Abstract
Free full text

Salmonella enterica Serovar Panama, an Understudied Serovar Responsible for Extraintestinal Salmonellosis Worldwide
ABSTRACT
In recent years nontyphoidal Salmonella has emerged as one of the pathogens most frequently isolated from the bloodstream in humans. Only a small group of Salmonella serovars cause this systemic infection, known as invasive nontyphoidal salmonellosis. Here, we present a focused minireview on Salmonella enterica serovar Panama, a serovar responsible for invasive salmonellosis worldwide. S. Panama has been linked with infection of extraintestinal sites in humans, causing septicemia, meningitis, and osteomyelitis. The clinical picture is often complicated by antimicrobial resistance and has been associated with a large repertoire of transmission vehicles, including human feces and breast milk. Nonhuman sources of S. Panama involve reptiles and environmental reservoirs, as well as food animals, such as pigs. The tendency of S. Panama to cause invasive disease may be linked to certain serovar-specific genetic factors.
INTRODUCTION
Salmonellosis is a disease caused by the enteric pathogen Salmonella enterica, a species that includes 2,637 different serovars (1). The various clinical presentations of Salmonella disease in humans include enteric fever, gastroenteritis, extraintestinal complications, and a chronic carrier state (2, 3). The clinical manifestation of Salmonella is dependent on a number of features, including host immune status (reviewed in reference 4), as well as factors specific to the Salmonella pathovariant that is causing the infection (5). Certain pathogen factors are associated with clinical presentation, including serovar and certain core and accessory genome components, such as the presence of plasmids, prophages, virulence factors, and antimicrobial resistance genes (6). In this review, we focus on Salmonella enterica serovar Panama, which has a strong association with invasive disease (7) and is a rarely discussed serovar that has global public health relevance. We review the global epidemiology, as well as the clinical picture, the transmission vehicles, and antimicrobial resistance, and put them into the context of our current genomic understanding.
GLOBAL DISEASE BURDEN AND EPIDEMIOLOGY
In 1931, an unknown bacterium caused widespread foodborne diarrheal disease among American soldiers stationed at the Panama Canal. A full microbiological investigation was conducted, and the organism was identified as a “not previously described Salmonella,” which was subsequently named S. Panama (8). Since initial isolation and serological characterization, S. Panama has been implicated in numerous geographically localized outbreaks of gastrointestinal and extraintestinal disease around the globe (9).
French territories in the Americas.
S. Panama is responsible for a significant proportion of the total Salmonella disease burden worldwide and is a leading cause of invasive nontyphoidal salmonellosis in French territories of America located in the Caribbean and South America (7, 10, 11). Between 1972 and 1974, S. Panama was the major Salmonella serovar isolated from human fecal samples in Martinique (10). Two decades later, a study focused on pediatric salmonellosis in Martinique identified S. Panama as the most commonly isolated Salmonella serovar, accounting for 35% of all cases between 1990 and 1994 (11). Similarly, in French Guiana, S. Panama was the most frequent Salmonella serovar acquired by humans, accounting for 12.9% of all cases of Salmonella infection in 2011 (12). More recently, S. Panama was listed as the Salmonella serovar most frequently isolated from pediatric blood samples in Guadeloupe, contributing to one-third of all cases of Salmonella infection between 2010 and 2014 (7), and univariate analysis showed S. Panama was associated with causing disease in children older than 6 months of age (P
months of age (P =
= 0.002) (7). These examples demonstrate the significant impact that S. Panama has on public health in French territories in the Americas and shows that S. Panama causes extraintestinal infection and gastrointestinal disease, particularly in children. Although more extensive work needs to be done, no evidence for antimicrobial resistance in S. Panama exists in these regions.
0.002) (7). These examples demonstrate the significant impact that S. Panama has on public health in French territories in the Americas and shows that S. Panama causes extraintestinal infection and gastrointestinal disease, particularly in children. Although more extensive work needs to be done, no evidence for antimicrobial resistance in S. Panama exists in these regions.
Latin America.
