Abstract
Free full text

ABCB1 Gene Is Associated With Clinical Response to SNRIs in a Local Chinese Han Population
Abstract
Background: The relation between the ATP-binding cassette subfamily B member 1 (ABCB1) gene and major depressive disorder (MDD) has been studied in a local Chinese Han population. MDD is associated with the rs2032582 (G2677T) and rs1128503 (C1236T) single-nucleotide polymorphisms (SNPs) of ABCB1 but not with rs1045642, rs2032583, rs2235040, and rs2235015. This study aims to explore the potential correlations of therapeutic responses with selective serotonin reuptake inhibitors (SSRIs) and serotonin-norepinephrine reuptake inhibitors (SNRIs) in a local Chinese Han population.
Methods: The study population included 292 patients with MDD. All patients were assessed at baseline and at first, second, fourth, and sixth weeks according to the 17-item Hamilton Rating Scale for Depression (HAM-D17) to determine their therapeutic responses to SSRIs and SNRIs.
Results: In the SSRI therapy group, the genotype or allele distribution of six SNPs was not significantly different between responders and nonresponders. In the SNRI therapy group, only rs2032583 was associated with a therapeutic response to SNRIs. The C allele of the ABCB1 rs2032583 polymorphism was negatively correlated with therapeutic responses according to logistic regression analysis.
Conclusion: The ABCB1 gene polymorphisms may not be associated with therapeutic responses to SSRIs but not with SNRIs. The TT genotype of rs2032583 could be a predictive factor of improved treatment responses to SNRIs in the Chinese population. These findings should be replicated in future studies with larger patient groups.
Introduction
The ATP-binding cassette subfamily B member 1 (ABCB1) gene, a multidrug resistance protein 1 (MDR1) gene, is located on the chromosomal region 7q21.1 and encodes p-glycoprotein (P-gp), which plays an important role in drug bioavailability and response to drugs. P-gp is a 1280-amino acid transporter and serves as a genetically polymorphic efflux transporter that removes foreign substances from cells. This protein is expressed in the blood–brain barrier and protects the brain from drugs or neurotoxic substances, such as glucocorticoids and amyloid beta (de Klerk et al., 2013).
After treatment with regular doses of antidepressants, several patients with major depressive disorder (MDD) fail to obtain satisfactory therapeutic effects, and some patients even incur serious side effects. Among patients with MDD treated with a single antidepressant medication, only 50% of patients received adequate clinical response, and 30% of patients achieved recovery. There was a delayed response to symptom relief, ranging from 4 to 8 weeks. During this period, the risk of suicide increased significantly, whereas another 10% of the patients were ineffective for any kind of antidepressant medication (Weizman et al., 2012). ABCB1 gene polymorphisms affect the ability of drugs to pass through the blood–brain barrier into the central nervous system, leading to inadequate drug concentrations in the brain (Uhr et al., 2008). ABCB1 gene polymorphisms can predict the effect of antidepressant drugs that are MDR1 substrates (Mihaljevic Peles et al., 2008; Sarginson et al., 2010).
weeks. During this period, the risk of suicide increased significantly, whereas another 10% of the patients were ineffective for any kind of antidepressant medication (Weizman et al., 2012). ABCB1 gene polymorphisms affect the ability of drugs to pass through the blood–brain barrier into the central nervous system, leading to inadequate drug concentrations in the brain (Uhr et al., 2008). ABCB1 gene polymorphisms can predict the effect of antidepressant drugs that are MDR1 substrates (Mihaljevic Peles et al., 2008; Sarginson et al., 2010).
Some drugs have been identified as substrates of P-gp. These drugs include nortriptyline, imipramine, escitalopram, amitriptyline, paroxetine, venlafaxine, and citalopram; and those with non-ABCB1 substrates include bupropion, mirtazapine, and fluoxetine (Mihaljevic Peles et al., 2008). The ABCB1 gene has single-nucleotide polymorphisms (SNPs) in the encoding regions. Variants such as C3435T (rs1045642), G2677T/A (rs2032582), and rs2032583 are the most commonly studied. Ameyaw et al. (2001) reported that ABCB1 gene knockout mice possessed insufficient P-gp, leading to high drug concentrations in the blood and weak ability to eliminate drugs. The plasma concentrations of drugs in mice without the ABCB1 gene were fivefold higher and seven- to 36-fold higher in the cerebrospinal fluid (Ameyaw et al., 2001).The concentrations of citalopram, venlafaxine, and d-venlafaxine in the brain of mutant mice were 3.0, 1.7, and 4.1 times higher than those in their wild-type littermates (Uhr et al., 2008). In vivo studies indicated that mdr1 ab (−/−) mutant mice possessed higher cerebral concentrations of paroxetine compared with those of mdr1ab (+/+) control mice. This finding suggests that P-gp could prevent paroxetine from entering into the brain (Uhr et al., 2003). Patients with the CC genotype of SNP rs2232583 in the ABCB1 gene exhibited a higher response rate than that of patients with the TT genotype after 4 weeks of antidepressant treatment. However, in the non-P-gp substrate mirtazapine group, no difference in genotype was observed between remission and nonremission groups (Uhr et al., 2007). A similar result was also found by Sarginson et al. (2010). Moreover, Dong et al. reported that ABCB1 gene polymorphisms rs4728697, rs2032583, and rs58898486 were associated with depression therapeutic response, and rs17064 was related to the curative effect of desipramine (Dong et al., 2009). ABCB1 haplotypes and SNPs rs1045642, rs2032582, and rs2032583 affect responses to antidepressant treatment (Menu et al., 2010; Rosenhagen and Uhr, 2011; Lin et al., 2011; Singh et al., 2012; de Klerk et al., 2013).
