Abstract
Free full text

Collagen fiber structure guides 3D motility of cytotoxic T lymphocytes
Abstract
Lymphocyte motility is governed by a complex array of mechanisms, and highly dependent on external microenvironmental cues. Tertiary lymphoid sites in particular have unique physical structure such as collagen fiber alignment, due to matrix deposition and remodeling. Three dimensional studies of human lymphocytes in such environments are lacking. We hypothesized that aligned collagenous environment modulates CD8+ T cells motility. We encapsulated activated CD8+ T cells in collagen hydrogels of distinct fiber alignment, a characteristic of tumor microenvironments. We found that human CD8+ T cells move faster and more persistently in aligned collagen fibers compared with nonaligned collagen fibers. Moreover, CD8+ T cells move along the axis of collagen alignment. We showed that myosin light chain kinase (MLCK) inhibition could nullify the effect of aligned collagen on CD8+ T cell motility patterns by decreasing T cell turning in unaligned collagen fiber gels. Finally, as an example of a tertiary lymphoid site, we found that xenograft prostate tumors exhibit highly aligned collagen fibers. We observed CD8+ T cells alongside aligned collagen fibers, and found that they are mostly concentrated in the periphery of tumors. Overall, using an in vitro controlled hydrogel system, we show that collagen fiber organization modulates CD8+ T cells movement via MLCK activation thus providing basis for future studies into relevant therapeutics.
Introduction
Lymphocytes are highly motile cells that experience diverse environments throughout the body. Naïve T lymphocytes are trafficked through the blood into the lymph node for antigen presentation followed by recruitment to a tertiary site usually within the interstitial space. Physically, the environment surrounding the lymphocytes changes drastically through this process, and is accompanied by phenotypic changes within the cells themselves.
T cells express β1 integrins allowing binding to ECM proteins such as collagen and fibronectin as well as the leukocyte-specific β2 and β7 integrins which permit binding to cell surfaces such as antigen presenting cell as well as endothelial cells through ICAMs [1]. Specifically, LFA-1 (αLβ2 integrin) and its ligand ICaM-1 mediate T cell trafficking through the lymph node [2].
T cell movement, unlike many other cell types, is classified as amoeboid migration due to the presence of flexible retractable projections, a lack of defined actin stress fibers, and transient contact with the ECM [3]. It was observed that T cells, unlike cells that utilize mesenchymal motility, do not degrade the ECM in order to migrate. Rather, they circumvent collagen fibers by contorting their cell body to squeeze through pores in the matrix [3]. T cell motility patterns are intimately connected to their activation status. Naïve T cells exhibit a “Brownian” random walk thereby increasing their chances of interaction with an antigen presenting cell [4]. In contrast, activated T cells in the lymph node adopt a directional migration usually guided by chemotactic cues. Subsequently, T cells within peripheral tissues can exhibit a super diffusive or Levy-type walk which optimizes scanning area by utilizing higher speeds and turning behavior [5].
Many studies on T lymphocyte motility have interrogated haptokinetic movement, or movement on a coated surface, to determine factors that influence T cell motility [6] such as chemotactic gradients [7]. While studying haptokinetic movement on coated surfaces has provided valuable insight into T cell movement especially in the context of soluble factors, it neglects an entire dimension and is not a good model of movement within a tertiary site within the interstitial space such as a tumor.
CD8+ T cells are central to cancer immunotherapeutic strategies. Moreover, our current understanding of how T cell motility is influenced by the ECM at “tertiary” lymphoid sites is lacking as many studies have focused on T cell movement within the lymph node. Tertiary lymphoid sites such as tumors have distinctly remodeled ECM with enhanced collagen deposition, crosslinking [8,9], and alignment. High grade tumors have been shown to exhibit collagen alignment [10–12] and have been suggested to be utilized by tumor cells as tracks to migrate and metastasize. Moreover, aligned ECM matrices have been shown to guide cell migration in multiple cell types [13,14].
One study shows that deposition of collagen surrounding tumors correlates with accumulation of CD8+ T cells within the tumor stroma [15]. These studies have remarked that T lymphocytes seem to migrate along collagen fibers in the lymph node and in tumor slices however, no controlled studies have been done to determine whether collagen fiber alignment itself can instruct T cell motility. In this study we aim to identify how collagen fiber alignment influences CD8+ T cell movement patterns as well as the mechanism by which collagen fiber alignment instructs this behavior. We have utilized a microfluidic device to generate collagen with aligned fibers and use this platform to understand why T cells preferentially move along collagen fibers in an attempt to increase availability of T cells to tumors.
Materials and methods
Microfluidic device fabrication and collagen hydrogel preparation
Polydimethylsiloxane (PDMS) microfluidic channels approximately 2.5 cm long, 250 μm tall, and 250 μm wide were fabricated and plasma bonded to glass slides as previously described [16] 1.5 mm ports were used for injecting collagen solution into devices. The collagen gel synthesis protocol was adapted from previously established protocols. Acid soluble rat tail collagen I [8] (Corning) was neutralized using 1 M NaOH and diluted in M199 1× medium (Thermo Fisher Scientific).
Alignment of collagen fibers and T cell encapsulation
Devices were coated with low molecular weight collagen (50 μg/mL), stored at 37 °C for 45 min, and stored at 4 °C for at least 30 min prior to gel loading. Following collagen solution ice incubation, activated CD8+ T cells were pelleted and resuspended in collagen solution to a final concentration of 1 × 106 cells/mL. 100 μL of cell solution was then loaded into the microfluidic device and allowed to solidifying for 15 min at room temperature following by 30 min at 37 °C. Following polymerization, injection ports were removed and media was added and allowed to freely diffuse and nourish the encapsulated T-cells.