S. Panama causes a significant proportion of the salmonellosis burden in Latin America, which in the 2000s was 3.5 cases confirmed by serotyping per 100,000 people (9). As early as the 1950s, 41 (12%) of 357 human Salmonella isolates collected in Maracaibo, Venezuela, were Salmonella serovar Panama. Interestingly, 15 isolates came from patients suffering from gastroenteritis, 4 came from individuals with enteric fever, and 22 came from healthy carriers, indicating that S. Panama could be carried asymptomatically (13).
Historically, an outbreak of S. Panama in Chile originated from river water in Santiago in 1975 (14). By 1978, the serovar had infiltrated almost the entire country, expanding southward to Punta Arenas and northward toward Arica. The resulting human epidemic across Chile lasted for 4 years and involved the isolation of S. Panama from food, animals, and water, demonstrating the ability of the serovar to spread rapidly and survive outside of the human host. The majority of clinical cases involved children under 15 months of age with self-limiting diarrheal disease. However, examples of bacteremia and meningitis were also reported (14).
months of age with self-limiting diarrheal disease. However, examples of bacteremia and meningitis were also reported (14).
S. Panama continues to be isolated periodically in Chile and other parts of Latin America. According to global Salmonella monitoring compiled by the World Health Organization between 2001 and 2007, S. Panama was the ninth most common serovar isolated in Latin America (9). In 2007, S. Panama was responsible for 1% of 3,439 cases of Salmonella infection across Argentina, Brazil, Chile, and Costa Rica (9). In Colombia, S. Panama was the fifth most common serovar isolated from patients between 2005 and 2011 (15). Rapid dissemination of S. Panama around Chile in the 1970s, and the consistent reporting of the serovar among the top 10 that cause human disease post-2000, highlight the persistent burden of S. Panama in Latin America.
Asia.
In Asia, S. Panama was the 11th most frequently isolated Salmonella serovar in humans between 2001 and 2007 (9). In 2001, 4% of salmonellosis cases in Thailand were caused by S. Panama, dropping to 3% in 2007 (9). In Tokyo, Japan, S. Panama was the third most common Salmonella serovar between 1974 and 1979, accounting for 5% of cases of Salmonella infection, and was commonly isolated from asymptomatic people (16). In Taiwan, where S. Panama causes 7% of the clinical cases of salmonellosis, S. Panama causes a higher rate of bacteremia in children under 5 years of age than other serovars, such as Salmonella enterica serovar Enteritidis (17). These findings demonstrate that S. Panama is an important public health issue in Asia.
years of age than other serovars, such as Salmonella enterica serovar Enteritidis (17). These findings demonstrate that S. Panama is an important public health issue in Asia.
Europe and the United States of America.
Historically, S. Panama has caused a significant proportion of the salmonellosis cases in Europe, particularly related to the pig industry, and in the United States, where S. Panama has been implicated in several hospital and statewide outbreaks associated with a variety of food sources (18, 30, 98). The serovar was introduced into the United Kingdom during World War II as a result of unsterilized dried eggs imported from the United States being fed to pigs (18). Humans have also been involved in the spread of S. Panama during hospital outbreaks in France and in other Western European countries during the 1960s and 1970s (19, 20). Over this period, there was a 3-fold increase in S. Panama cases in the United Kingdom, which led to a doubling of the number of salmonellosis cases (18). Subsequently, between 1969 and 1984, S. Panama was one of the top five serovars responsible for invasive disease in the United Kingdom (21). It is thought that these isolates were exposed to high antibiotic selective pressure in humans or food animals and consequently became resistant to antibiotics via acquisition of many types of plasmids (22,–28). Elsewhere in the European Union, S. Panama was reported among the top 10 most frequently isolated serovars during 2012, following 706 confirmed cases of S. Panama salmonellosis associated with outbreaks in Germany and Italy (29). Sporadic outbreaks of S. Panama salmonellosis also occurred in Switzerland (1972), Hungary (1979), Spain (1998), and the Netherlands (2008) (30,–33). S. Panama maintained its ranking in the top 20 serovars associated with salmonellosis in the European Union until 2017, when it was replaced by other serovars (Salmonella enterica serovar Brandenburg, Salmonella enterica serovar Kottbus, and Salmonella enterica serovar Coeln) (34).
