ERK activation that is independent of upstream pathway activity. Activator mutants strongly activate ERK and lead to negative feedback suppression of upstream signaling. Amplifier mutants augment the downstream signal to ERK and commonly co-occur with other activating mutations upstream in the pathway. They lead to modest activation of ERK and consequently cause minimal negative feedback inhibition of upstream signaling.
RAF ALTERATIONS
BRAF is by far the most frequently altered gene in the MAPK pathway downstream of RAS, altered in 7-10% of all cancers(11). Point mutations are the most common mode of alteration in BRAF, but fusions and in-frame deletions are also observed in some cancers. Recent studies have demonstrated that different alterations in BRAF can produce a spectrum of functionally distinct variants with markedly different signaling properties. As normally RAF proteins require an interaction with RAS for their dimerization and subsequent activation, the functional variability is, at its core, driven by how mutants modify this interaction. How each specific alteration affects the formation and function of RAF dimers is critical for understanding the functional consequences of each alteration and the different strategies needed to target each variant. Overall, BRAF alterations can be characterized into three general functional classes, with Class I and II mutants constituting “activators”, and Class III mutants representing “amplifiers” (FIGURE 2).
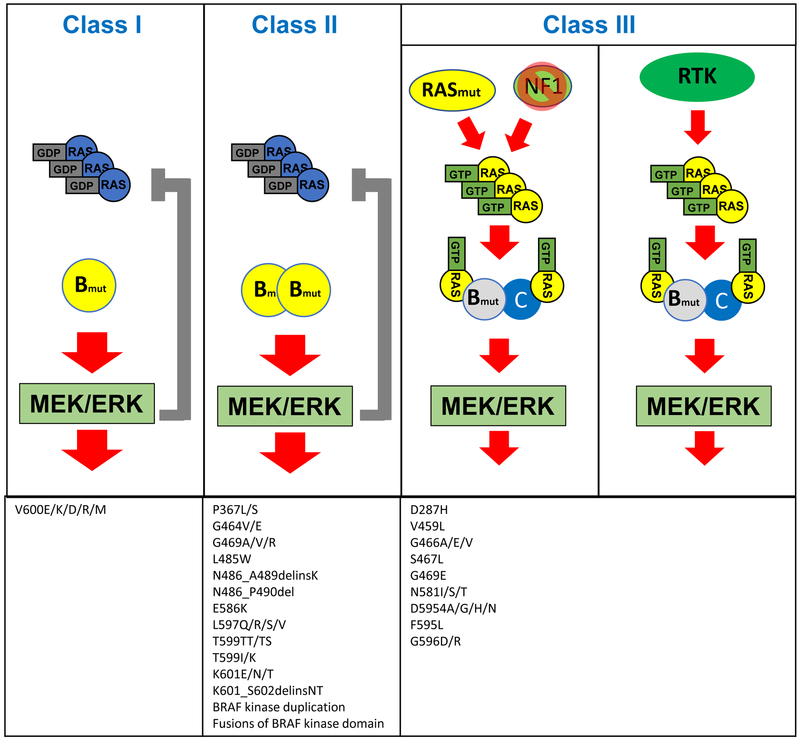
Class I BRAF mutants can signal as monomers, independent of RAS activation. They lead to high ERK activation, which causes negative feedback inhibition of upstream signaling. RAS-GTP levels are therefore low in tumors with class I BRAF mutants. Class II BRAF mutants signal as RAS independent, mutant-mutant BRAF dimers. They strongly activate ERK, causing negative feedback inhibition of upstream signaling and low levels of RAS-GTP. Class III BRAF mutants exhibit enhanced binding to RAS and CRAF to signal as mutant BRAF-wild-type CRAF dimers. They amplify the signaling downstream of RAS and thus require upstream activation to increase ERK signaling, either through genomic alterations (RAS mutations or NF1 loss, as shown on the left) or receptor tyrosine kinase (RTK) signaling (as shown on the right). Tumors with these mutations therefore exhibit high RAS-GTP levels.
Class I BRAF mutations: RAS-independent kinase activation, signal as monomers
Class I BRAF mutations thus far include only BRAFV600 mutations, which represent over 90% of BRAF alterations observed in cancer. BRAFV600 mutations are found in ~50% of melanoma, ~40% of papillary thyroid cancer, and ~10% of colorectal cancer(11). BRAFV600E is the most common amino acid substitution, accounting for >90% of BRAFV600 mutations, though substitutions of valine-600 to other amino acids, such as lysine, arginine, and aspartic acid are sometimes observed(11). Additionally, these mutations have been detected upon acquired resistance to EGFR inhibitors in lung and colorectal cancers(12,13). BRAFV600 mutations are unique in that they produce a constitutively active BRAF kinase that is capable of signaling as a monomer(14). BRAFV600 mutations result in high BRAF kinase activity and high levels of phosphorylated and activated ERK. The ability of BRAFV600 mutations to activate MAPK signaling is independent of RAS activity(15). In fact, RAS activity levels are found to be suppressed in BRAFV600 mutant cells due to strong negative feedback signals downstream of activated ERK(16).
Class II BRAF mutations: RAS-independent kinase activation, signal as dimers
Non-V600 BRAF mutants can be divided into two groups(17,18) based on their signaling properties and dependence on RAS activation. The first group, Class II activating mutants, include K601E, L597Q, and G469A. Most BRAF fusions and BRAF in-frame deletions (discussed below) also share many characteristics of Class II mutations. These variants signal as constitutively activated mutant dimers independent of RAS activation; they do not need RAS activation to dimerize. In these mutants, high ERK activation drives feedback suppression of RAS activation(17), and these mutants do not commonly co-occur with other MAPK pathway alterations. In patients with EGFR mutant non-small cell lung cancer, class II BRAF mutations have been identified as a mechanism of acquired resistance to EGFR inhibitors(19).