Murine CD8+ lymphocyte isolation and activation
Murine cells were obtained from adult (8–12 week old) C57BL/6J mouse lymph nodes and spleens. C57BL/6J mice were maintained per guidelines approved by the Johns Hopkins University’s Institutional Review Board Cells were macerated through cell filters using a syringe and treated with ACK lysing buffer for erythrocyte lysis and to isolate splenocytes. CD8+ T lymphocytes were isolated from splenocytes by negative selection using CD8+ isolation kits and magnetic columns from Miltenyi Biotech (Auburn, CA, USA) according to the manufacturer’s protocol. To isolate only naïve CD8+ T cell populations, 1 μg of biotin anti-mouse/human CD44 antibody, clone IM7 (Biolegend) was added per 100 × 106 splenocytes when also adding CD8+ T cell kit antibody solution to deplete CD44+ CD8+ T cells. CD8+ T cells were activated in 96-u-bottom plates at a 1:1 ratio with Dynal anti-CD3, anti-CD28 aAPCs produced as described previously. CD8+ T cells were cultured in the T cell culture media (RPMI supplemented with L-glutamine, non-essential amino acids, vitamin solution, sodium pyruvate, β-mercaptoethanol, 10% fetal bovine serum, ciproflaxin, and a cocktail of T cell growth factors as described previously [32]). On day 3 or 4 of culture, cells were fed with half the volume of the initial T cell culture media with twice the concentration of T cell growth factor cocktail. Viable stimulated cells were isolated with a Ficoll-Paque PLUS gradient centrifugation (GE Healthcare).
Reflective confocal imaging of collagen fibers
To visualize collagen fibers within a three-dimension (3D) collagen gel reflection, confocal images were collected using LSM 780 (Zeiss) microscopes. The microscopes are equipped with a 40× water-immersion objective (Zeiss). For reflection imaging, the microscope was configured to capture 561 nm light reflected during illumination with at 561 nm. Reflection microscopy was used for live samples and fiber dimensions [8].
Collagen fiber density and orientation analysis
Collagen fiber density and orientation analysis was adapted from a previously developed method using MATLAB (Mathworks) software MatFiber [17]. The software segments the image and then calculates intensity gradients within subregions of images and uses them to track the overall directions of individual fibers. Subregions of 20, 40, 80, 120 square pixels were used to explore micro and macro alignment. Fiber angle distribution and probability distribution were plotted. In order to study cell aspect ratio, code was developed to fit an ellipse around the cell [8,18].
Cell expansion and encapsulation
Human CD8+ T cells isolated from peripheral blood were purchased from Astarte biosciences. T cells were cultured in RPM11640 medium containing 10% FBS and IL-2. T cells were activated using human T cell stimulator (CD3/CD28) Dynabeads for 24 h at 37 °C 5% CO2 prior to encapsulation in collagen gels. For inhibition studies, mL-7 (10 μm) was diluted in collagen mixture prior to cell encapsulation and solidification of the gel. Tracking was performed 24 h after cells were encapsulated.
Cell tracking
Cells were tracked via brightfield microscopy in 3D aligned and unaligned hydrogels over a period of 20 min using a Zeiss (AxioObserver) epifluorescent microscope with an incubation chamber with temperature and CO2 control. Tracked using live cell brightfield microscopy equipped with a controlled incubator (37 °C and 5% cO2).
Cell tracking analysis
Cell tracking videos were analyzed using manual tracking in FIJI (NIH) or the spots package in Imaris 8.2.1 (Bitplane). Velocity, speed profiles, angular velocity, and mean square displacements (MSDs) were calculated using code adapted from previously established methods. Trajectory plots were created using MATLAB (Mathworks, Inc.) [19] which plots the cell position in 3D space after it has been normalized to start at 0. Track length was calculated as the total distance that cell moved over the course of the timelapse. Motility coefficient was calculated as using the following equation:
where the motility coefficient, M, is equal to the displacement, D, squared divided by 4 times the total time in frame [20]. Straightness was calculated as the displacement divided by the track length. Finally turning angle is defined as the angle the cell moved between each frame. Statistical analysis was performed using MATLAB (Mathworks, Inc.) to calculate the mean, SD, and SE of the mean. A t-test was performed where appropriate to determine significance (GraphPad Prism 4.02; GraphPad Software, Inc.). Graphed data are presented as average ± SD.
Xenograft tumor growth
TRAMPC2 cells were injected (1 × 106 in 100 μL HBSS) in the flank of 9–12 week old C57BL/6 male mice and allowed to grow for 30 days prior to excision. Excised tumors were embedded in OCT and subsequently frozen for cryosectioning and staining. All experimental animal protocols were approved by the Institutional Animal Care and Use Committee at Johns Hopkins Medical School.
Immunostaining and quantification
Tumor cryo-sections (5 μm) and hydrogels containing T cells were fixed using 4% PFA prior to permeabilization, blocking, and staining with Phalloidin-488 (Life Tech) and anti-myosin light chain kinase (Abcam) for the hydrogels or collagen I (Novus Biologicals) CD8 (Novus Biologicals), and CD3 (Proteintech) for tumors, and DAPI nuclear stain. Secondary anti-Rabbit Alexafluor488 and Alexafluor594 (Thermo Fisher Scientific) as well as ant-rat NL637 (R&D) were used for staining. For immunohistochemical analysis, tissues were dehydrated through graded ethanols followed by antigen retrieval and detection using the ImmPRESS HRP anti-rabbit IgG polymer detection kit (Vector Laboratories). Images were captured using the Zeiss 780 confocal microscope. 3D circularity was calculated using surfaces in Imaris 8.2.1. Second harmonic generation was performed on a Zeiss AxioExaminer microscope equipped with a Coherent Chameleon Visionll multi-photon laser. Number and size of projections were counted in cell tracking videos using FIJI.
