Abstract
Free full text

Precision Oncology: Three Small Steps Forward
Abstract
Only a subset of cancer patients currently benefit from personalized treatment approaches. Detection of actionable drug targets in cell-free DNA, more comprehensive molecular profiling integrating transcriptional analyses, and personalized combination regimens all hold promise in expanding the benefits of personalized oncology to larger numbers of cancer patients.
Over the past two decades, several dozen cancer therapies have been developed that selectively inhibit molecular alterations that promote cancer initiation and/or progression. In large part, these drugs are effective only in patients whose tumors harbor specific molecular alterations. The need to identify the subset of patients most likely to respond has necessitated the development of diagnostic assays that rapidly and accurately identify those patients whose tumors harbor the predictive biomarkers of interest. Initial diagnostic platforms were based on polymerase chain reaction, immunohisto-chemistry (IHC), or fluorescence in situ hybridization and capable of identifying alterations in only one or a few cancer-associated genes. More recently, next-generation sequencing (NGS) has enabled rapid and lower-cost interrogation of hundreds or thousands of cancer-associated genes using small quantities of DNA derived from formalin-fixed, paraffin-embedded tumor tissue (Zehir et al., 2017). The rapid adoption of NGS-based clinical diagnostics has led to the hope that a greater fraction of patients will soon benefit from personalized treatment approaches.
While targeted cancer therapies can have profound and durable clinical activity, only a minority of patients currently benefit. This can be attributed to several factors. Foremost, current tumor profiling panels identify genomic alterations that robustly predict for drug response in only a minority of cancer patients. In others, druggable biologic processes mediated by protein overexpression or loss or epigenetic alterations may go undetected by DNA-based profiling assays.
In such cases, multiplatform tumor profiling incorporating transcriptomic or proteomic analyses may benefit individual patients. Second, not all patients who receive matched therapies respond. Variability of response may be the result of additional genomic complexity, for example PTEN mutations in EGFR mutant tumors (Sos et al., 2009), or lineage-specific factors (Solit and Jänne 2012) that abrogate dependence on the drug target. In such instances, molecular profiling may identify potentially effective personalized drug combinations. Finally, in some patients, adequate tumor tissue may not be readily available for genomic analysis. In this clinical context, analysis of circulating tumor DNA (ctDNA) may facilitate the non-invasive identification of actionable genomic alterations. Here, we summarize three recently reported clinical studies that sought to expand the proportion of patients who benefit from molecularly based therapies by addressing limitations of current precision oncology paradigms.
As outlined above, access to adequate tumor tissue for molecular analyses remains a significant challenge in a subset of advanced cancer patients. To address this challenge, the TARGET study (Rothwell et al., 2019) examined the feasibility of using ctDNA as a substitute for tissue-based testing. In a cohort of 100 patients, the investigators demonstrated that a 641-gene ctDNA NGS panel could be successfully performed in 99% of patients. Actionable alterations were identified in 41 patients, of which 11 received matched therapy, with partial responses achieved in 4. This and other recent studies of ctDNA confirm that non-invasive detection of actionable alterations is feasible but that only a minority of patients ultimately receive or derive benefit from a matched therapy.
As accessibility to more comprehensive clinical tumor profiling becomes more widespread, the challenge of rationally targeting the complex co-mutational pattern found in most cancers remains. The I-PREDICT trial (Sicklick et al., 2019) was based on the hypothesis that molecular complexity limits the utility of targeted therapies and that personalized combinations matched to a patient’s specific tumor mutational profile could result in improved clinical outcomes. To prospectively identify actionable genomic alterations, patients were offered NGS of tumor tissue or ctDNA. Of 149 patients consented, 73 were administered a personalized matched therapy. For each patient, a matching score was calculated, defined as the number of alterations targeted with an administered drug divided by the total number of actionable alterations detected in the patient’s tumor. A high matching score was an independent predictor of longer progression-free survival after adjusting for other variables.
The WINTHER trial (Rodon et al., 2019) sought to test whether expanded tumor profiling incorporating RNA analysis could increase the match rate and improve patient outcomes. Of the 303 patients that consented, 69 were enrolled to Arm A (DNA-guided matching) and 38 to Arm B (RNA-guided matching). As in I-PREDICT, a matching score was calculated for each patient. For RNA-guided matching, an algorithm was developed that utilized a knowledge base of genes known to be associated with drug efficacy that was then cross-referenced to gene-expression changes in the patient’s tumor. As in I-PREDICT, a higher matching score was associated with longer progression-free survival, and the authors concluded that both genomic and transcriptomic profiling were useful for informing personalized therapy recommendations and improving patient outcome.