Class III BRAF mutations: kinase impaired, RAS-dependent, signal as dimers
Conversely, Class III mutants, including D594 and G466 mutants, have impaired BRAF kinase activity(20) and increase ERK signaling by amplifying signaling through wild-type RAF through mutant/wild-type RAF heterodimers(18). Thus, abnormal signaling by Class III BRAF mutants is dependent on heterodimerization with wild-type RAF protomers. Class III mutants bind more tightly than wild-type BRAF to RAS and exhibit enhanced binding and activation of wild-type CRAF(18). ERK activation in tumors with these mutants thus requires upstream RAS activation. As a result, these alterations occur in tumors with high RTK activity, leading to RAS activation, and often co-occur with activating RAS or NF1 loss-of-function mutations(18). The frequency of these concurrent mutations varies by tissue of origin; in melanoma, where there is low endogenous basal RAS activity, these mutants almost always coexist with mutant NF1 or RAS, while in colorectal and lung cancers, which have higher basal RTK activation, the resulting RAS activity is sufficient to support activation of these mutants, and only a minority of cases coexist with RAS/NF1 mutations(18,21). In the absence of concurrent genomic alterations in RAS or NF1, growth of tumors with these mutants is sensitive to inhibition of the dominant RTK driving RAS activity(18). Consequently, impaired BRAF mutants have been associated with improved survival and increased sensitivity to EGFR inhibitors in colorectal cancer (18,22,23), in stark contrast to activating BRAF mutations, such as V600E, which is associated with poor survival and lack of response to EGFR inhibitors.
BRAF fusions and in-frame deletions
Recently, recurrent in-frame deletions removing ~5 amino acids in the β3-αC region of BRAF near the P-loop have been identified at low frequencies in several cancers, including ~0.5-1% of pancreatic and thyroid cancers(24,25). These alterations are mutually exclusive with other MAPK activating alterations. Indeed, these alterations are present in ~5% of KRAS wild-type pancreatic cancers(25). These small in-frame deletions lead to a shortened β3-αC loop, which constrains the αC helix of BRAF kinase in the active “in” conformation, leading to increased kinase activity(25). These variants signal as active dimers, similar to class II BRAF alterations.
BRAF fusions are found in a number of cancers, particularly in pediatric low-grade gliomas, including >50% of pilocytic astrocytomas(26-29). They are present in ~0.3% of cancers overall, but in ~3-4% of melanoma (enriched in the Spitzoid subtype), and ~0.3% of pancreatic cancers, but are enriched in KRAS wild-type pancreatic cancer, and pancreatic acinar cell carcinoma (~20% of cases). These rearrangements produce an abnormal fusion product coupling the C-terminal BRAF kinase domain with an N-terminal dimerization domain. This leads to constitutive dimerization and activation of BRAF kinase activity independent of RAS or other upstream signals.
Non-BRAF alterations in RAF genes
Mutations in ARAF and CRAF are extremely rare, though some activating mutations in RAF genes other that BRAF have been reported(30). RAF1 fusions are observed in some cancers, typically resulting (much like BRAF fusions) in an aberrant gene product fusing the C-terminal CRAF kinase domain to an N-terminal fusion partner with a dimerization domain that leads to RAS-independent dimerization and activation of CRAF kinase activity. These fusions are found in low-grade pediatric gliomas, prostate cancer, melanoma, and pancreatic cancer(26,31-33).
MEK ALTERATIONS
In contrast to RAS and BRAF where mutations occur in hotspots, alterations affecting MEK1 or MEK2 can occur across the MAP2K1 or MAP2K2 genes, respectively. The function of many recurrent MEK1 mutants was recently elucidated by Gao et al(34) and they suggest these mutants can be functionally divided into three classes: RAF-independent, RAF-regulated, and RAF-dependent (FIGURE 3).
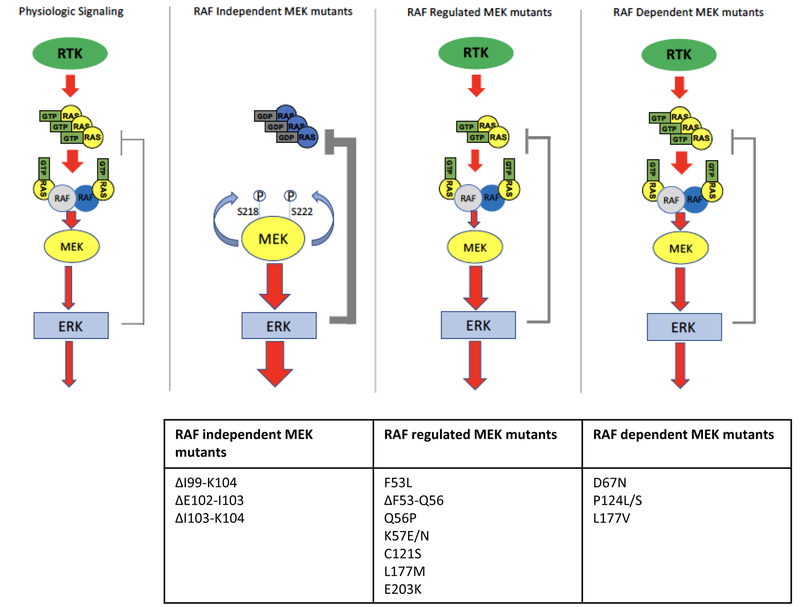
Schema showing physiologic MAPK signaling (left panel) or signaling in cells with MEK mutants. RAF-independent MEK mutants strongly activate ERK and induce negative feedback regulation of upstream signaling. These mutants are able to auto-phosphorylate the key regulatory sites S218 and S222 on MEK in cis. RAF-regulated MEK mutants exhibit some independent kinase activity, but this activity can be increased further in the presence of activated RAF, augmenting signaling from RAF. They do not activate ERK signaling to the same degree as the RAF-independent MEK mutants and therefore exhibit more modest feedback inhibition of upstream signaling. RAF-dependent mutants increase ERK activation only in the setting of active RAF; they bind more tightly to RAF, augmenting ERK activation. They modestly activate ERK and lead to minimal feedback inhibition of upstream signaling and often co-occur with upstream, activating alterations.
