Abstract
Free full text

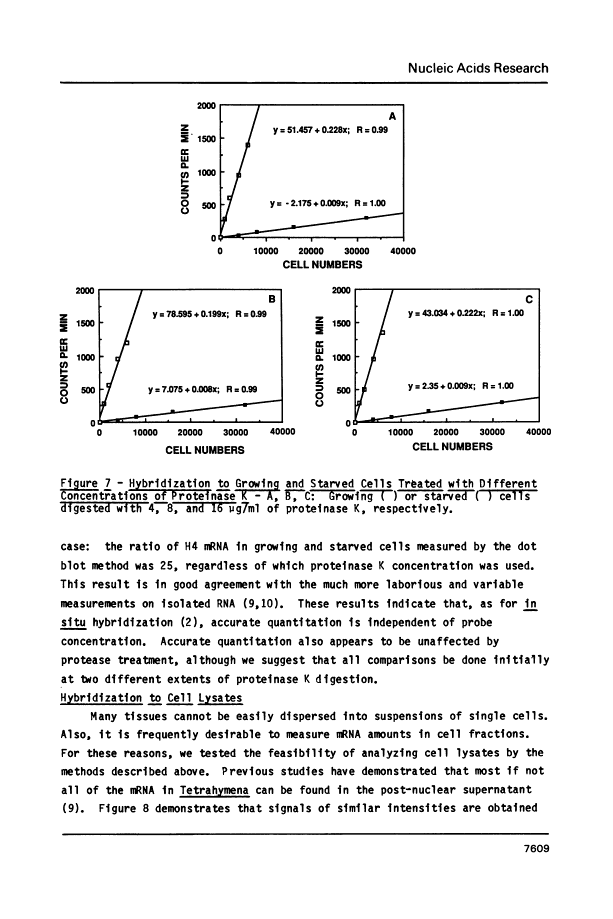
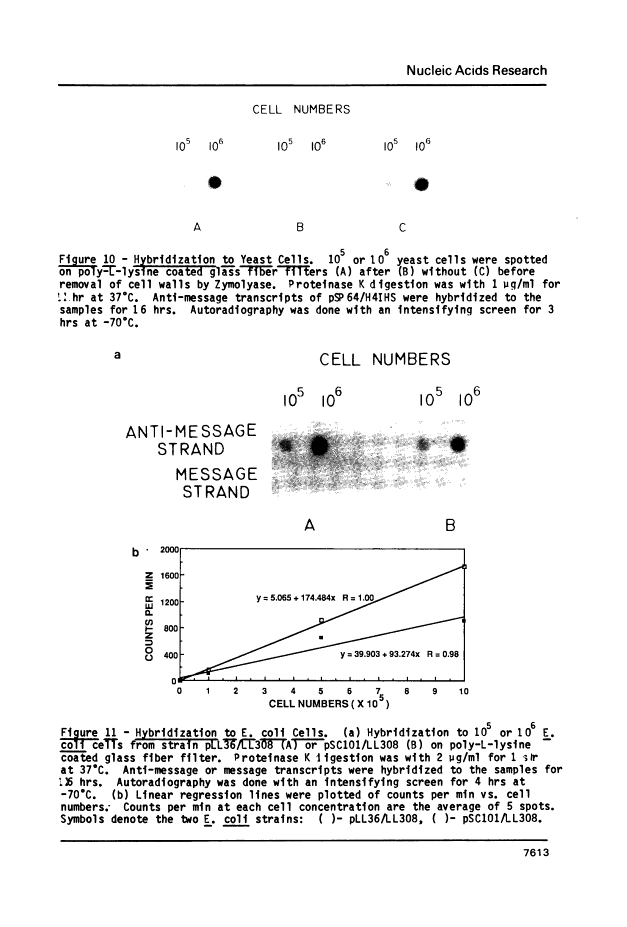
In situ dot blots: quantitation of mRNA in intact cells.