Traction force microscopy
Collagen gel solution (3 mg/mL) was prepared as stated above. One micron sized FluoSpheres that are Carboxylated modified (Thermo Fisher) were rinsed with phosphate buffered saline 2 times and then mixed with the cell pellet prior to hydrogel formation. Using a confocal microscope (LSM 780, Zeiss) z-stacks of beads were taken in 1 min intervals for 20 min to observe bead movement. To analyze bead movement in collagen gels, we analyzed according to a previously established method [21]. Tracking analysis was performed as stated above using IMARIS and MATLAB to calculate displacement, max displacement, track length, velocity, speed, and mean squared displacement (MSD) of beads. Contour plots of bead displacement were generated using the PIV plugin for FIJI (NIH) [22].
Statistical analysis
Statistical analysis was performed using MATLAB (Mathworks, Inc.) or Prism 4 (GraphPad Software, Inc.) to calculate the mean, standard deviation, and standard error mean. t-Test and one-way ANOVA were perfumed where appropriate to determine significance (GraphPad). Biological and technical replicates are indicated throughout the figure captions. All graphical data are reported as mean ± SEM. * p < 0.05, **p < 0.01, *** p < 0.001, and *** p < 0.0001.
Results
Alignment of collagen fibers in microfluidic devices
To test our hypothesis we needed to generate aligned and unaligned collagen matrices in which we can perform live analysis of T cell motility. Shear forces have previously been shown to align collagen fibers [24]. Microfluidic devices are ideal to generate high shear force during polymerization due to their small channel dimensions. A single channel device with two sets of dimensions was constructed to allow for aligned (250 μm wide x 250 μm tall) and un-aligned collagen (22 mm wide × 250 μm tall) matrices to be present within one device (Fig. 1A).
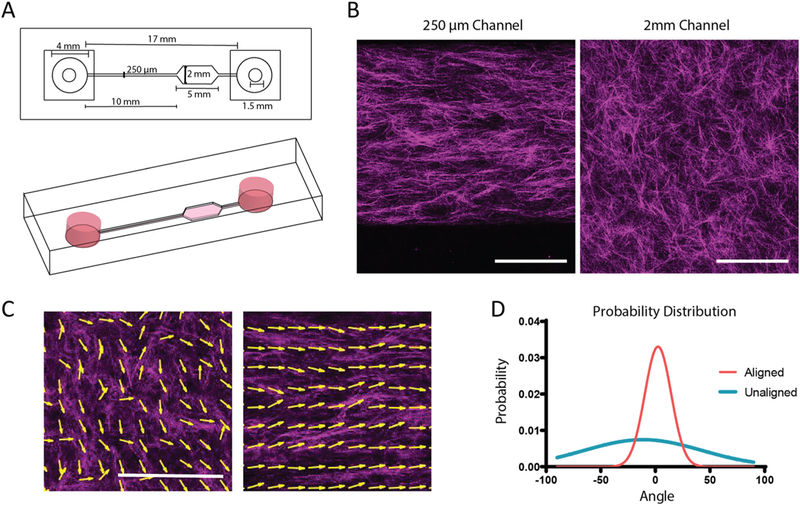
Alignment of collagen fibers in microfluidic devices. A. Schematic of microfluidic device for fiber alignment. Channel widths are 250 × 250 μm for the small channel and 250 μm × 2 mm for the large channel. Drawing to scale. B. Representative reflective confocal image of collagen ultrastructure in large and small channels, scale bar = 100 μm. (C) MatFiber output with arrows tracing collagen fibers to indicate directionality in unaligned and aligned channels. Scale bar = 200 μm. (D) Probability distribution of angles of collagen fibers in aligned and unaligned conditions. These experiments were independently repeated three times.
To increase shear forces on the collagen, devices were coated with low molecular weight collagen to enhance adherence of the collagen gel to the microfluidic wall during gel loading. To develop a significant fibrous structure within the collagen hydrogel, 3 mg/mL collagen solution was allowed to nucleate on ice for 2 h prior to injection into devices.
To confirm alignment of collagen fibers, loaded devices were imaged using reflectance confocal microscopy (Fig. 1B–C). Alignment of collagen fibers was quantified using the MatFiber code suite [23], where angles of all fibers are normalized to the median angle over the distribution. Probability distribution of the fiber angles was then plotted (Fig. 1D). Microscopic and macroscopic alignment of the collagen fibers were assessed by changing the window size that was being interrogated by the Matfiber code (Supplementary Fig. 1). Significant alignment was observed in the small channels as the distribution of angles of the collagen fibers only varied within 50° where as a wide range of angles was present in the unaligned region of the device (Fig. 1D; Supplementary Fig. 1).