RAF-independent MEK alterations
RAF-independent mutants strongly activate MEK and ERK independent of upstream signaling. This group of mutants typically harbors in-frame deletions within the stretch of amino acids from 98-104 that remove a potent negative regulatory segment of MEK1(34). This region corresponds to a similar β3-αC loop in the MEK kinase domain, resulting in a shortened loop that constrains the kinase in the active “αC-in” conformation. Loss of this negative regulatory domain drives autophosphorylation of the activating serine residues at positions 218 and 222 and a marked increase in MEK kinase activity. Indeed, expression of these MEK1 mutants can drive strong MAPK signaling and cellular transformation in “RAF-less” cells—which bear conditional ARAF, BRAF, and CRAF (RAF1) alleles that can be deleted by CRE recombinase–confirming their independence from RAF activity(34). These mutants are found infrequently (<0.1%) in human cancers and thus far do not co-occur with other oncogenic MAPK alterations. Thus, RAF-independent MEK alterations function as strong “activators”.
RAF-regulated MEK alterations
RAF-regulated mutants exhibit some basal increase in ERK activation but can activate signaling further in the presence of activated RAF. Indeed, RAF-regulated MEK1 mutants can still produce some increase in ERK phosphorylation in “RAF-less” cells relative to wild-type MEK1, but levels of ERK phosphorylation are substantially lower than in the presence of functional RAF and are insufficient to drive cellular transformation, unlike the RAF-independent MEK1 mutants(34). Furthermore, mutation of the critical activating RAF phosphorylation sites on MEK1 (S218A and S222A) also reduces, but does not eliminate, MEK kinase activity in these mutants. These mutants exhibit a range of basal activity toward ERK. Because kinase activity of these mutants can be further increased in the presence of activated RAF, RAF-regulated mutants can, but do not always, co-occur with other activating MAPK alterations. Notably, these mutants have been observed in patient samples to emerge at acquired resistance to upstream inhibitors. For example MEK1K57 mutants have been identified in colorectal cancer patients with acquired resistance to EGFR antibodies(35,36), and MEK1K57 and MEK1F53L mutations have been observed in BRAFV600E colorectal cancer patients with acquired resistance to RAF/MEK and/or EGFR inhibitor combinations(37,38). Thus, RAF-regulated MEK mutations have properties of both “activators” and “amplifiers”.
RAF-dependent MEK alterations
Finally, RAF-dependent mutants increase ERK activation only in the setting of active RAF; they bind more tightly to RAF, augmenting ERK activation(34). Indeed, these mutants do not lead to ERK phosphorylation in RAF-less cells and fail to drive transformation. Similarly, mutation of the critical activating RAF phosphorylation sites on MEK1 (S218A and S222A) abolishes MEK kinase activity in these mutants. Thus, these mutants are particularly sensitive to feedback inhibition of RAF, which limits their functional output, and nearly universally co-occur with upstream MAPK alterations, such as BRAF or RAS mutations. Accordingly, these mutants act as amplifiers of RAF signaling.
INHIBITORS OF MAPK SIGNALING
RAF inhibitors
Currently approved RAF inhibitors selectively inhibit RAF monomers (i.e., Class I, BRAFV600 mutants)(14). While these inhibitors, such as vemurafenib, dabrafenib, and encorafenib, are often referred to as “BRAF inhibitors”, they in fact do not have selectivity for BRAF and are capable of inhibiting ARAF and CRAF with similar potency. However, the efficacy of these inhibitors in BRAFV600 mutant cancers is due to inhibition of monomeric mutant BRAF that is present and active in BRAFV600 mutant cells (FIGURE 4). Indeed, the effect of these inhibitors in cells lacking BRAFV600 mutations is different and complex. Binding of these drugs to one protomer within a RAF dimer pair causes allosteric transactivation of the other protomer, while at the same time reducing the affinity of the drug at the other protomer(14,17). These inhibitors bind to RAF in the αC helix “out” inactive conformation; drug binding stabilizes the first protomer in this inactive conformation and shifts the partner protomer into an “in” active conformation, that cannot be bound by drug because of steric hindrance(39). Thus, binding of the inhibitor to one protomer in the RAF dimer can actually lead to paradoxical activation of RAF signaling through transactivation of the protomer in the dimer. It takes a much higher dose of the RAF inhibitor, which cannot be achieved in patients, to overcome this negative cooperativity and bind both RAF protomers in the dimer to completely block RAF dimer signaling. Thus, these drugs are not effective against tumors where RAF signals as dimers.