CLINICAL PICTURE IN HUMANS
Although S. Panama can cause gastrointestinal infection in humans (9), the serovar is more widely known for its ability to cause invasive disease and to colonize extraintestinal sites. For most salmonellae, extraintestinal colonization refers to bloodstream infection (2). However, S. Panama can also invade specific body sites, causing atypical presentations, including throat infection, brain abscess, and Bartholin’s abscess (35,–37) (summarized in Fig. 1). These unexpected symptoms of S. Panama infection can impede diagnosis and delay treatment.
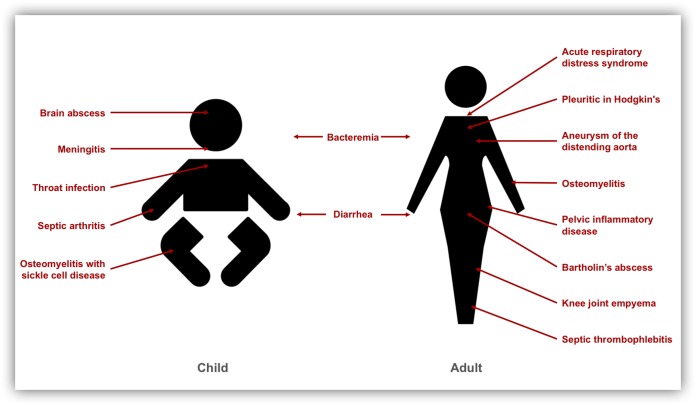
Overview of the clinical presentations caused by S. Panama in adults and children according to the published literature, as follows: baby, brain abscess (36), meningitis (8, 14, 36, 38,–43, 45), throat infection (35), septic arthritis (91), and osteomyelitis with sickle cell disease (92); adult, acute respiratory distress syndrome (93), pleuritic in Hodgkin’s disease (94), aneurysm of the distending aorta (95), osteomyelitis (40, 92), pelvic inflammatory disease (96), Bartholin’s abscess (37), knee joint empyema (40), and septic thrombophlebitis (97).
The clinical presentation of S. Panama disease varies between adults and children (Fig. 1). A common complication of neonatal S. Panama infection is the development of Salmonella meningitis (8, 14, 36, 38,–45), a lethal disease that has previously been linked to localized outbreaks in hospital maternity wards (8, 31). For example, S. Panama was recovered from 138 babies, new mothers, and staff during an outbreak of salmonellosis in a neonatal nursery in Michigan in 1934 to 1944 that resulted in 18 fatalities due to Salmonella meningitis (8). Similar outbreaks have historically occurred in other countries, including Germany, where a hospital outbreak in a maternity unit caused prolonged contamination despite radical disinfection of the entire ward (46).
S. Panama causes more cases of clinically invasive disease in humans than most Salmonella serovars. Historically, S. Panama infections have been 11 times more likely to cause invasive disease than those by other serovars in Martinique (10, 11). In England, 7% of all S. Panama isolates were isolated from extraintestinal sites compared to 2% of Salmonella enterica serovar Typhimurium and 3% of S. Enteritidis isolates (21). In Taiwan, 70% of S. Panama isolates were isolated from invasive disease compared to 12% of S. Enteritidis isolates (47). In addition to these epidemiologically suggestive data, multivariate analysis has recently confirmed the association of S. Panama with clinically invasive infection (P <
< 0.001) as part of a retrospective study of Salmonella infections in children living in Guadeloupe (7). A gnotobiotic-mouse model has been described for S. Panama (48), which could help to elucidate the mechanisms behind the increased invasiveness.
0.001) as part of a retrospective study of Salmonella infections in children living in Guadeloupe (7). A gnotobiotic-mouse model has been described for S. Panama (48), which could help to elucidate the mechanisms behind the increased invasiveness.