CD8+ T cells move faster and more persistently in aligned collagen matrices
To understand how T cell motility patterns were influenced by alignment of collagen in a 3D environment, CD8+ T cells isolated from peripheral blood were activated and embedded in the collagen pre-polymer. Subsequently, T cells were tracked following gel formation using brightfield microscopy. We monitored CD8+ T cells 3D migration over short periods of time (20 min). Paired experiments were performed to reduce variability due to T cell batch and stimulation timing. We have performed preliminary experiments comparing naïve and effector CD8+ T cells into our system to potentially gain some insight into whether aligned collagen fibers could activate naïve T cells. We found, as expected, that naïve CD8+ T cells barely move in the collagen gel, thus making it difficult to assess any potential effect of alignment on them (Supplemental Fig. 2). Thus, we continued all our experiments with activated CD8+ T cells (Fig. 2A–B). Initial tracking of T cells in aligned and unaligned collagen revealed a significant difference in mean squared displacement (MSD), track length, and overall speed between cells encapsulated in the two conditions (Fig. 2C–E). Speed over the entire time lapse was increased while instantaneous velocity was also enhanced when cells were encapsulated in aligned fiber gels. Both of these parameters are considered to calculate a motility coefficient of the population of cells, which was significantly increased in the T cells exposed to aligned collagen matrices (Fig. 2E). To determine whether T cells were preferentially migrating along the axis of collagen alignment, tracks were broken into individual vector components, with the X axis being the axis of alignment of the collagen fibers. Although the T cells in aligned collagen fiber gels were covering greater distances, most of the distance was along the X axis (Fig. 2F). Along the Y axis (the axis perpendicular to alignment) the difference in MSD between aligned and unaligned conditions was significantly less (Fig. 2G). In addition, the linearity of each individual cell’s track was compared, and as hypothesized, the tracks of the cells encapsulated in aligned fiber gels were straighter than in unaligned fiber gels (Fig. 2H). Although, the tracks were straighter in aligned conditions, the tracks were not entirely straight suggesting that the CD8+ T cells were not physically confined to a particular path. The turning angle of T cells in both conditions was then compared to determine the persistent movement along the alignment axis. T cells within aligned matrices exhibited smaller turning angles than their unaligned counterparts (Fig. 2I). Moreover, polarity plots were calculated to determine the anisotropy of the T cell movement. In unaligned fiber gels, T cells were found to exhibit faster turning movements and more variability of turning speeds, while the T cells in aligned fiber gels had less variability in the speed of turning behavior, indicative of more anisotropic movement.
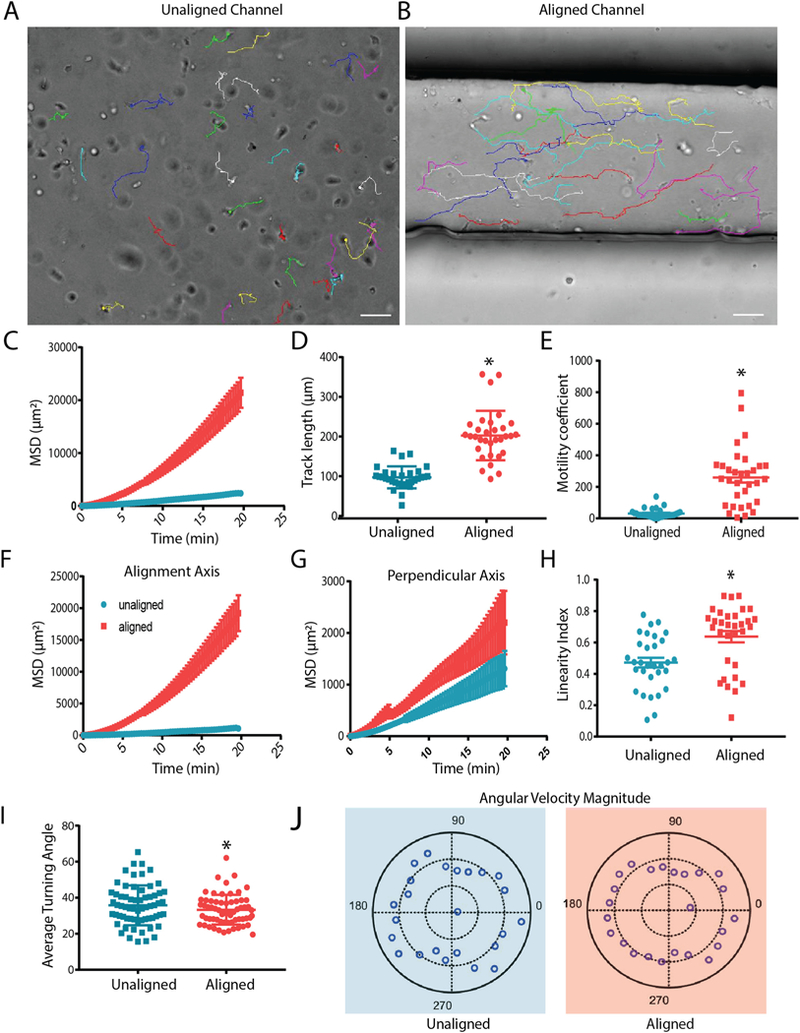
CD8+ T cells move faster and more persistently in aligned collagen matrices. (A) Brightfield still images from the final frame of time-lapse videos of CD8+ T cell tracking in unaligned and (B) aligned collagen matrices over a 20 minute time period. Lines represent path of movement throughout the 20 min recording period Scale bars = 50 μm. (C) Overall MSD of CD8+ T cells within unaligned and aligned matrices. (D) Cumulative length of T cell tracks after 20 minute timelapse. (E) Motility coefficient was calculated for T cells in unaligned and aligned matrices. (F) Mean squared displacement along the horizontal axis (axis of alignment in devices). (G) Mean squared displacement of cells in the axis perpendicular to alignment of collagen fibers. (H) Straightness of T cell tracks as determined by the ratio of the track length and overall displacement. (I) The average angle at which T cells change direction. (J) Polar plots of turning angle in combination with velocity at which T cells complete turning behavior. Experiments were independently repeated three times and at least 50 cells were analyzed in each experiment. * indicates p < 0.05.