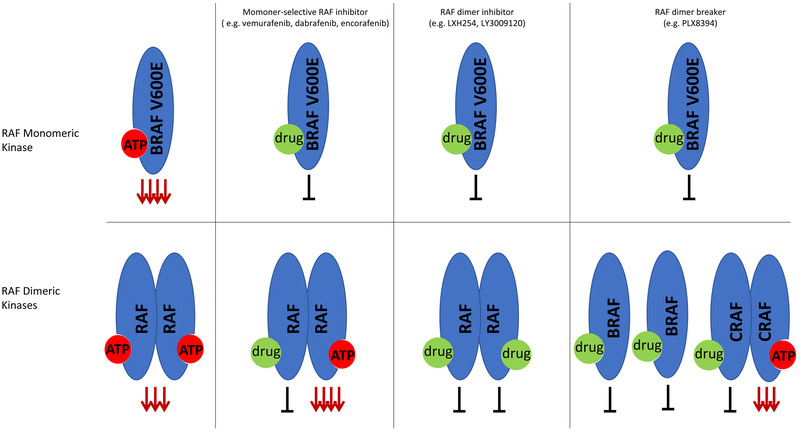
Schema showing effect of different RAF inhibitors in monomeric RAF kinases (i.e. BRAFV600E) (top section) or dimeric RAF kinases (bottom section). ERK activation is strongly activated downstream of BRAFV600E, even more so than seen for RAF dimeric kinase signaling. Monomer-selective RAF inhibitors bind to the ATP site in BRAF monomers and inhibit downstream signaling. In RAF dimeric kinases, binding of drug inhibits the bound RAF protomer, but leads to a conformational change in the other protomer in the dimer pair and strong transactivation of this protomer, leading to overall increased ERK activation. Drug binding to one site of the RAF dimer pair leads to a negative cooperativity for binding to the other site, and therefore at clinical doses, only one protomer in the dimer pair binds drug. RAF dimer inhibitors are able to bind to mutant RAF monomers and dimers at equipotent doses without negative cooperativity for the second site, and therefore can inhibit mutant RAF monomers and dimers at the same dose. RAF dimer breakers bind to and inhibit BRAF monomers. In RAF dimeric kinases, these drugs act by directly disrupting dimerization, rather than binding to both protomers in the dimer pair and exhibit negative cooperativity for binding a second protomer in the dimer pair. Dimer breakers disrupt BRAF-containing dimers, but do not disrupt CRAF homodimers, where they cause transactivation of the unbound CRAF protomer.
Newer classes of RAF inhibitors vary from RAF monomer inhibitors in their selectivity. All RAF inhibitors exhibit some degree of preferential inhibition of BRAFV600E compared to wild-type BRAF because BRAFV600E has a weaker ATP binding affinity than wild-type BRAF(40). Newer classes of RAF inhibitors, described as “RAF dimer inhibitors,” bind both sites in RAF dimers at equipotent doses(17,39). These drugs should have a therapeutic index because they inhibit mutant RAF dimers and monomers with greater potency than they inhibit wild-type dimers, and thus would not inhibit MAPK signaling to the same degree in normal cells(17). Mechanistically, these RAF inhibitors would be expected to inhibit tumors with mutant RAF dimers, including non-V600 BRAF activating mutants and BRAFV600 tumors that have become resistant to RAF inhibitors through genomic alterations, such as RAS mutation/amplification, BRAFV600E amplification, or intragenic deletion or splice variants of BRAF, that lead to BRAFV600 dimerization(17,39,41). Next generation RAF inhibitors that do not induce paradoxical ERK activation in RAF wild-type cells have also been developed(42) and these drugs (e.g., PLX8394), which are often referred to as “paradox breakers”, are currently in phase 1 studies. These drugs inhibit signaling by specifically disrupting BRAF-containing dimers, including BRAF-BRAF homodimers and BRAF-heterodimers(43). These drugs are not effective against CRAF homodimers or ARAF-containing dimers likely due to amino acid residues situated at the N-terminus of the RAF kinase domain that leads to enhanced stabilization of the dimer interface in these dimers. RAF dimer inhibitors show less selectivity between the different RAF isotypes, and thus can inhibit RAF signaling even in cells with strong upstream RAS activation. As these inhibitors may inhibit MAPK signaling in normal cells as well as in tumor cells, the therapeutic window of these agents may be more narrow than the “RAF dimer breaker”(44). In a phase I trial of the dimer inhibitor LY3009120, there was a limited dose escalation and the maximally tolerated dose of this agent was associated with minimal effect on tumor phospho-ERK levels and expression of ERK target genes(45). However, the majority of patients participating in this trial had tumors harboring RAS mutations, rather than RAF alterations.
MEK inhibitors
Most MEK inhibitors, such as selumetinib and the three FDA-approved agents—trametinib, cobimetinib, and binimetinib—are allosteric kinase inhibitors. These compounds bind to an allosteric pocket adjacent to the catalytic site of MEK, leading to a conformational change that constrains MEK kinase activity. Specifically, drug binding causes MEK to adopt a “closed” conformation and leads to a series of conformational changes that shift away a highly conserved glutamate residue (Glu114) from the active site, so that it is unable to form a critical ion pair with a conserved catalytic lysine residue (Lys97)(46). Allosteric MEK inhibitors have some clear advantages—because they bind to a unique allosteric pocket on MEK, rather than to the catalytic site, which bears higher homology to other kinases, these inhibitors are highly specific for MEK and are thus less likely to drive off-target toxicity. However, allosteric MEK inhibitors also have key vulnerabilities. In particular, many allosteric MEK inhibitors exhibit a reduced ability to inhibit MEK kinase activity in the presence of increased upstream MAPK signaling, leading to increased activation of MEK. This may occur through a confluence of two potential mechanisms. First, many MEK inhibitors bind preferentially to the inactive form of MEK and may exhibit reduced binding affinity to MEK when it is in its phosphorylated and activated form(47). Thus, if increased upstream signaling leads to increased levels of phosphorylated and activated MEK, the ability of the inhibitor to bind and inhibit MEK may be reduced. Second, marked increases in upstream MAPK signaling can lead to levels of activated MEK that are in excess of what is required for maximal ERK induction(48). Thus, a greater percent inhibition of MEK is needed to reduce the absolute levels of MEK activity to a degree that will translate into a reduction in ERK phosphorylation, requiring a greater concentration of inhibitor relative to baseline conditions.