Abstract
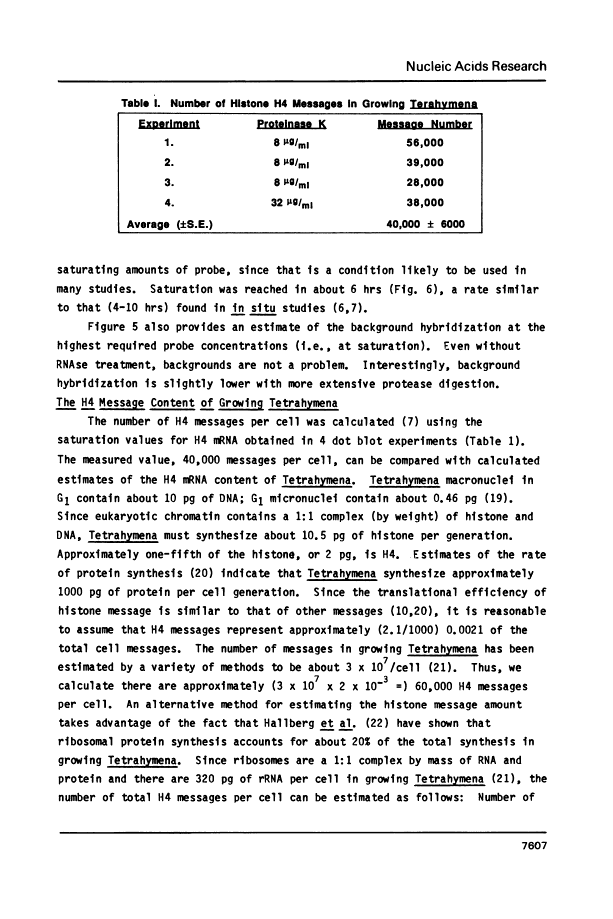
A rapid, simple and reproducible dot blot method is described for quantitating the amounts of specific messages in small numbers of intact cells. The method has been used to accurately determine the number of histone H4 mRNA molecules in growing (approximately 40,000) and in starved (approximately 1600) Tetrahymena thermophila, and to measure the amount of message contributed by an E. coli plasmid containing part of the S10 ribosomal operon. Use of the method is illustrated to optimize in situ hybridization protocols and to measure mRNA amounts in cell lysates. Preliminary studies also indicate that the method can be used to detect mRNA in intact yeast cells.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (2.0M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Click on the image to see a larger version.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brahic M, Haase AT. Detection of viral sequences of low reiteration frequency by in situ hybridization. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6125–6129. [Europe PMC free article] [Abstract] [Google Scholar]
- Angerer LM, Angerer RC. Detection of poly A+ RNA in sea urchin eggs and embryos by quantitative in situ hybridization. Nucleic Acids Res. 1981 Jun 25;9(12):2819–2840. [Europe PMC free article] [Abstract] [Google Scholar]
- Singer RH, Ward DC. Actin gene expression visualized in chicken muscle tissue culture by using in situ hybridization with a biotinated nucleotide analog. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7331–7335. [Europe PMC free article] [Abstract] [Google Scholar]
- Edwards MK, Wood WB. Location of specific messenger RNAs in Caenorhabditis elegans by cytological hybridization. Dev Biol. 1983 Jun;97(2):375–390. [Abstract] [Google Scholar]
- McAllister LB, Scheller RH, Kandel ER, Axel R. In situ hybridization to study the origin and fate of identified neurons. Science. 1983 Nov 18;222(4625):800–808. [Abstract] [Google Scholar]
- Cox KH, DeLeon DV, Angerer LM, Angerer RC. Detection of mrnas in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol. 1984 Feb;101(2):485–502. [Abstract] [Google Scholar]
- Lawrence JB, Singer RH. Quantitative analysis of in situ hybridization methods for the detection of actin gene expression. Nucleic Acids Res. 1985 Mar 11;13(5):1777–1799. [Europe PMC free article] [Abstract] [Google Scholar]
- Calzone FJ, Angerer RC, Gorovsky MA. Regulation of protein synthesis in Tetrahymena: isolation and characterization of polysomes by gel filtration and precipitation at pH 5.3. Nucleic Acids Res. 1982 Mar 25;10(6):2145–2161. [Europe PMC free article] [Abstract] [Google Scholar]
- Bannon GA, Bowen JK, Yao MC, Gorovsky MA. Tetrahymena H4 genes: structure, evolution and organization in macro- and micronuclei. Nucleic Acids Res. 1984 Feb 24;12(4):1961–1975. [Europe PMC free article] [Abstract] [Google Scholar]
- McMaster GK, Carmichael GG. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. [Europe PMC free article] [Abstract] [Google Scholar]
- Carmichael GG, McMaster GK. The analysis of nucleic acids in gels using glyoxal and acridine orange. Methods Enzymol. 1980;65(1):380–391. [Abstract] [Google Scholar]
- Thomas PS. Hybridization of denatured RNA transferred or dotted nitrocellulose paper. Methods Enzymol. 1983;100:255–266. [Abstract] [Google Scholar]