T cells decrease protrusions and interact with the matrix less in aligned collagen gels
Based on our observations regarding T cell speed and trajectories, we analyzed CD8+ T cell morphology in unaligned and aligned collagen. The morphology of T cells within aligned collagen was elongated and much more elliptical than T cells encapsulated in unaligned collagen (Fig. 3A). Interestingly, T cells in aligned collagen extended far fewer projections that were longer and cover less area than cells encapsulated in unaligned fiber gels (Fig. 3B–C). This is consistent with previous findings in breast cancer cells, where fewer projections were seen in aligned collagen [24]. To further investigate how T cells were interacting with aligned and unaligned matrices, traction force microscopy was performed. One μm carboxylated beads were encapsulated alongside T cells in aligned and unaligned fiber gels to more easily indicate forces applied by T cells to the surrounding matrix. Carboxylated beads were chosen as they are large enough to neglect Brownian displacements and are effectively tethered to the collagen matrix fibers due to the reactive carboxyl groups [21]. Beads were tracked in 3D and the magnitude of displacement as well as directionality of movement of beads with respect to cells was plotted (Fig. 3D). The maximum displacement of the beads is indicative of the amount which the T cells deformed the matrix through their interaction (Fig. 3E), while the speed represents magnitude of interaction during the time lapse (Fig. 3F). Overall, the T cells encapsulated in unaligned fiber gels pulled on the matrix more which was indicated by the significantly increased max displacement and speed of traction force beads during the time lapse. These results suggest that T cells encapsulated in unaligned collagen fibers extend more projections which pull on the matrix to a greater extend in all directions when compared with cells encapsulated in aligned collagen fibers.
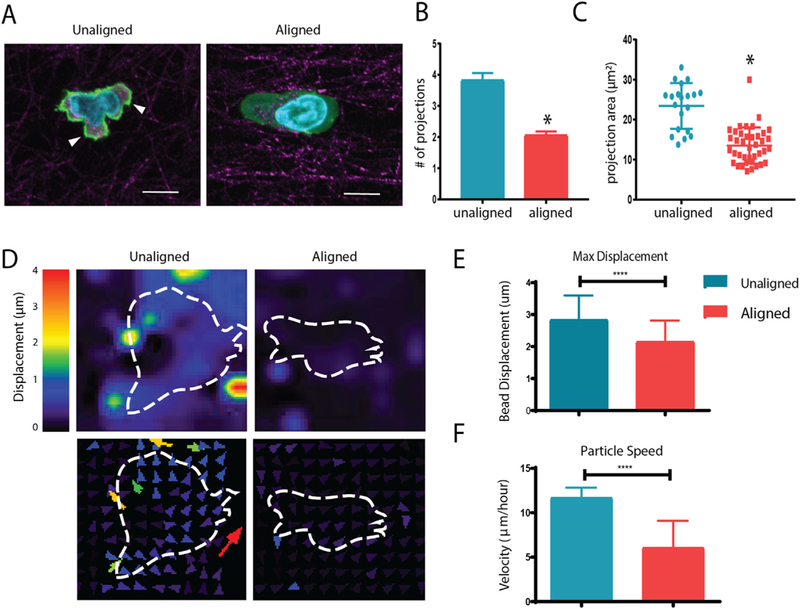
Aligned collagen gels decrease T cell protrusions and interact with the matrix less. (A) Reflective confocal images of T cell morphology within aligned and unaligned collagen gels (phalloidin in green, collagen fibers in red; nuclei in blue). Arrowheads indicate cellular protrusions (B) Number of projections extended per cell during 20 minute time-lapse videos in unaligned and aligned fiber gels. (C) Area of projections extended by T cells during time lapse. (D) Deformation plot showing magnitude and direction of displacement with respect to embedded cells (outline). (E) Maximum overall displacement of beads indicative of deformation of the collagen matrix by T cells. (F) Speed of traction force beads over time lapse. At least 20 cells were analyzed for each condition. This experiment was independently repeated three times * indicates p < 0.05. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
MLCK inhibition abrogates T cell motility difference in aligned and unaligned collagen gels
Unlike cancer cells and fibroblasts, T cells do not exhibit strong integrin binding and formation of focal adhesions as mechanisms for migration. Rather, non-muscle myosin facilitates T cell movement on coated surfaces as well as in 3D environments [25]. Furthermore, myosin light chain kinase (MLCK) activity has been shown to permit D011.10 blast cell climbing up ramp-like structures on ICAM coated surfaces [26]. Therefore, we queried expression patterns of MLCK in CD8+ T cells encapsulated in unaligned and aligned collagen matrices. Interestingly, MLCK exhibited a diffuse staining pattern in unaligned fiber gels where T cells were extending more projections and exhibiting high degrees of turning behavior (Fig. 4A). In contrast, T cells encapsulated in aligned collagen gels showed punctate expression of MLCK (Fig. 4B). Overall, the number of T cells that exhibited punctate MLCK staining was higher in aligned collagen (Fig. 4C) as was the average number of puncta per cell (Fig. 4D).