It is for these reasons that many mechanisms of both primary and secondary (e.g., acquired) resistance to MEK inhibitors involve increased upstream signaling to MEK(48-51). This is in stark contrast to the typical mechanisms of resistance to other targeted agents, which tend to involve alterations at the level of or downstream of the drug target. Indeed, MEK inhibitors are notably susceptible to adaptive feedback reactivation of MAPK signaling(52). In this setting, transient inhibition of ERK phosphorylation by MEK inhibitors leads to reduced negative feedback on the upstream MAPK pathway. This results in increased RAS activation, often through engagement by RTKs, and ultimately to increased RAF-mediated MEK phosphorylation. In fact, markedly increased MEK phosphorylation is observed over time following MEK inhibitor treatment, and this feedback ultimately leads to increases in ERK phosphorylation despite the continued presence of MEK inhibitor(15). Similarly, multiple upstream alterations in the MAPK pathway are observed in the setting of clinical acquired resistance in BRAFV600 mutant cancers treated with RAF and MEK inhibitor combinations, including KRAS and NRAS mutations, amplification of the BRAFV600 allele, and RTK amplification(37,53). These events lead to increased phosphorylation and activation of MEK, and to reduced efficacy of MEK inhibitor.
However, key differences also exist among the different allosteric MEK inhibitors that can affect their susceptibility to upstream pathway activity(47,52). Trametinib, for example can limit RAF mediated phosphorylation of MEK, likely by reducing the formation of RAF-MEK protein complexes. However, feedback induction of MEK phosphorylation is still observed with trametinib, and multiple upstream mechanisms of acquired resistance (i.e., RAS mutations) are observed clinically in patients treated with trametinib(38). The MEK inhibitor CH5126766 inhibits both MEK and RAF because its binding to MEK results in a conformational change that interferes with the MEK phosphorylation by RAF and thus its release from RAF-MEK complexes(54), and early clinical data suggest this MEK inhibitor may have increased activity against RAS mutant tumors, particularly non-small cell lung cancers(55).
Finally, newer MEK inhibitors, such as MAP855, have been developed that are ATP-competitive and bind to the catalytic site(34). These inhibitors may also be less susceptible to differences in the activation state of MEK.
ERK inhibitors
More recently, direct inhibitors of ERK have been developed and have entered early clinical testing. Some ERK inhibitors, such as SCH772984, lead to reduced ERK kinase activity and to a reduction in MEK-mediated ERK phosphorylation (at least for a time)(56). Other ERK inhibitors, such as ulixertinib (BVD-523) and the structurally-related tool compound Vx-11e, inhibit ERK kinase activity, but result in marked increases in MEK-mediated ERK phosphorylation driven by release of negative feedback signaling(57). For this reason, ERK phosphorylation is not a reliable measure of the effectiveness of ERK inhibitors, and downstream markers of ERK activity, such as phosphorylation of ERK substrates, such as p90RSK, or levels of ERK-regulated transcripts, such as DUSP6, have been utilized.
Notably, initial studies of ERK inhibitors have suggested that these inhibitors are less susceptible to MAPK pathway reactivation due to increased upstream signaling, relative to MEK inhibitors(56,57). Indeed, several studies have suggested that ERK inhibitors are better able to maintain suppression of MAPK output even in the presence of upstream resistance alterations that drive MAPK reactivation in the presence of MEK inhibitors(38), because of reduced binding of MEK inhibitors to the active conformation of MEK, attenuation of the effect of MEK inhibitors by decreased levels of ERK phosphatases (DUSP proteins) from loss of ERK-dependent transcription, or “excess” MEK activation, as discussed above. As ERK inhibitors block ERK activity directly, they are less susceptible to a preponderance of MEK in its active conformation, decrease in ERK phosphatases, or excess levels of MEK activity. Preclinical data suggest that alterations at the level of ERK, such as amplification or mutations, may be able to cause resistance to these inhibitors(58). While further study is needed, this newer class of MAPK inhibitors may become a key part of the therapeutic arsenal to target cancers with MAPK pathway alterations.
TARGETING RAF AND MEK ALTERATIONS
Pharmacodynamic analysis of paired pre-treatment and on-treatment tumor biopsies from clinical trials of BRAFV600 cancer patients have revealed that profound inhibition of MAPK signaling is required for clinical response. Indeed, pharmacodynamic studies from the earliest clinical trials of vemurafenib in BRAFV600 melanoma suggested that >80% suppression of ERK activity is required to achieve a therapeutic response(59). Thus, an overarching goal in targeting cancers with MAPK alterations is to produce robust and sustained MAPK pathway inhibition. In particular, the therapeutic index of MAPK pathway inhibitors, alone or in combination, must be carefully considered such that robust pathway inhibition can be accomplished at the concentration of drug that is achievable in patients. Indeed, understanding key characteristics of signaling biology and identifying and intercepting mechanisms by which tumor cells might maintain MAPK output are critical to the design of effective therapeutic strategies (FIGURE 5).

The efficacy of specific inhibitors (green “+”: active; red “-”, inactive) and a potential rational therapeutic approach (blue boxes) are shown for each class of BRAF or MEK mutation. In general, the therapeutic approach for each class of mutation is based on its classification as an “activator” or “amplifier”, with activators requiring targeting downstream of or at the level of the mutation, and with amplifiers requiring upstream inhibition in combination with downstream inhibition. The level at which the pathway is targeted in each scenario is marked in orange. For MEK mutants, upstream inhibition would include RAF inhibition in BRAFV600 cancers or RTK inhibition in RAS/BRAF wild type cancers. In some cases, upstream inhibition may be helpful even when targeting activator mutations as a means of disrupting adaptive feedback reactivation of MAPK signaling, for example as when adding an RTK inhibitor (e.g., anti-EGFR antibodies) in BRAFV600 colorectal cancers (CRC).
RAF Alterations
Class I BRAF mutations (BRAFV600)
To date, RAF monomer inhibitors—including the three FDA-approved agents vemurafenib, dabrafenib, and encorafenib—have been the core of therapeutic strategies against BRAFV600 mutant cancers. These drugs exhibit a relatively wide therapeutic window because of their different effect in BRAFV600 tumor cells, where BRAF signals as a monomer, versus normal cells, where RAF signals as dimers. The paradoxical activation of ERK signaling in normal tissues(60) underlies the development of proliferative skin lesions, such as keratoacanthomas, with these drugs, and supports the improved therapeutic profile of combining RAF inhibitors with MEK inhibitors(61). Opposing effects or RAF and MEK inhibitors on ERK activation in skin attenuates skin toxicity with the combination and reduces the incidence of these hyperproliferative skin lesions(62).
