Abstract
Free full text

Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees
Abstract
Evolview is an interactive tree visualization tool designed to help researchers in visualizing phylogenetic trees and in annotating these with additional information. It offers the user with a platform to upload trees in most common tree formats, such as Newick/Phylip, Nexus, Nhx and PhyloXML, and provides a range of visualization options, using fifteen types of custom annotation datasets. The new version of Evolview was designed to provide simple tree uploads, manipulation and viewing options with additional annotation types. The ‘dataset system’ used for visualizing tree information has evolved substantially from the previous version, and the user can draw on a wide range of additional example visualizations. Developments since the last public release include a complete redesign of the user interface, new annotation dataset types, additional tree visualization styles, full-text search of the documentation, and some backend updates. The project management aspect of Evolview was also updated, with a unified approach to tree and project management and sharing. Evolview is freely available at: https://www.evolgenius.info/evolview/.
INTRODUCTION
Tree visualizations are increasingly used in multidisciplinary studies, partly driven by technological advancements in data driven research. The growing availability of large-scale genetic sequence data in a variety of fields such as medicine (1), ecology (2) and evolutionary biology (3) has resulted in a rising number of phylogenetic tree visualization tools. Specifically, visualization of large trees with 500 or more taxa has driven the development of many different approaches for tree analysis and visualization (4–6). In recent years, several online phylogenetic tree visualizations programs have been published (6–14) to overcome the challenges posed by the massive increased of dataset sizes.
Evolview is an online tree visualization and management tool, first released in 2012 (15) and last updated in 2016 (16). It takes phylogenetic trees in various formats, such as Newick/Phylip, Nexus, Nhx and PhyloXML, and visualizes them as phylograms and cladograms, each in either rectangular or circular layout. Evolview allows users to upload fifteen types of tree annotation datasets, to visualize them as pie charts, bar plots, heatmaps etc., and to export them in various graphical formats, including SVG, PDF, TFF, JPEG and textual formats such as Newick, Nexus, Nhx and PhyloXML. The new update is aimed at improving the overall interface experience of the user, with new tree visualization styles and added functionalities. In total, Evolview now supports fifteen annotation types. With the carefully re-designed user interface (UI), users can view, manage and manipulate the uploaded trees and datasets easily and efficiently. Consolidations related to the UI have been done on upload options to deliver a simpler user interaction. The quick access controls to export trees in various image and textual formats have been redesigned to also improve the tree saving experience. The ‘dataset-system’ can be used to provide additional visualization information, such as plots, and can also help users to manipulate the visual representations of leaves and internal branches of the tree. With the expanded functionalities, we observed a need for a clearer and simpler documentation. In this update, we thus introduce a complete set of detailed user help pages with the ability to search for topics.
About 5541 new users have registered since 2016; users have uploaded 29 411 new trees and 33 729 new datasets since the last release. In 2017, on average ~750 trees and more than 800 datasets were uploaded per month. The ratio of active versus new users has also increased steadily; in 2018, the numbers of trees and of datasets uploaded reached over 900 per month.
USER INTERFACE: NEW UI AND LATEST IMPROVEMENTS
Over the years, we have made many improvements to Evolview and added new functionalities based on user feedback. The new version includes a complete UI redesign to accommodate the current web design standards and to provide the user with an improved experience. The access to the various tree controls was grouped into sections; the ability to toggle between show/hide statuses provides a simple interaction (Figure (Figure1).1). The controls were grouped into quick access controls, tree tweaking controls, export controls and upload controls (Figure (Figure11).