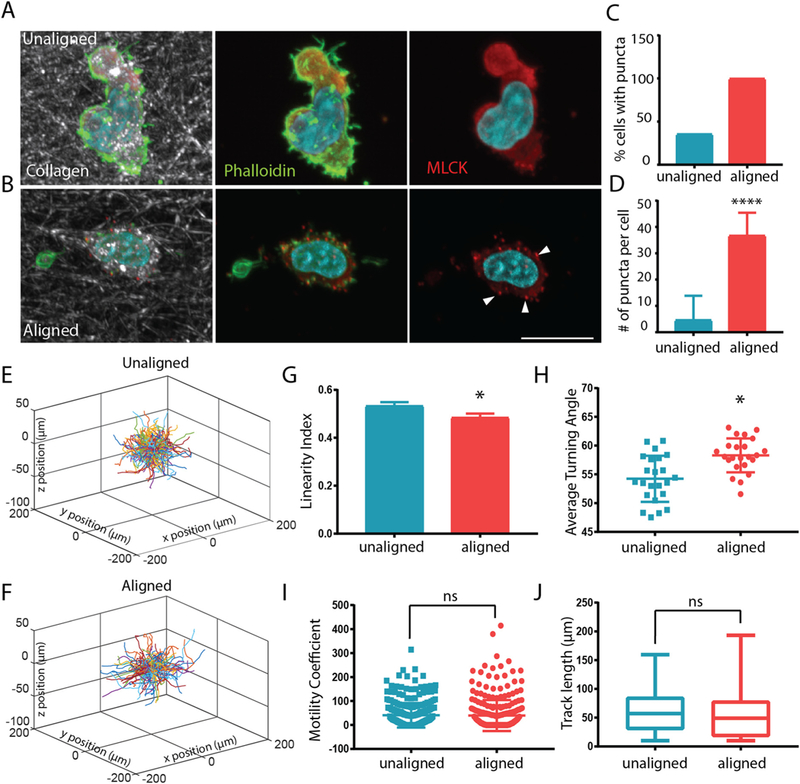
MLCK inhibition abrogates T cell motility difference in aligned and unaligned collagen gels. (A) Staining of MLCK (red) and phalloidin (in green, nuclei in blue) staining in migrating T cells within unaligned and aligned collagen. Arrowheads indicate punctate MLCK stain. Scale bar is equal to 10 μm. (C) The number of cells with punctate MLCK expression and (D) the number of MLCK puncta per cell were quantified in cells within unaligned and aligned gels. At least 25 cells were analyzed for each condition (E–F) Trajectory plot of migrating T cells in unaligned and aligned collagen treated with ML-7 inhibitor for 24 h (10 μm). (G) Linearity of migratory tracks of ML-7 treated cells (H) Average turning angle of ML-7 treated cells in aligned and unaligned collagen. (I) Motility coefficient of ML-7 treated cells within aligned and unaligned gels. (J) Cumulative track length of ML-7 treated cells over a 20 min time lapse in unaligned and aligned collagen. *** indicates p < 0.0001 * indicates p < 0.05. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To determine whether MLCK was partly responsible for the difference in migratory behavior in the unaligned and aligned collagen fiber gels we treated CD8+ T cells with ML-7, an MLCK inhibitor and tracked their movement after encapsulation. We hypothesized that because MLCK activity is important for T cell turning behavior, inhibition would affect motility patterns of T cells encapsulated within the unaligned regions of the gel. Interestingly, the difference in the trajectories of cells in aligned and unaligned fiber gels was practically abolished (Fig. 4E–F). Surprisingly, the linearity of the trajectories was slightly higher in the unaligned fiber gel compared to aligned fiber gel although not significant (Fig. 4G). Concomitantly, the angle at which the T cells turned was also higher in the aligned fibers (Fig. 4H), while the overall track length and motility coefficients (Fig. 4I–J) remained the same between the two conditions. We suggest that ML-7 treatment and blocking of MLCK activity, effectively inhibited turning behavior of the T cells in the unaligned collagen, which then led to them migrating in straighter paths.
CD8+ T cells colocalize with aligned collagen I fibers in tumors
To understand the relevance of our findings in vivo we employed a syngeneic xenograft tumor model of prostate cancer by injecting TRAMPC2 cells into the flank of adult male C57BL/6 mice [27]. The degree of collagen fiber alignment within established tumors was determined using second harmonic generation (SHG) imaging of tumor sections followed by quantification using MatFiber (Fig. 5A–C). To determine the distribution of CD8+ T cells with respect to collagen I deposition in the stroma and periphery of the tumor, excised tumors were cryo-sectioned and stained. As expected, collagen I made up a significant part of the peritumoral stroma. CD8+ T cells were seen infiltrating from blood vessels located within the stroma as well as extended along collagen fibers (Fig. 5D) within regions adjacent to the tumor. CD8 + T cells in the tumor mass were seen colocalized with aligned collagen I deposits within the tumor peripheral to the stroma (Fig. 5E). Additionally, SHG was performed along with CD3 staining to identify T cell morphology along more distinct collagen fiber structures within the tumor (Fig. 5F). To determine the potential migratory behaviors of the CD8+ T cells in the stroma and tumors, the major axis angle of T cells proximal to collagen fibers was measured. In addition, the angle of the adjacent collagen fiber was calculated (Fig. 5G). Angles of the major axes of T cells was positively correlated with the angles of the collagen fibers indicating that the T cells were extended parallel to the collagen fibers (Fig. 5H). This suggests that the T cells present in the stroma, periphery and center of the tumor may have been migrating along the collagen fibers. Lastly, we sought to interrogate the overall distribution of T cells within TRAMPC2 tumors. Upon quantification of CD8+ cells in the periphery, as well as central regions of tumors, we found that significantly more CD8+ cells were located closer to the invasive margin of the tumor (Fig. 5I–J). Overall, our data show that TRAMPC2 xenograft tumors exhibit highly aligned collagen matrix structure. T cells within these tumors are associated with the aligned collagen fibers, however, they are concentrated on the periphery of tumors and are not evenly distributed throughout the tumor mass.