However, incomplete pathway suppression or pathway reactivation is a major issue with RAF inhibitors. Release of RTK signaling from feedback inhibition after RAF inhibitor treatment leads to the rapid generation of RAF inhibitor-resistant RAF dimers and results in adaptive resistance to RAF inhibitors(16). Improved efficacy is seen with combined RAF and MEK inhibitors as MEK inhibition can suppress the resultant ERK reactivation. Adaptive resistance is most pronounced in tissues with high basal RTK signaling. In colorectal cancer, reactivation of EGFR signaling has been shown to limit activity of RAF inhibitors(63,64), and combinations of EGFR inhibitors with RAF or RAF/MEK inhibitors exhibit higher efficacy.
In patients who undergo resection of high risk BRAFV600 melanoma, combined RAF and MEK inhibitors have been shown to decrease the risk for recurrence and thus this regimen appears able to eradicate micrometastases in some patients(65). However, in the metastatic setting, nearly all patients eventually develop resistance to RAF inhibitor therapy. Resistance is often mediated by alterations that increase RAF dimerization, such as RAS mutations or amplification, BRAFV600 amplification, splice variants of BRAF, or increased RTK signaling, or, less commonly, mutations in MEK that increase ERK activation(53). The emergence of alterations that reactivate ERK signaling despite MEK inhibitor treatment suggest that it may not be optimal to combine RAF inhibitors with current MEK inhibitors. Recent data suggest that ERK inhibitors may yield improved suppression of ERK signaling and the ability to overcome commonly observed acquired resistance mechanisms(38,66). Whether new generation RAF inhibitors, likely in combination with MEK or ERK inhibitors, will play a key role in treatment of BRAFV600 tumors will necessitate further clinical study, and a key issue will be the therapeutic index of these combinations. While the combination of MEK and ERK inhibitors proved toxic(67), the first studies of RAF and ERK combinations are currently ongoing (NCT02974725).
Class II BRAF mutations
Unlike Class I BRAF mutations, Class II mutations are not sensitive to RAF monomer inhibitors like vemurafenib. Since these drugs, at clinically achievable doses, only bind one protomer in the dimer pair, they do not substantially inhibit ERK signaling in these variants where signaling consists of activated mutant dimers. However, as the mutant BRAF dimers are already fully activated and binding of the RAF inhibitor to the first protomer in the dimer does not lead to transactivation of the other protomer, RAF monomer inhibitors lead to a modest inhibition of phosphorylated ERK in these mutants. One alternative clinical strategy to target Class II mutations involves the use of newer RAF inhibitors that function as RAF dimer inhibitors or RAF dimer breakers(17,43). These inhibitors can disrupt signaling from active BRAF dimers and can suppress MAPK signaling in these models. As newer generation RAF inhibitors have only recently entered the clinic, it is not yet clear whether these agents can achieve clinical responses in tumors with Class II BRAF mutations.
A second potential strategy involves blocking MAPK signaling downstream of Class II BRAF mutations. Indeed, MEK inhibitors maintain the ability to suppress MAPK signaling in preclinical models of Class II BRAF mutations, but clinical experience with this approach is limited. Similarly, ERK inhibitors represent an alternative approach to downstream MAPK blockade in this setting. The recent trial of the ERK1/2 kinase inhibitor ulixertinib enrolled 28 patients with non-V600 BRAF mutants(68). Responses were seen in patients with activating non-V600 BRAF mutants(18,25), including a BRAFL597Q lung cancer, a BRAFL458W gallbladder cancer, a BRAFG469A head and neck cancer, and a BRAFG469A small bowel cancer. Overall, 33% (5 of 15) of patients with Class II BRAF mutations achieved partial response to ulixertinib. Interestingly, no responses were observed in 10 patients with Class III BRAF mutations. While further clinical study is needed, these early data suggest that downstream inhibition of MAPK signaling may be an effective clinical strategy for some cancers harboring Class II BRAF mutations, while feedback reactivation in cells with Class III BRAF mutants, which have high RAS activation, attenuates the effect of this approach.
Class III BRAF mutations
Similarly, Class III BRAF mutations are also not sensitive to RAF monomer inhibitors, like vemurafenib. These mutants signal as mutant BRAF/wild-type RAF dimers and binding of RAF inhibitors to the mutant BRAF in the dimer leads to transactivation of the wild-type RAF(18). Thus, RAF monomer inhibitors are unable to suppress MAPK signaling in the presence of these mutations.
Downstream inhibitors of MAPK signaling (i.e. MEK and ERK inhibitors) retain the ability to suppress MAPK signaling in the presence of Class III mutations(18). However, it is not clear if downstream inhibition of MAPK signaling will be sufficient to achieve clinical tumor responses or co-targeting RTK signaling in the tumor will be needed. As noted above, no patients whose tumors harbored Class III BRAF mutations responded to the ERK inhibitor ulixertinib(68), likely because high RAS activation in these tumors attenuates the effect of the ERK inhibitor.
Thus, one promising strategy for tumors with Class III BRAF mutations is to target the upstream signal for which the Class III mutations serves as an “amplifier”. This is most readily accomplished in tumors with wild-type RAS and dominant RTK signaling. For example, colorectal cancers with Class III BRAF mutations have been associated with increased sensitivity and improved survival with anti-EGFR antibodies. Thus, targeting the dominant RTK may represent an effective and tractable strategy for tumors with Class III mutations. As tumors do not always harbor a single dominant RTK and may receive signals from multiple RTKs, an alternative approach to blocking upstream signaling involves the use of SHP2 inhibitors, which block a key common effector target employed by multiple RTKs(69). SHP2 inhibitors have recently entered the clinic. Finally, combination therapy approaches that targets both downstream ERK activation (i.e. with a MEK or ERK inhibitor) and upstream signaling (i.e. with an RTK or SHP2 inhibitor) may represent potential therapeutic strategies for cancers harboring these alterations.