TRANSMISSION VEHICLES
Wild reptiles are the natural reservoir for S. Panama in Latin America (12, 49,–52). A study focusing on the frequency and host distribution of Salmonella serovars in reptiles and amphibians captured in the Republic of Panama between 1965 and 1967 showed that 2.6% of 78 Salmonella isolates were serovar Panama (49). In a subsequent study (1966 to 1969), 6.8% of Salmonella organisms isolated from neotropical lizards in Panama were S. Panama (50). In the past decade, a high prevalence of Salmonella has been found in the largest lizards in South America (Tegu lizards), and 3% of the isolates were classified as S. Panama (51). In French Guiana, where S. Panama was the most frequently isolated human-associated serovar in 2011, the serovar was also isolated from wild reptiles (12). Reptiles are likely to be an important source for transmission of S. Panama in regions of the world where many lizards and other reptiles are present in and around households. A recent survey of Salmonella strains carried by African venomous snakes did not isolate S. Panama (53).
In addition to reptiles, S. Panama has also been isolated from other wildlife species and companion animals. A study on pouched wild birds found S. Panama in cloacal swabs of chestnut-capped blackbirds in Rio de Janeiro, Brazil (54). In regard to companion animals, S. Panama was isolated from a household dog in Taiwan (55). S. Panama contamination has been found in birds and fish tanks sampled from pet shops and households in Trinidad (56). Wildlife, therefore, represent a potential reservoir for S. Panama dissemination.
In Europe, S. Panama infection is primarily a foodborne disease, with the main transmission vehicles being pork-derived products, including cured meat, minced pork, and sausages (57). The transmission pathway for S. Panama begins in animal feed, from where it can enter porcine and poultry animal reservoirs and move into animal food products, eventually infecting humans (18).
At the animal level, S. Panama was found in 2.08% of 200 abattoir pigs sampled in Budapest, Hungary (58), and has been found in cattle and swine in Germany (59). Outside Europe, S. Panama has been identified in beef and dairy herds in Argentina (60) and is the second most common Salmonella serovar to be isolated from swine finishing herds in Brazil (61).
S. Panama is also recognized as a contaminant in food-processing facilities and retail establishments globally, including butcher shops (62), public markets (63), meat vans (64), and slaughterhouses (65). The process of manufacturing pork-derived products includes several steps designed to result in a microbiologically safe, shelf-stable product by tightly controlling physicochemical conditions, such as salt and nitrate concentrations, pH, water activity, and temperature (66). However, Salmonella viability throughout this curing process has been reported, including the presence of S. Panama in salami (67, 68). In the Netherlands, S. Panama has additionally been implicated in the contamination of cattle-derived food products and was one of the three Salmonella serovars most frequently isolated from mincemeat over a 13-month period. Interestingly, mincemeat from slaughterhouses was more likely to contain Salmonella than mincemeat derived from slaughtering completed at butcher shops (69). Food-processing facilities themselves can play a role in the contamination of animal food products with S. Panama.
The impact of S. Panama entering the human food chain can be seen in an outbreak of salmonellosis that affected 300 people who had eaten contaminated roast pork in the United Kingdom in 1970. S. Panama was implicated as the etiological agent (18). S. Panama has also caused several foodborne outbreaks between 1990 and 1999 in Asturias, Spain, and isolates were collected from gastroenteritis and septicemia patients who had consumed contaminated fish puddings, cooked octopus, and cream cakes (32). Other studies have linked S. Panama infections to consumption of goat cheese, vegetables, beef, poultry, eggs, fruit juice, and shellfish (14, 33, 70).
In addition to the usual fecal-oral transmission route of Salmonella in humans, breast milk has also been suggested as a vector for S. Panama (71). A study demonstrated that S. Panama can infect the human mammary duct, can be shed for at least 2 weeks, and can remain stable during storage of breast milk at 4°C (71). Furthermore, it is possible that a case of meningitis in an exclusively breastfed 4-month-old patient was contracted from breast milk that was contaminated with an antimicrobial-susceptible S. Panama isolate (41).