- DeLeon DV, Cox KH, Angerer LM, Angerer RC. Most early-variant histone mRNA is contained in the pronucleus of sea urchin eggs. Dev Biol. 1983 Nov;100(1):197–206. [Abstract] [Google Scholar]
- Novick P, Botstein D. Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell. 1985 Feb;40(2):405–416. [Abstract] [Google Scholar]
- Lindahl L, Archer R, Zengel JM. Transcription of the S10 ribosomal protein operon is regulated by an attenuator in the leader. Cell. 1983 May;33(1):241–248. [Abstract] [Google Scholar]
- Lindahl L, Zengel JM. Operon-specific regulation of ribosomal protein synthesis in Escherichia coli. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6542–6546. [Europe PMC free article] [Abstract] [Google Scholar]
- Gorovsky MA. Genome organization and reorganization in Tetrahymena. Annu Rev Genet. 1980;14:203–239. [Abstract] [Google Scholar]
- Calzone FJ, Angerer RC, Gorovsky MA. Regulation of protein synthesis in Tetrahymena. Quantitative estimates of the parameters determining the rates of protein synthesis in growing, starved, and starved-deciliated cells. J Biol Chem. 1983 Jun 10;258(11):6887–6898. [Abstract] [Google Scholar]
- Hallberg RL, Bruns PJ. Ribosome biosynthesis in Tetrahymena pyriformis. Regulation in response to nutritional changes. J Cell Biol. 1976 Nov;71(2):383–394. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/14.19.7597
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc311783?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
Gene expression analysis of human renal biopsies: recent developments towards molecular diagnosis of kidney disease.
Curr Opin Nephrol Hypertens, 13(3):313-318, 01 May 2004
Cited by: 15 articles | PMID: 15073490
Review
Cloning and characterization of the major histone H2A genes completes the cloning and sequencing of known histone genes of Tetrahymena thermophila.
Nucleic Acids Res, 24(15):3023-3030, 01 Aug 1996
Cited by: 11 articles | PMID: 8760889 | PMCID: PMC146061
Mapping the 5' and 3' ends of Tetrahymena thermophila mRNAs using RNA ligase mediated amplification of cDNA ends (RLM-RACE).
Nucleic Acids Res, 21(21):4954-4960, 01 Oct 1993
Cited by: 112 articles | PMID: 8177745 | PMCID: PMC311412
Regulation and evolution of the single alpha-tubulin gene of the ciliate Tetrahymena thermophila.
Cell Motil Cytoskeleton, 27(3):272-283, 01 Jan 1994
Cited by: 13 articles | PMID: 8020112
Replication-dependent and independent regulation of HMG expression during the cell cycle and conjugation in Tetrahymena.
Nucleic Acids Res, 20(24):6525-6533, 01 Dec 1992
Cited by: 5 articles | PMID: 1480473 | PMCID: PMC334567
Go to all (15) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Multiple, independently regulated, polyadenylated messages for histone H3 and H4 in Tetrahymena.
Nucleic Acids Res, 11(12):3903-3917, 01 Jun 1983
Cited by: 56 articles | PMID: 6135196 | PMCID: PMC326015
Unusual features of transcribed and translated regions of the histone H4 gene family of Tetrahymena thermophila.
Nucleic Acids Res, 15(1):141-160, 01 Jan 1987
Cited by: 39 articles | PMID: 3822803 | PMCID: PMC340402
Regulation of protein synthesis in Tetrahymena. RNA sequence sets of growing and starved cells.
J Biol Chem, 258(11):6899-6905, 01 Jun 1983
Cited by: 18 articles | PMID: 6189832
Tetrahymena H4 genes: structure, evolution and organization in macro- and micronuclei.
Nucleic Acids Res, 12(4):1961-1975, 01 Feb 1984
Cited by: 54 articles | PMID: 6322129 | PMCID: PMC318633