RAF fusions and activating in-frame deletions
BRAF and CRAF (RAF1) fusions predominantly lead to C-terminal RAF kinase domains fused to an N-terminal partner with a dimerization domain, leading to constitutive RAS-independent dimerization and activation of RAF kinase activity. Accordingly, RAF monomer inhibitors are not effective against BRAF or CRAF fusions(31,70). In fact, RAF monomer inhibitors can induce paradoxical activation of BRAF and CRAF fusions constructs, likely through transactivation of the dimer partner, similar to what is observed with wild-type RAF dimers in the presence of activated RAS. “Paradox-breaker” RAF inhibitors, such as PLX8394 do not lead to paradoxical activation of BRAF fusions, and can inhibit kinase activity—likely through the disruption of BRAF-containing dimers(43,70). However, PLX8394 cannot inhibit signaling from CRAF fusion dimers, and actually leads to paradoxical activation of CRAF signaling in this setting(31). However, some RAF dimer inhibitors can disrupt signaling from both BRAF and CRAF fusions in preclinical models(31). Thus, RAF dimer inhibitors warrant further exploration in this setting.
Furthermore, anecdotal success has been observed in multiple case reports by utilizing downstream blockade with MEK inhibitors. Indeed, clinical responses have been observed in melanoma patients harboring BRAF fusions treated with the MEK inhibitor trametinib(28,71). Downstream MAPK inhibition with ERK inhibitors would also merit consideration for BRAF fusions.
Similarly, BRAF in-frame deletions in the β3-αC loop also produce activated BRAF kinases that are insensitive to RAF monomer inhibitors(24). In particular, RAF inhibitors such as vemurafenib—which preferentially bind the inactive αC-out conformation—exhibit impaired binding to these mutants, which are constrained in the active αC-in conformation(25,39). However, RAF dimer inhibitors and RAF inhibitors that can bind to the αC-in conformation retain efficacy in these models(24). Furthermore, downstream inhibition with MEK (or ERK) inhibitors can effectively suppress MAPK signaling driven by these variants. Indeed, a recent report detailed a KRAS wild-type pancreatic cancer patient harboring a BRAF in-frame deletion in the β3-αC loop who was treated with trametinib and achieved a clinical response(72). Interestingly, when the patient developed acquired resistance to trametinib, the emergence of three MEK2 alterations were detected in cell-free DNA (cfDNA), suggesting that this cancer was highly dependent on MAPK signaling and became resistant through pathway reactivation. Notably subsequent treatment with an ERK inhibitor led to a dramatic reduction in the levels of these MEK2 mutations in cfDNA. Thus, while further clinical experience is clearly needed, these data support downstream inhibition with MEK or ERK inhibitors as a promising strategy for cancers harboring these alterations.
MEK Alterations
Different MEK mutants exhibit different sensitivities to currently approved MEK inhibitors, all of which are allosteric MEK inhibitors(34). All the MEK mutants, however, appear to retain sensitivity to a new class of ATP-competitive MEK kinase inhibitors that bind to the catalytic site or to ERK inhibitors. These data suggest that efforts to target tumors with mutant MEK in the clinic will need to take into account the mechanism of ERK activation.
RAF-independent MEK mutations
RAF-independent mutants are insensitive to allosteric MEK inhibitors, such as trametinib, cobimetinib, binimetinib, and selumetinib, because these drugs preferentially bind to the inactivated “αC-out” conformation of MEK1(34). The short in-frame deletions in the β3-αC loop in these MEK variants constrain the kinase in the active “αC-in” conformation, reducing the binding affinity of allosteric MEK inhibitors. However, these mutants retain sensitivity to a newer class of ATP-competitive MEK inhibitors that bind to the catalytic site. These variants also retain sensitivity to downstream inhibition with ERK inhibitors.
RAF-regulated MEK mutations
Preclinical studies have suggested that RAF-regulated MEK mutations may retain sensitivity to MEK inhibitors, as these inhibitors can suppress MAPK signaling in preclinical models harboring these mutants(34,73). In fact, there is anecdotal clinical evidence that certain RAF-regulated MEK mutations (i.e. MEK1K57 mutations) may exhibit clinical responsiveness to MEK inhibitors(35). Indeed, in one RAS wild-type colorectal cancer patient who developed a MEK1K57T mutation as a mechanism of acquired resistance following a prolonged response to the anti-EGFR antibody cetuximab, addition of the MEK inhibitor trametinib to anti-EGFR therapy led to a regression in the tumor lesion that harbored the MEK1K57T mutation and a marked reduction in the detectable variant allele frequency of MEK1K57T in serial cell-free DNA(35).
However, there is also clinical evidence that MEK inhibitors may not be able to overcome RAF-regulated MEK mutations. Indeed, several of these mutations have been observed to emerge in the setting of acquired resistance to RAF and MEK inhibitor combination therapies in BRAFV600 melanoma and colorectal cancer(38,53,74-76). As the activity of these variants can be augmented by increasing RAF-mediated phosphorylation, it is possible that adaptive feedback signaling may thus render MEK inhibitors less effective in the setting of persistent upstream signaling.
As an alternative approach, ERK inhibitors may be a promising strategy to overcome these alterations. Preclinical studies have demonstrated that ERK inhibitors retain efficacy in the presence of these MEK mutations(38). A recent study showed that RAF and MEK inhibitor combinations could not suppress the outgrowth of BRAFV600 colorectal cancer cells harboring K57 and F53 MEK1 mutations, but that ERK inhibitors alone or RAF and ERK inhibitor combinations could completely suppress their outgrowth(38). Thus, ERK inhibitors represent a promising class for targeting these alterations.