Relevant to both the I-PREDICT and WINTHER trials, the lack of comparator arms, the small sample sizes, and the heterogeneity of the trial populations dictate that additional studies will be needed to change clinical practice. While both trials suggest that a higher matching score may be associated with clinical benefit, as measured by comparing PFS on the investigational therapy to that on the prior line of therapy (Von Hoff et al., 2010), the observed clinical benefit of RNA expression analysis and investigational combinations of customized agents were modest at best, and few durable responders were reported. A major limitation of the matching score algorithm employed is the presumption that actionable alterations have equal predictive value. In contrast, clinical experience suggests that the likelihood of clinical response varies widely among actionable drug targets and in different tumor lineages (Figure 1). This variability in clinical actionability among drug targets has prompted the development of more nuanced knowledge bases, such as OncoKB (Chakravarty et al., 2017), which assign levels of evidence to molecular alterations as a function of the specific mutant allele and tumor type. Future refinements of the matching score should seek to incorporate levels of evidence for each alteration, giving varying weights to mutant alleles based on the strength of the available clinical and biological data and as a function of specific lineage or co-mutational contexts. Furthermore, as most mutations identified by NGS, even in well-studied cancer genes, are variants of unknown significance, achieving the full promise of precision oncology will require comprehensive characterization of a significantly larger number of mutational events than has been performed to date.
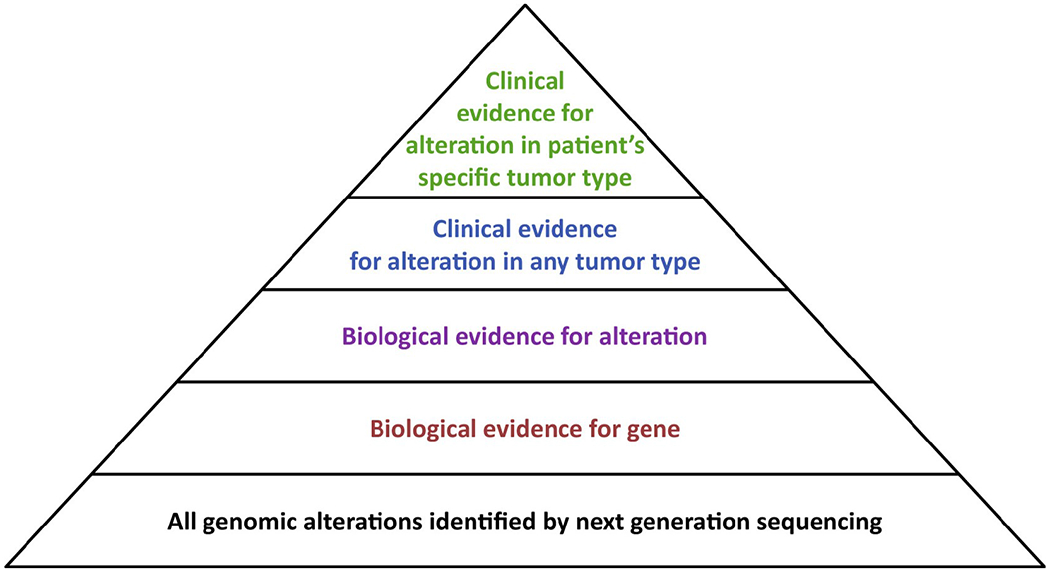
For most targeted therapies, drug sensitivity varies as a function of both cancer type and the specific mutant allele present. Future matching algorithms should give varying weight to alterations based on these factors, with compelling clinical evidence in the patient’s specific disease type and mutant allele conferring the greatest weight. A primarly limitation of current cancer precision paradigms is that few alterations identified by clinical tumor profiling are predictive of drug response based on compelling clinical data generated in the context of the patient’s specific mutant allele and disease type.
In sum, next-generation sequencing is rapidly transforming the clinical management of cancer patients. While technological improvements and reductions in sequencing costs will inevitably result in an expansion of the breadth of clinical tumor-profiling assays, these diagnostic innovations will need to be integrated with biological and clinical studies designed to elucidate allele- and lineage-specific factors that influence drug sensitivity. The precision medicine field also needs to be more nuanced as to how it defines clinical actionability. Despite frequent claims to the contrary, effective therapies are lacking for many cancer-associated genes, including KRAS, TP53, RB1, NF1, and PTEN, among others. We thus believe that expanding the fraction of cancer patients who benefit from precision medicine will be driven more by the development of drugs that are effective in the context of these currently non-druggable molecular alterations rather than by incremental improvements in tumor-profiling assays. As tumors typically harbor more than one actionable molecular driver, we should prioritize the development of agents that are highly selective to facilitate the testing of personalized drug combinations. Future studies should also seek to address the fundamental question of whether the fraction of targets matched or the quality of those matches is most important in dictating drug sensitivity. In essence, biology and pharmacology, and not just genetics, matter for improving the treatment of cancer patients.