ANTIMICROBIAL RESISTANCE
Burden of antimicrobial resistance in S. Panama.
Antimicrobial resistance (AMR) is an important public health concern (72). There are conflicting reports in the literature relating to the AMR status of the S. Panama serovar, with studies in Italy and Brazil reporting low levels of antibiotic resistance (41, 73). They are supported by further reports from Martinique, where 91% of S. Panama isolates were susceptible to beta-lactams (11), and Guadeloupe, where all Salmonella serovars demonstrated high overall susceptibility to antibiotics (7). In contrast, other studies have seen higher levels of resistance in S. Panama, particularly against tetracycline (e.g., 67%) and chloramphenicol (e.g., 67%) since the 1980s (24, 47, 59, 74,–76). Antibiotic stewardship promises to be an effective tool for decreasing antimicrobial resistance in the S. Panama serovar. For example, following a ban on tetracycline use in the pork industry in the Netherlands, S. Panama tetracycline resistance dropped from 90% to 1% (24).
In Asia, S. Panama has been associated with high levels of AMR since 1980, when 58% of the S. Panama isolates from Tokyo were resistant to at least one antibiotic agent (77). This figure appears to be on the rise. By the turn of the millennium, 83% of domestic and imported S. Panama isolates from cases in Tokyo were multidrug resistant. Similarly, in Taiwan, the serovar also exhibited resistance to multiple antibiotics, including cotrimoxazole (67%), ampicillin (56%), streptomycin (56%), kanamycin (56%), and gentamicin (45%) (74). The high proportion of S. Panama isolates that show AMR should be considered by clinicians working in Asia and by health care practitioners globally when treating Asian-travel-associated salmonellosis cases caused by S. Panama.
Genomic markers and trends in antimicrobial resistance.
A large proportion of S. Panama antimicrobial resistance has been associated with plasmid carriage (P =
= 0.012), class 1 integron presence, and transmissible drug resistance (R) factors (22, 47, 74, 78). Resistance to tetracycline, for example, has often been mediated by the R factor R1 in S. Panama (26). Such R factors have been implicated in the transfer of multiple antimicrobial resistance genes, usually simultaneously, between S. Panama strains and other bacteria. However, an isolate from an epidemic of S. Panama infection in Paris showed unusual patterns of transferable resistance, which may extend to other strains in the S. Panama serovar. The isolate was able to transfer genes involved in antimicrobial resistance singly or in pairs, rather than as one antibiotic resistance cassette. The proposed mechanism involved the simultaneous transfer of several discrete genetic elements that were able to coexist stably and to replicate noncompetitively in S. Panama. The authors suggested that frequent cotransfer of genetic elements may be propagated by conjugative-transfer machinery (27).
0.012), class 1 integron presence, and transmissible drug resistance (R) factors (22, 47, 74, 78). Resistance to tetracycline, for example, has often been mediated by the R factor R1 in S. Panama (26). Such R factors have been implicated in the transfer of multiple antimicrobial resistance genes, usually simultaneously, between S. Panama strains and other bacteria. However, an isolate from an epidemic of S. Panama infection in Paris showed unusual patterns of transferable resistance, which may extend to other strains in the S. Panama serovar. The isolate was able to transfer genes involved in antimicrobial resistance singly or in pairs, rather than as one antibiotic resistance cassette. The proposed mechanism involved the simultaneous transfer of several discrete genetic elements that were able to coexist stably and to replicate noncompetitively in S. Panama. The authors suggested that frequent cotransfer of genetic elements may be propagated by conjugative-transfer machinery (27).
INVASIVE DISEASE—GENOMIC INFERENCES IN S. PANAMA
Evolutionary history and virulence.
The study of evolutionary history may explain why S. Panama is associated with invasive disease. The majority of salmonellae that cause disease in humans belong to S. enterica subsp. enterica, which is further divided into two main clades, A and B, and a number of smaller clades (79). Phylogenetically, S. Panama is in clade B, which is associated with increased levels of clinically invasive disease (53, 80, 81). Another review of the population structure within S. enterica found that S. Panama is in lineage 3 (equivalent to the above-mentioned clade B) (82).