RAF-dependent MEK mutations
MEK and ERK inhibitors retain the ability to suppress MAPK signaling driven by this class of MEK mutations and thus represent a potential therapeutic strategy in this setting. However, these MEK alterations act as “amplifiers” and co-exist in the majority of cases with an upstream activating alteration, commonly BRAFV600 mutations. Therefore, targeting the dominant upstream signal in combination with a downstream blockade of MEK or ERK may be an optimal approach. For example, in BRAFV600 cancers co-harboring this class of alterations, RAF inhibitor combinations with MEK or ERK inhibitors may be optimal.
SUMMARY
The MAPK pathway plays a critical role in human cancer and is inappropriately activated in a large fraction of cancers through a variety of different mechanisms. However, due to the complex signaling biology of the MAPK pathway, distinct alterations in downstream pathway components, like RAF and MEK, can have dramatically disparate signaling properties. Careful biochemical and functional studies in recent years have been key to elucidating the critical nuances of MAPK signaling to create key opportunities for therapeutic development. In parallel, future clinical trials of novel strategies targeting RAF or MEK alterations should include careful pharmacodynamic assessment, through paired pre-treatment and on-treatment tumor biopsies, to ascertain the specific signaling effects of each therapy on its target. Overall, a detailed understanding of the unique signaling characteristics of specific RAF and MEK alterations will be critical to guide the design of effective therapies.
ACKNOWLEDGEMENTS
The authors acknowledge Drs. Zhan Yao and Yijin Gao for help in the design of the figures.
Grant Support:
R.B.C. is supported by NIH/NCI Gastrointestinal Cancer SPORE P50 CA127003, R01CA208437, U54CA224068, and R.Y. is supported by R01 CA233736 and the cancer center core grant P30 CA008748. The content of this review is the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. R.Y. and R.B.C are also supported by a Stand Up To Cancer Colorectal Dream Team Translational Research Grant (SU2C-AACR-DT22-17). Stand Up To Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the Scientific Partner of SU2C.
Footnotes
Disclosure of Potential Conflicts of Interest:
R.B.C. is a consultant/advisory board member for Amgen, Array Biopharma, Astex Pharmaceuticals, Avidity Biosciences, BMS, Chugai, Fog Pharma, Genentech, LOXO, Merrimack, N-of-one, nRichDx, Roche, Roivant, Shire, Spectrum Pharmaceuticals, Symphogen, Taiho, and Warp Drive Bio; holds equity in Avidity Biosciences and nRichDx; and has received research funding from AstraZeneca and Sanofi. R.Y. has received research funding from Array BioPharma, Genentech, GlaxoSmithKline, and Novartis, and has served as an advisory board member for GlaxoSmithKline.
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1158/2159-8290.cd-18-1321
Read article for free, from open access legal sources, via Unpaywall:
https://cancerdiscovery.aacrjournals.org/content/candisc/9/3/329.full.pdf
Citations & impact
Impact metrics
Article citations
Histology Agnostic Drug Development: An Updated Review.
Cancers (Basel), 16(21):3642, 29 Oct 2024
Cited by: 0 articles | PMID: 39518080 | PMCID: PMC11544807
Review Free full text in Europe PMC
Expert consensus on the diagnosis and treatment of solid tumors with BRAF mutations.
Innovation (Camb), 5(6):100661, 18 Oct 2024
Cited by: 0 articles | PMID: 39529955
Review
Mediating kinase activity in Ras-mutant cancer: potential for an individualised approach?
Front Pharmacol, 15:1441938, 20 Sep 2024
Cited by: 0 articles | PMID: 39372214 | PMCID: PMC11450236
Review Free full text in Europe PMC
Traditional Chinese herbal medicine: harnessing dendritic cells for anti-tumor benefits.
Front Immunol, 15:1408474, 19 Sep 2024
Cited by: 0 articles | PMID: 39364399 | PMCID: PMC11446781
Review Free full text in Europe PMC
First line therapy in stage IV BRAF mutated colorectal cancer.
Heliyon, 10(17):e36497, 22 Aug 2024
Cited by: 0 articles | PMID: 39263130 | PMCID: PMC11388748
Go to all (208) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth.
Nature, 464(7287):431-435, 03 Feb 2010
Cited by: 996 articles | PMID: 20130576
The Pan-RAF-MEK Nondegrading Molecular Glue NST-628 Is a Potent and Brain-Penetrant Inhibitor of the RAS-MAPK Pathway with Activity across Diverse RAS- and RAF-Driven Cancers.
Cancer Discov, 14(7):1190-1205, 01 Jul 2024
Cited by: 0 articles | PMID: 38588399 | PMCID: PMC11215411
Clinical Acquired Resistance to RAF Inhibitor Combinations in BRAF-Mutant Colorectal Cancer through MAPK Pathway Alterations.
Cancer Discov, 5(4):358-367, 11 Feb 2015
Cited by: 183 articles | PMID: 25673644 | PMCID: PMC4390490
Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer.
Oncogene, 26(22):3291-3310, 01 May 2007
Cited by: 1660 articles | PMID: 17496923
Review
Funding
Funders who supported this work.
Cancer Center Core (1)
Grant ID: P30 CA008748
Cancer Colorectal Dream Team Translational Research (1)
Grant ID: SU2C-AACR-DT22-17
NCI NIH HHS (5)
Grant ID: R01 CA208437
Grant ID: U54 CA224068
Grant ID: P50 CA127003
Grant ID: P30 CA008748
Grant ID: R01 CA233736
NIH NCI (4)
Grant ID: R01CA208437
Grant ID: P50 CA127003
Grant ID: U54CA224068
Grant ID: R01 CA233736