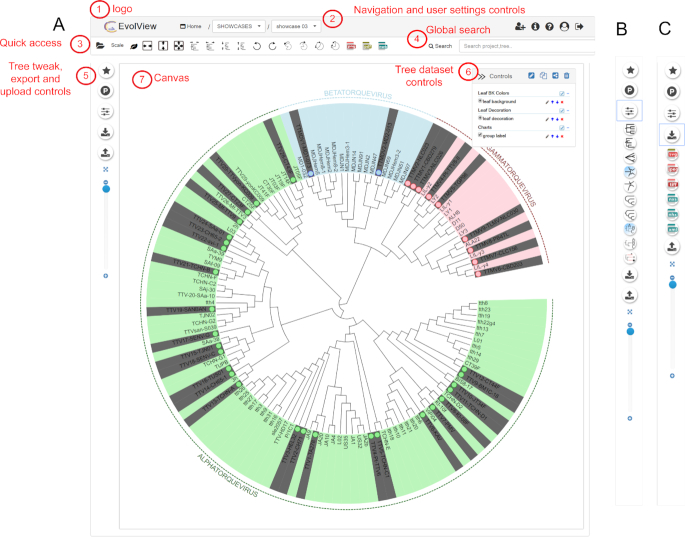
The new and improved interface. (A) The main interface consist of seven parts. (1) Logo. Clicking on the logo shows statistics on Evolview usage in a popup panel. (2) Header bar, consisting of project/tree navigation controls along with quick help and user account settings. (3) Quick access tool bar, providing a comprehensive list of quick tree tweaking controls for easy access. (4) Search box, allowing the user to search across projects and trees. (5) Tree style, tweak, upload and export controls. (6) Tree dataset controls. Datasets can be easily manipulated using the icons. Users can choose to hide the user data panel to give more space to the canvas by clicking the widget to the right of this panel. The panel also provides controls to share, edit, copy, and delete the currently displayed tree information. (7) Tree canvas, displaying the visualization results. (B) Expanded menu of Tree visualization controls, which are displayed when clicking on the tree style control button. The menu allows to choose between different phylogram and cladogram display styles. (C) Expanded menu for Tree exports, allowing users to save the tree data in various formats, including Newick, Nexus, NHX, PhyloXML, and image formats such as SVG, PNG, TIFF and PDF.
The new Evolview UI supports larger icons, and the grouping of controls provides more area for tree viewing. The navigation between projects and trees was made easy through project/tree drop-downs in the header bar section of the web page. Navigation can also be performed through the global search functionality (Figure (Figure1).1). For more information on new and old functionalities, users can use the inbuilt search function located in the help pages.
User interface design
Figure Figure11 shows a complete overview over the various new aspects of the UI re-design. The circular controls on the left provide more comprehensive functions such as managing projects, displaying tree information, switching tree display between phylogram and cladogram, exporting tree information, and setting dataset upload options.
Global search for projects and trees
With an increasing number of projects and trees, navigating across trees has become tedious for some users. To address this problem, Evolview now supports searching for trees and projects. The search functionality provides suggestions based on the user input in the search area. Suggestions differentiate between project suggestions and tree suggestions, which are displayed along with the corresponding project information.
Improved annotation upload
The annotation upload functionality has been consolidated into a single control center (Figure (Figure2A),2A), which offers various dataset upload types along with links to help information. The dataset file formats remains the same as in the previous version, but additional annotation dataset type has been included. The rich-text editor now highlights different elements of a dataset with different colors and font styles, and allows users to validate a dataset before uploading it to Evolview.
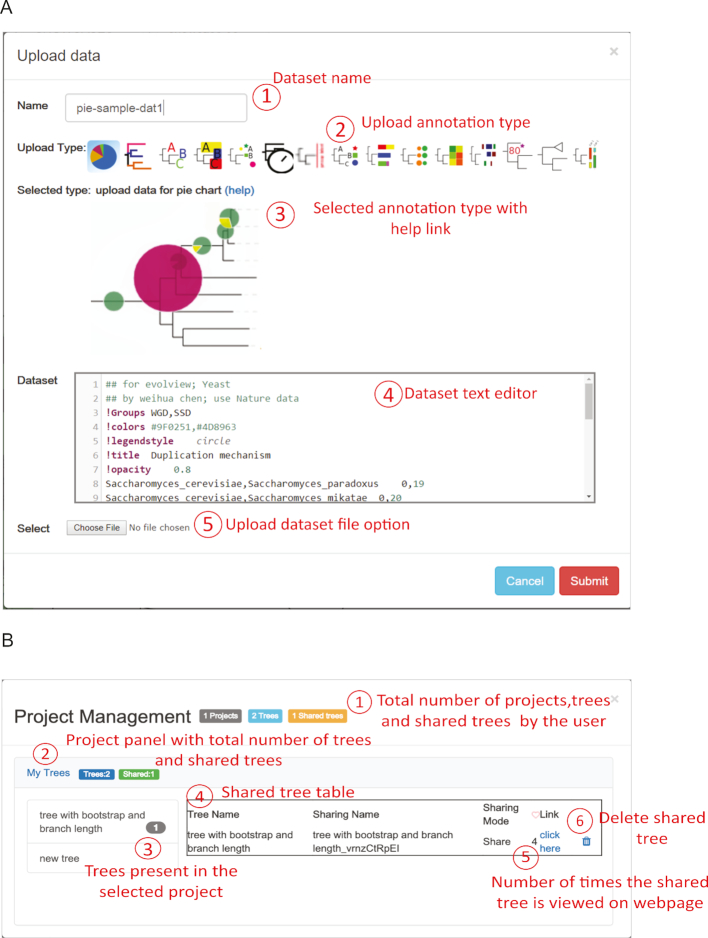
New sections of the user interface. (A) A unified Upload dataset window displays options to select and upload data for all supported annotation types. (B) The Project Management window assists users in managing the shared projects, trees, and datasets and allows them to assess their viewing statistics.