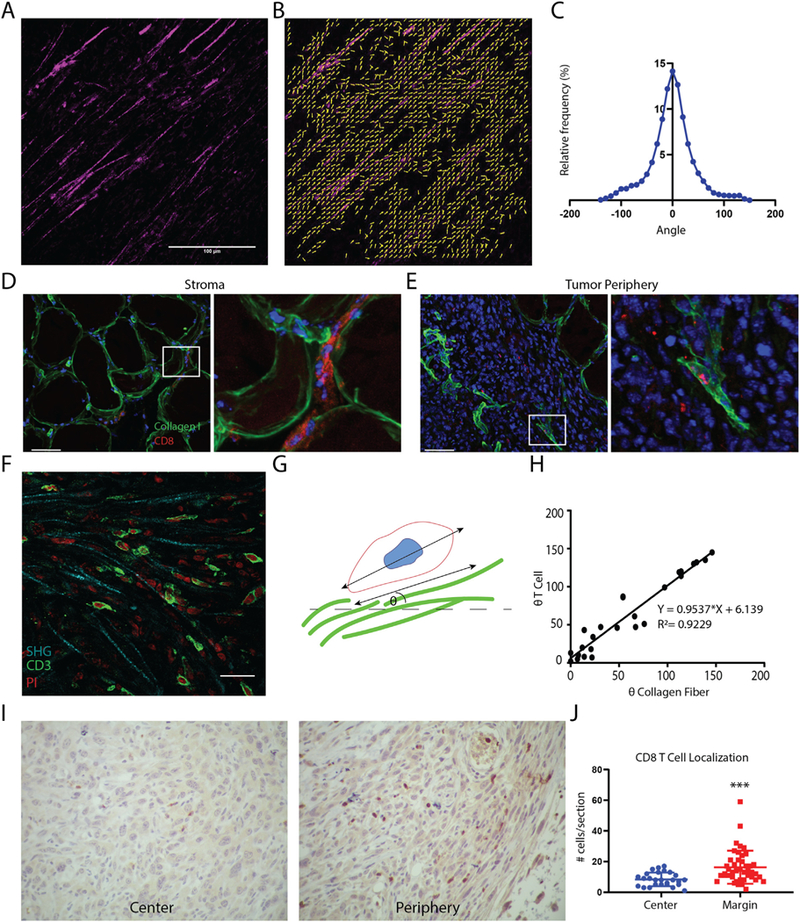
CD8+T cells colocalize with aligned collagen I fibers in tumors. (A) Representative SHG image of TRAMPC2 xenograft tumors. (B) Matfiber alignment vectors of SHG image. (C) Histogram of collagen alignment profiles in TRAMPC2 tumors (n = 8 tumors) Xenograft tumors (n = 4) stained for collagen I (green) and CD8 (red; nuclei in blue): (D) Left-Stromal regions adjacent to TRAMPC2 subcutaneous xenograft tumors and right- high magnification of the boxed area on the right, showing CD8+ T cells localized between collagen fibers. (E) Right - periphery of TRAMPC2 tumors depicting infiltrating CD8+ T cells and right- high magnification of the boxed area on the right, showing CD8+ T cell located along aligned collagen fibers. Scale bars = 50 μm. (F) Photomicrograph depicting T cells crawling along collagen fibers using SHG imaging (G) Schematic depiction of method for quantifying T cell association with collagen fibers. (H) Scatter plot of collagen fiber and T cell angles (n = 26 cells) showing positive correlation. (I) CD8 immunostaining of central regions and peripheral regions of TRAMPC2 tumors. (J) Quantification of CD8+ cells within different regions of the tumor. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Discussion
Success of immunotherapy in solid tumors has proven challenging potentially due to exclusion of CD8+ T cells from tumor beds. Studies have shown that matrix density contributes to exclusion of CD8+ T cells [28]. Although highly aligned collagen matrices have been shown in aggressive solid tumors [11,29,30], whether the alignment contributes to lymphocyte infiltration remains to be seen. In the current study, we show that solely by aligning collagen fibers CD8+ T cell migration behavior can be augmented. Our study has allowed a direct comparison of CD8+ T cell migration in unaligned and aligned collagen. We show much straighter paths of CD8+ T cells in aligned collagen along with decreased turning behavior. Additionally, our results show that CD8+ T cells migrate faster in aligned collagen. These results indicate that the directionality collagen fiber alignment may impact the path of infiltrating lymphocytes into or around tumor beds, and that not only is matrix deposition an important factor for the control of lymphocyte motility, but matrix alignment is as well.
It is currently unknown how matrix remodeling and collagen fiber alignment could impact activation or polarization of T lymphocytes within tertiary lymphoid sites. Activation and polarization of T lymphocytes is critical to their cytotoxic activity and is heavily influenced by soluble factors and the presence of antigen presenting cells. Incorporation of these factors into our system would be an excellent future study to identify how matrix remodeling could contribute to T lymphocyte activation and polarization.
Our results are consistent with the contact guidance hypothesis, that collagen fiber orientation can direct cellular migration paths. CD4+ T cell blasts encapsulated in low concentration collagen gels were observed to exhibit forward movement when collagen fibers were in parallel to the cell body [3]. Our study has allowed a direct comparison of CD8+ T cell migration in unaligned and aligned collagen. We show much straighter paths of CD8+ T cells in aligned collagen along with decreased turning behavior. Additionally, our results show that CD8+ T cells migrate faster in aligned collagen. In our system, extension of projections in unaligned collagen gels slows down the migration of CD8+ T cells. It is also likely that more projections in T cells contribute to accelerated turning behavior leading to less linear paths.