The evolutionary history of S. Panama was studied by Selander et al. (83), who used multilocus enzyme electrophoresis to assess the relationships among Salmonella serovars that cause invasive disease. It was proposed that S. Panama evolved from the same ancestors that gave rise to Salmonella enterica serovar Paratyphi, Salmonella enterica serovar Sendai (which causes enteric fever), and Salmonella enterica serovar Miami (83). In the current era of genomically informed epidemiological analysis, phylogenetic methods can be used to understand the evolutionary history of Salmonella. However, no large-scale phylogenetic study has yet been conducted on S. Panama, and only one complete S. Panama genome sequence (from strain ATCC 7378; GenBank accession no. CP012346) is available (84). As part of the current review, virulence genes were identified in the complete genome of S. Panama strain ATCC 7378 using the program ABRicate v0.8.10 (https://github.com/tseemann/abricate) against a virulence factor database (85) with default parameters. In total, 131 virulence-associated genes were identified. The analysis confirmed the presence of typical Salmonella virulence determinants, including type III secretion systems, type III effector proteins, fimbriae, and flagella. Of interest, S. Panama was also found to carry the cytolethal distending toxin B gene (cdtB), which is characteristic of S. enterica clade B and the highly invasive Salmonella enterica serovar Typhi (53, 80, 81). A more detailed, epidemiologically representative analysis is required to further elucidate the uniqueness of the S. Panama serovar.
Accessory genome and virulence.
Generally, plasmids play a key role in systemic Salmonella infection, but little is known about the plasmid complement of the S. Panama serovar. In the small number of available studies, it is reported that S. Panama, including the above-mentioned S. Panama ATCC 7378, does not commonly carry the large plasmids that have previously been associated with virulence in other Salmonella serovars (41). Rather, S. Panama strains carry a heterogeneous population of plasmids (86). Prophages can also make significant contributions to Salmonella virulence (87, 88), but only one study has reported the presence of prophages in S. Panama (84). The Salmonella RE-2010 prophage was identified in the genome of S. Panama ATCC 7378. The prophage (also known as ElPhiS) has also been found in S. Enteritidis, where it has been associated with specific phylogenetic clusters (89, 90). The importance of S. Panama for public health globally necessitates that a concerted comparative genomic analysis be conducted in the future.
PERSPECTIVES
S. Panama is a globally relevant pathogen that has consistently been reported as one of the most frequently isolated Salmonella serovars over the past 70 years. The proportion of clinical cases caused by S. Panama is particularly high in French territories in the Americas, where it is associated with invasion of extraintestinal sites, particularly in infants. Reptiles act as natural reservoirs for Salmonella in these regions, and it has been speculated that the large numbers of reptiles found in and around homes in tropical regions of America lead to high levels of S. Panama transmission to humans. The serovar was also introduced into Europe, where it spread through the pork industry and caused hospital outbreaks in the 1960s and 1970s. S. Panama continues to contribute to the global disease burden caused by salmonellae.
years. The proportion of clinical cases caused by S. Panama is particularly high in French territories in the Americas, where it is associated with invasion of extraintestinal sites, particularly in infants. Reptiles act as natural reservoirs for Salmonella in these regions, and it has been speculated that the large numbers of reptiles found in and around homes in tropical regions of America lead to high levels of S. Panama transmission to humans. The serovar was also introduced into Europe, where it spread through the pork industry and caused hospital outbreaks in the 1960s and 1970s. S. Panama continues to contribute to the global disease burden caused by salmonellae.
It is important to highlight the unusual clinical presentation of S. Panama in different patient populations to avoid delays in patient treatment. Clinicians and researchers should remain aware of the potential for increasing levels of antimicrobial resistance in the serovar, as has been described in Asia. Unraveling the molecular epidemiology and evolutionary history of S. Panama is the obvious next step in understanding more about this rarely studied serovar that continues to cause invasive salmonellosis worldwide.