Project management
While previous versions structured trees and datasets into projects, Evolview v3 introduces a comprehensive project management system (Figure (Figure2B).2B). This system lets users monitor and control projects and tree data that is shared with collaborators or other users. It also lets the user track the shared content usage/view count, providing an overall view about the access to project datasets.
NEWLY ADDED AND UPDATED ANNOTATION TYPES
Evolview v3 supports in total fifteen annotation types, including one newly added. We also updated three existing annotation types and added two new tree visualization styles.
New annotation type
Timeline plot is a newly added annotation type. It allows the user to put trees and associated data into a temporal context (Figure (Figure3A),3A), emphasized through customization options such as grid line backgrounds.
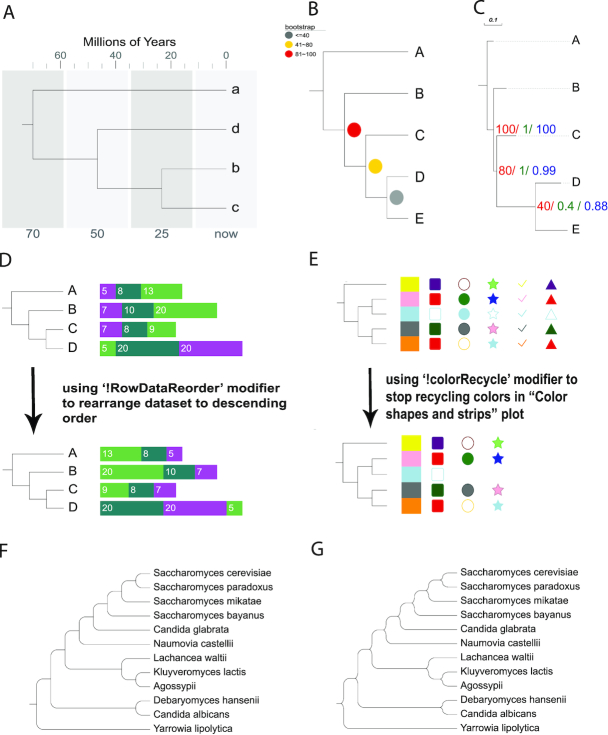
Tree annotation updates since Evolview v2. In addition to the UI changes and performance improvements, Evolview v3 provides the following updates to its tree annotation functionalities. (A) Timeline plot, a new annotation dataset type allowing users to put trees into a temporal context. (B) New bootstrap styles, allowing users to customize colors of bootstrap values based on their value range, and even replace them with various symbols. (C) Multiple bootstrap values display, supporting the display of multiple bootstrap values on the same branches, with styling options. (D) Bar plot data can now be displayed in ascending and descending order. (E) Color shapes and strips plot, illustrating the option to stop recycling colors. The ‘!colorRecycle’ modifier lets user have empty or no value rows for some columns. (F and G) New tree visualization styles, displayed for phylograms. The styles can be applied to both phylograms and cladograms.
Other updates
The Bootstrap annotation dataset type now supports multiple sets of bootstrap values for the same tree, providing additional bootstrap styles (Figure (Figure3B3B and C). With the Bar plot annotation dataset, now users can use the modifier ‘!RowDataReorder’ to change the display order of the stacked bars according to their values in either ascending or descending orders (Figure (Figure3D).3D). The Color shapes and strips annotation now has an additional option to control color recycling (using the modifier ‘!colorRecycle’; Figure Figure3E).3E). Finally, two new tree styles were added to the tree visualization controls (Figure (Figure3F3F and G).
See Table Table11 for key features comparison between the version 2 and version 3 of Evolview.
Table 1.