Footnotes
DECLARATION OF INTERESTS
D.B.S. has consulted with or received honorarium from Pfizer, Loxo Oncology, lllumina, Vivideon Oncology, and Lilly Oncology.
REFERENCES
- Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MH, et al. (2017). OncoKB: a precision oncology knowledge base. JCO Precis. Oncol 2017 Published online May 16, 2017. 10.1200/PO.17.00011. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Rodon J, Soria JC, Berger R, Miller WH, Rubin E, Kugel A, Tsimberidou A, Saintigny P, Ackerstein A, Braña I, et al. (2019). Genomic and transcriptomic profiling expands precision cancer medicine: the WINTHER trial. Nat. Med 25, 751–758. [Europe PMC free article] [Abstract] [Google Scholar]
- Rothwell DG, Ayub M, Cook N,Thistlethwaite F, Carter L, Dean E, Smith N, Villa S, Dransfield J, Clipson A, et al. (2019). Utility of CtDNAto support patient selection for early phase clinical trials: the TARGET study. Nat. Med 25, 738–743. [Abstract] [Google Scholar]
- Sicklick JK, Kato S, Okamura R, Schwaederle M, Hahn ME, Williams CB, De P, Krie A, Piccioni DE, Miller VA, et al. (2019). Molecular profiling of cancer patients enables personalized combination therapy: the I-PREDICT study. Nat. Med 25, 744–750. [Europe PMC free article] [Abstract] [Google Scholar]
- Solit DB, and Jänne PA (2012). Primed for resistance. Nature 483, 44–45. [Abstract] [Google Scholar]
- Sos ML, Koker M, Weir BA, Heynck S, Rabinovsky R, Zander T, Seeger JM, Weiss J, Fischer F, Frommolt P, et al. (2009). PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR. Cancer Res. 69, 3256–3261. [Europe PMC free article] [Abstract] [Google Scholar]
- Von Hoff DD, Stephenson JJ, Rosen P, Loesch DM, Borad MJ, Anthony S, Jameson G, Brown S, Cantafio N, Richards DA, et al. (2010). Pilot study using molecular profiling of patients’ tumors to find potential targets and select treatments for their refractory cancers. J. Clin. Oncol 28, 4877–4883. [Abstract] [Google Scholar]
- Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, et al. (2017). Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med 23, 703–713. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.ccell.2019.05.009
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S153561081930251X/pdf
Citations & impact
Impact metrics
Article citations
Horizons in Veterinary Precision Oncology: Fundamentals of Cancer Genomics and Applications of Liquid Biopsy for the Detection, Characterization, and Management of Cancer in Dogs.
Front Vet Sci, 8:664718, 23 Mar 2021
Cited by: 18 articles | PMID: 33834049 | PMCID: PMC8021921
Review Free full text in Europe PMC
The Potential of a Digital Twin in Surgery.
Surg Innov, 28(4):509-510, 08 Dec 2020
Cited by: 15 articles | PMID: 33290181 | PMCID: PMC8381595
Large-Scale Characterization of Drug Responses of Clinically Relevant Proteins in Cancer Cell Lines.
Cancer Cell, 38(6):829-843.e4, 05 Nov 2020
Cited by: 27 articles | PMID: 33157050 | PMCID: PMC7738392
Repositioning Lidocaine as an Anticancer Drug: The Role Beyond Anesthesia.
Front Cell Dev Biol, 8:565, 17 Jul 2020
Cited by: 20 articles | PMID: 32766241 | PMCID: PMC7379838
Review Free full text in Europe PMC
Deep Digital Phenotyping and Digital Twins for Precision Health: Time to Dig Deeper.
J Med Internet Res, 22(3):e16770, 03 Mar 2020
Cited by: 24 articles | PMID: 32130138 | PMCID: PMC7078624
Go to all (6) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The Personalization of Therapy: Molecular Profiling Technologies and Their Application.
Semin Oncol, 42(6):775-787, 30 Sep 2015
Cited by: 4 articles | PMID: 26615125
Review
Precision oncology: neither a silver bullet nor a dream.
Pharmacogenomics, 18(16):1525-1539, 24 Oct 2017
Cited by: 14 articles | PMID: 29061079
Review
Precision cancer medicine: the future is now, only better.
Am Soc Clin Oncol Educ Book, 61-69, 01 Jan 2014
Cited by: 19 articles | PMID: 24857061
Review
SwissMTB: establishing comprehensive molecular cancer diagnostics in Swiss clinics.
BMC Med Inform Decis Mak, 18(1):89, 29 Oct 2018
Cited by: 11 articles | PMID: 30373609 | PMCID: PMC6206832