ACKNOWLEDGMENTS
Caisey V. Pulford is supported by a Fee Bursary Award from the Institute of Integrative Biology at the University of Liverpool and by a John Lennon Memorial Scholarship from the University of Liverpool. Kate S. Baker is funded by a Wellcome Trust Clinical Research Career Development Fellowship (106690/A/14/Z). Jay C. D. Hinton is funded by a Wellcome Trust Senior Investigator Award (106914/Z/15/Z).
Biographies

Caisey V. Pulford, B.Sc. (Hons), is a senior Ph.D. student at the University of Liverpool, Liverpool, United Kingdom. After graduating at the top of her year with a first-class degree in tropical disease biology, she became fascinated with bacterial genomic applications in public health. For the past 3 years, she has focused on understanding the molecular epidemiology of nontyphoidal Salmonella that causes bloodstream infection in Africa and the French Caribbean. She has made a significant contribution to global Salmonella sequencing efforts, working as part of the 10,000 Salmonella Genomes project. Her research has received numerous awards, including the NOVA for outstanding early contributions in biology and the John Lennon Memorial Scholarship in recognition of global health research. As an early career researcher, she has contributed a first author research paper focused on Salmonella diversity in venomous snakes. Her ambition is to pursue a career in the surveillance of bacterial pathogens during global epidemics.

Blanca M. Perez-Sepulveda, Ph.D., is a molecular microbiologist working on invasive nontyphoidal Salmonella (iNTS). After completing an M.Sc. (Res) in biochemistry at the University of Chile, she moved to the United Kingdom, where she obtained a Ph.D. at the University of Warwick studying the molecular mechanisms of phage resistance. She moved to the University of Liverpool in 2016 to join Jay Hinton’s laboratory as a postdoctoral research associate. She currently focuses on understanding the virulence determinants of novel invasive Salmonella Enteritidis clades identified in sub-Saharan African regions, using a combination of phenotypic characterization, comparative genomics, and transcriptomics. In collaboration with the Earlham Institute, she has been leading the 10,000 Salmonella Genomes project, a worldwide collaborative effort to understand the transmission and virulence of iNTS. Her interests lie in understanding how bacteria survive in the environment. Her focus has been to determine the environmental reservoirs and transmission of Salmonella and its phages by studying molecular mechanisms of virulence and phage resistance.

Ella V. Rodwell, B.Sc. (Hons), is a Master of Research student at the University of Liverpool, Liverpool, United Kingdom. Her research interests lie in the molecular microbiology of bacterial pathogens during global epidemics. As an early career researcher, she has published her first research paper, which was focused on understanding reservoirs for Salmonella infections in Africa. Between 2015 and 2018, she completed her undergraduate studies in biological sciences at the University of West England, from which she obtained a first-class degree with honors. In 2017, she held a Microbiology Society-funded studentship within the University of Liverpool, where she focused on Salmonella metabolism in venomous snakes. Her current work focuses on the molecular epidemiology and characterization of prophages and their importance in driving bacterial epidemics. She has a keen interest in public health, epidemiology, and disease survelliance. Her goal is to develop treatments and control initiatives for infectious diseases.

François-Xavier Weill, M.D., Ph.D., is a clinical microbiologist and a research director at the Pasteur Institute, Paris, France. For the last 10 years, he has been heading the Enteric Bacterial Pathogens Research and Expertise Unit, which hosts two French National Reference Centers and one World Health Organization Collaborative Center. Between 2014 and 2016, he was a visiting scientist in the Bacterial Genomics and Evolution group at the Wellcome Sanger Institute, Cambridge, United Kingdom. His research interests are the population structure and transmission dynamics of emerging, epidemic, and antimicrobial drug-resistant enteric bacterial pathogens, as well as molecular and genomic epidemiology, and the development of new diagnostic tools for these pathogens. He has published over 150 peer-reviewed papers, including 55 as first or last author, in journals such as Science, Nature, Nature Microbiology, and Lancet Infectious Diseases.