Evolview version 2 and version 3 comparison
| Features | Version 2 | Version 3 |
|---|---|---|
| Web protocol | HTTP | HTTPS |
| UI components | 1. Tree tweaking controls, export controls, upload controls were divided into tabs; | 1. Controls related to tree manipulation, data upload, and tree export are grouped into individual control options. |
| 2. Tree and project management were shown from the same window, with all options at one place, thus dividing the tree view space, reducing the viewing area. | 2. New UI design has been adopted to provide more tree viewing space and easy access controls. | |
| Search | Not available | Tree and project search function is present. |
| Project management | Not available | New Tree and project management window to manage user data and check viewing statistics of shared trees. |
| User documentation | Help content provided through GitHub pages | New and improved help pages with additional search functionality, hosted from same Evolview domain. |
| Total number of supported annotation types | 14 | 15 |
| FLASH dependency | YES, site usage statistics used Google annotation timeline to show the information. | NO, site usage statistics now uses a javascript based Google charts to show the information. |
CONCLUSION
In this article, we introduced Evolview v3, a comprehensive web application for visualizing, annotating and managing phylogenetic trees. Updates since the previous version include a re-designed user interface, additional annotation types and tree visualizing styles, and an overall improved user experience. In the future, we aim to keep Evolview up-to-date and state-of-the-art by supporting new annotation types that reflect the needs of the users.
DATA AVAILABILITY AND REQUIREMENTS
Evolview is accessible using modern browsers such as Firefox, Chrome, Opera and Safari. Due to some compatibility issues, Internet Explorer (IE) and Microsoft Edge are not supported. However, Chromium-based Microsoft Edge will be supported in near future. The Evolview website is free and open to all users and there is no login requirement. If users want to save their data on our server, an account is required. Registration and the account are free.
Evolview is freely available at: https://www.evolgenius.info/evolview/. Its source codes are available upon request. A zip file contains all trees and datasets used to create Figure Figure33 of this study is available for download at: https://evolgenius.info/supplimentary-tree-files.zip. Users who wish to install Evolview locally can find the compiled codes and instructions at: https://evolgenius.info/helpsite/deploy-local.html. The documentation is hosted at: https://evolgenius.info/evolview/helpsite/quick-start.html. The previous version of Evolview is available at: https://evolgenius.info/evolview-v2/.
FUNDING
National Key Research and Development Program of China [2018YFC0910502].
Conflict of interest statement. None declared.
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/gkz357
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/nar/article-pdf/47/W1/W270/28880085/gkz357.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/nar/gkz357
Article citations
Characterization of the gibberellic oxidase gene <i>SdGA20ox1</i> in <i>Sophora davidii</i> (Franch.) skeels and interaction protein screening.
Front Plant Sci, 15:1478854, 16 Oct 2024
Cited by: 0 articles | PMID: 39479549 | PMCID: PMC11521860
Origin, Evolution, and Diversification of the Expansin Family in Plants.
Int J Mol Sci, 25(21):11814, 03 Nov 2024
Cited by: 0 articles | PMID: 39519364 | PMCID: PMC11547041
Genome-Wide Identification of MKK Gene Family and Response to Hormone and Abiotic Stress in Rice.
Plants (Basel), 13(20):2922, 18 Oct 2024
Cited by: 0 articles | PMID: 39458871 | PMCID: PMC11510841
Introgression of Herbicide-Resistant Gene from Genetically Modified <i>Brassica napus</i> L. to <i>Brassica rapa</i> through Backcrossing.
Plants (Basel), 13(20):2863, 13 Oct 2024
Cited by: 0 articles | PMID: 39458810 | PMCID: PMC11510986
Diverse rhizosphere-associated Pseudomonas genomes from along a Wadden Island salt marsh transition zone.
Sci Data, 11(1):1140, 17 Oct 2024
Cited by: 0 articles | PMID: 39419992 | PMCID: PMC11487137
Go to all (381) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees.
Nucleic Acids Res, 44(w1):W236-41, 30 Apr 2016
Cited by: 315 articles | PMID: 27131786 | PMCID: PMC4987921
EvolView, an online tool for visualizing, annotating and managing phylogenetic trees.
Nucleic Acids Res, 40(web server issue):W569-72, 13 Jun 2012
Cited by: 231 articles | PMID: 22695796 | PMCID: PMC3394307
Interactive Tree Of Life (iTOL) v4: recent updates and new developments.
Nucleic Acids Res, 47(w1):W256-W259, 01 Jul 2019
Cited by: 2992 articles | PMID: 30931475 | PMCID: PMC6602468
Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees.
Nucleic Acids Res, 44(w1):W242-5, 19 Apr 2016
Cited by: 2689 articles | PMID: 27095192 | PMCID: PMC4987883
Funding
Funders who supported this work.
National Key Research and Development Program of China (1)
Grant ID: 2018YFC0910502