Studies of haptokinetic T cell migration have shown that myosin light chain accumulates in the head-to-tail region long the side to which the T cell is about to turn [26]. Non-muscle myosin II has been shown to mediate integrin binding to ICAM-1 in T cells. We observed diffuse MLCK expression patterns (i.e. inactive MLCK) in T cells encapsulated in unaligned while T cells embedded in aligned collagen matrices showed punctate MLCK localization (i.e. activated MLCK). Moreover, when the same tracking metrics were analyzed, after MLCK inhibition via ML-7 treatment, the differences seen previously were no longer significant, indicating that T cells in unaligned matrices exhibited less turning behavior and therefore straighter paths. We thus propose that the MLCK pathway is activated when T cells are turning and therefore are more active in unaligned gels causing the cells to turn more (Fig. 6). This is in contrast to previous observations in breast cancer cells where ML-7 treatment did not affect the trajectories of breast cancer cells in aligned and unaligned collagen gels [24]. This is not unlikely as breast cancer cell movement has been characterized as mesenchymal movement dependent on focal adhesions, actin stress fiber formation, and matrix degradation while T cell movement does not exhibit any of the same features. It is possible that MLCK punctate localization in aligned collagen gels is indicative of enzyme sequestration in order to reduce turning behavior due to MLCK phosphorylation. Indeed, spatial redistributions of MLCK have been observed in migrating and dividing cells. Specifically, activated MLCK has been shown to localize to the lamella of migrating epithelial cells in 2 dimensional culture as well as cleavage furrows during cytokinesis [31]. Overall, we have shown that activated CD8+ T cell migratory behavior can be modulated solely by modifying collagen fiber alignment. We have found that MLCK activity contributes to turning behavior of CD8+ T cells in unaligned collagen matrices, and that inhibition of this enzyme results in similar migratory patterns in unaligned and aligned collagen.
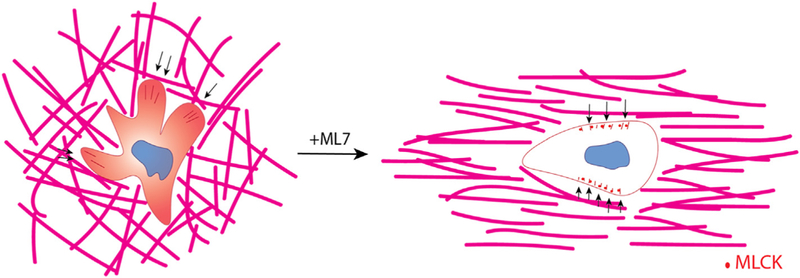
Collagen fiber structure guides 3D motility of CD8+ T cells. Schematic describing the effect of collagen fiber alignment on T cell motility. In unaligned collagen matrices (left) T cells extend numerous protrusions that pull on the collagen fibers (force indicated by black arrows) and exhibit diffuse MLCK expression patterns within the cytoplasm. These cells drastically change their mode of migration when MLCK is inhibited thereby resembling cells encapsulated in aligned collagen matrices. Although the forces exhibited on the matrix from these cells is significantly less it is mostly exerted perpendicular to the axis of alignment (indicated by black arrows). Moreover, MLCK localizes to the periphery of the cell in puncta.
Acknowledgements
This work was funded by the Johns Hopkins Physical Science - Oncology Center 5U54CA210173-03 and the Patrick C. Walsh Prostate Cancer Research Fund and the Johns Hopkins Prostate Cancer SPORE P30CA006973 (to SG) and fellowship from the Nanotechnology for Cancer Research Training Grant 5T32CA153952-09 (to HCP). The authors acknowledge services provided by the Johns Hopkins University Oncology Tissue Services core Sydney Kimmel Cancer Center (P30 CA006973) as well as the Johns Hopkins University Integrated Imaging Center.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.matbio.2019.02.003.
References
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.matbio.2019.02.003
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6697628
Citations & impact
Impact metrics
Article citations
Spatial interactions modulate tumor growth and immune infiltration.
NPJ Syst Biol Appl, 10(1):106, 30 Sep 2024
Cited by: 0 articles | PMID: 39349537 | PMCID: PMC11442770
CAF-induced physical constraints controlling T cell state and localization in solid tumours.
Nat Rev Cancer, 24(10):676-693, 09 Sep 2024
Cited by: 0 articles | PMID: 39251836
Review
Phenotypic and spatial heterogeneity of CD8+ tumour infiltrating lymphocytes.
Mol Cancer, 23(1):193, 09 Sep 2024
Cited by: 1 article | PMID: 39251981 | PMCID: PMC11382426
Review Free full text in Europe PMC
Altered extracellular matrix correlates with an immunosuppressive tumor microenvironment and disease progression in younger adults with oral cavity squamous cell carcinoma.
Front Oncol, 14:1412212, 18 Jun 2024
Cited by: 1 article | PMID: 38957320 | PMCID: PMC11217481
Oncogene-induced matrix reorganization controls CD8+ T cell function in the soft-tissue sarcoma microenvironment.
J Clin Invest, 134(11):e167826, 23 Apr 2024
Cited by: 0 articles | PMID: 38652549 | PMCID: PMC11142734
Go to all (28) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Rapid Quantification of 3D Collagen Fiber Alignment and Fiber Intersection Correlations with High Sensitivity.
PLoS One, 10(7):e0131814, 09 Jul 2015
Cited by: 15 articles | PMID: 26158674 | PMCID: PMC4497681
Contact guidance diversity in rotationally aligned collagen matrices.
Acta Biomater, 66:248-257, 28 Nov 2017
Cited by: 21 articles | PMID: 29196116 | PMCID: PMC5750117
Microtissue size and cell-cell communication modulate cell migration in arrayed 3D collagen gels.
Biomed Microdevices, 20(3):62, 30 Jul 2018
Cited by: 5 articles | PMID: 30062494
Directing fibroblast self-assembly to fabricate highly-aligned, collagen-rich matrices.
Acta Biomater, 81:70-79, 27 Sep 2018
Cited by: 8 articles | PMID: 30267883
Funding
Funders who supported this work.
Johns Hopkins Physical Science-Oncology Center (1)
Grant ID: 5U54CA210173-03
Johns Hopkins Prostate Cancer SPORE (1)
Grant ID: P30CA006973
NCI NIH HHS (4)
Grant ID: P30 CA006973
Grant ID: U54 CA210173
Grant ID: T32 CA130840
Grant ID: T32 CA153952