Jay C. D. Hinton, B.Sc. (Hons), M.A., Ph.D., is based at the University of Liverpool, Liverpool, United Kingdom. He did his first degree in microbiology, when he was inspired to think genetically by George Salmond. After receiving his Ph.D., he moved to the University of Oxford to work on the regulation of virulence gene expression in Salmonella and subsequently moved to Norwich, United Kingdom, as Head of Molecular Microbiology at the Institute of Food Research. He has done bacterial functional genomics for the past 20 years. He pioneered a transcriptomic approach that revealed a “snapshot” of Salmonella gene expression during the process of infection of mammalian cells in 2003 and codiscovered the H-NS-mediated mechanism of silencing gene expression in bacteria in 2006. After moving to Liverpool in 2012, he now uses a combination of genomics and functional transcriptomics to bring new insights to the lethal epidemic of bloodstream infections caused by Salmonella in sub-Saharan Africa.
REFERENCES
Articles from Infection and Immunity are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/iai.00273-19
Read article for free, from open access legal sources, via Unpaywall:
https://iai.asm.org/content/iai/87/9/e00273-19.full.pdf
Citations & impact
Impact metrics
Article citations
The evolutionary diversification of the Salmonella artAB toxin locus.
Front Microbiol, 13:1016438, 25 Nov 2022
Cited by: 4 articles | PMID: 36504768 | PMCID: PMC9732031
Genome-based analysis of infrequent Salmonella serotypes through the Thai pork production chain.
Front Microbiol, 13:968695, 25 Aug 2022
Cited by: 3 articles | PMID: 36090074 | PMCID: PMC9453559
Antibiotic resistance and associated resistance determinants in different Salmonella enterica serovars isolated from pigs in Argentina.
Vet World, 15(5):1215-1220, 20 May 2022
Cited by: 1 article | PMID: 35765497 | PMCID: PMC9210835
Stepwise evolution of Salmonella Typhimurium ST313 causing bloodstream infection in Africa.
Nat Microbiol, 6(3):327-338, 21 Dec 2020
Cited by: 55 articles | PMID: 33349664 | PMCID: PMC8018540
Genomic characterization of Salmonella Uzaramo for human invasive infection.
Microb Genom, 6(7), 01 Jul 2020
Cited by: 30 articles | PMID: 32589568 | PMCID: PMC7478631
Go to all (6) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences
- (1 citation) ENA - CP012346
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Integrative analysis of Salmonellosis in Israel reveals association of Salmonella enterica Serovar 9,12:l,v:- with extraintestinal infections, dissemination of endemic S. enterica Serovar Typhimurium DT104 biotypes, and severe underreporting of outbreaks.
J Clin Microbiol, 52(6):2078-2088, 09 Apr 2014
Cited by: 11 articles | PMID: 24719441 | PMCID: PMC4042803
First report of Salmonella enterica serotype panama meningitis associated with consumption of contaminated breast milk by a neonate.
J Clin Microbiol, 43(10):5400-5402, 01 Oct 2005
Cited by: 12 articles | PMID: 16208031 | PMCID: PMC1248486
Pathogenicity and phenotypic analysis of sopB, sopD and pipD virulence factors in Salmonella enterica serovar typhimurium and Salmonella enterica serovar Agona.
Antonie Van Leeuwenhoek, 107(1):23-37, 14 Oct 2014
Cited by: 10 articles | PMID: 25312847
Host-pathogen interaction in invasive Salmonellosis.
PLoS Pathog, 8(10):e1002933, 04 Oct 2012
Cited by: 145 articles | PMID: 23055923 | PMCID: PMC3464234
Review Free full text in Europe PMC
Funding
Funders who supported this work.
University of Liverpool (2)
Grant ID: Institute of Integrative Biology Fee Bursary Award
Grant ID: John Lennon Memorial Scholarship
Wellcome Trust (4)
Grant ID: Senior Investigator Award 106914/Z/15/Z
Grant ID: Clinical Research Development Fellowship 106690/A/14/Z
Grant ID: 106690/A/14/Z
Grant ID: 106914/Z/15/Z

 a,
a,