weeks of antidepressant treatment. However, in the non-P-gp substrate mirtazapine group, no difference in genotype was observed between remission and nonremission groups (Uhr et al., 2007). A similar result was also found by Sarginson et al. (2010). Moreover, Dong et al. reported that ABCB1 gene polymorphisms rs4728697, rs2032583, and rs58898486 were associated with depression therapeutic response, and rs17064 was related to the curative effect of desipramine (Dong et al., 2009). ABCB1 haplotypes and SNPs rs1045642, rs2032582, and rs2032583 affect responses to antidepressant treatment (Menu et al., 2010; Rosenhagen and Uhr, 2011; Lin et al., 2011; Singh et al., 2012; de Klerk et al., 2013).
A total of 292 Chinese patients with MDD and 208 unrelated control individuals from a local Chinese Han population were studied. There are few studies on the relationship between gene polymorphism of ABCB1 and depression in the Chinese Han population; only a team of Taiwan scholars studied the relationship between the efficacy of escitalopram and gene polymorphism of ABCB1 in patients with depression (Lin et al., 2011). The purpose of this study was to further understand the relationship between the two in the Chinese Han population and to provide a theoretical basis for individualized treatment.
Materials and Methods
Subjects
A total of 292 patients with MDD and 208 healthy controls were included in this study. All participants were biologically unrelated and of Chinese Han ethnicity. The patients were diagnosed as having MDD as defined in the Axis I of the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, Text Revision (DSM-IV-TR) and obtained scores ≥18 by the 17-item Hamilton Depression (HAM-D17) Rating Scale. A consensus diagnosis by at least two psychiatrists was made for each patient according to the DSM-IV criteria. Patients were not eligible to participate in the study if they have any other mental disorder according to DSM-IV-TR Axis I criteria, had major physical and neurological illnesses and sequelae of serious illness, or serious suicide attempts and behavior. Patients were also excluded if they had used electroconvulsive therapy or antipsychotic drugs with long-lasting effects within the last 6 months or any antipsychotic drugs within the last 4
months or any antipsychotic drugs within the last 4 weeks. The mean ages ± standard deviations of the patients and controls were 30.89 ± 10.92
weeks. The mean ages ± standard deviations of the patients and controls were 30.89 ± 10.92 years and 31.71 ± 8.25
years and 31.71 ± 8.25 years, respectively. Among the 292 patients with MDD, 71.6% had a single episode and 28.4% had recurrent episodes. The study protocol was approved by the Medical Ethics Committee of Second Xiangya Hospital, Central South University. Written informed consent was obtained from each patient after the study was explained.
years, respectively. Among the 292 patients with MDD, 71.6% had a single episode and 28.4% had recurrent episodes. The study protocol was approved by the Medical Ethics Committee of Second Xiangya Hospital, Central South University. Written informed consent was obtained from each patient after the study was explained.
Study Design
Eligible patients were treated with one of the five antidepressants (escitalopram, paroxetine, sertraline, duloxetine, and venlafaxine) for 6 weeks. All patients affirmed a regular dose intake of antidepressant drug per day during the study. The primary efficacy measurement was the change in the HAM-D17 total score from baseline until the end of the study period. Patients were evaluated at screening, baseline, and on the first, second, fourth, and sixth weeks of treatment. Response was defined as changes in the HAM-D17 total score of ≥50%.
weeks. All patients affirmed a regular dose intake of antidepressant drug per day during the study. The primary efficacy measurement was the change in the HAM-D17 total score from baseline until the end of the study period. Patients were evaluated at screening, baseline, and on the first, second, fourth, and sixth weeks of treatment. Response was defined as changes in the HAM-D17 total score of ≥50%.
Sample Collection and DNA Extraction
Peripheral blood samples were collected from ethylene diamine tetraacetic acid (EDTA)-containing tubes following the standard venipuncture technique. Genomic DNA was extracted from whole blood according to standard procedures. In this study, we selected SNPs by the following three methods: literature reviewing, searching for Tag-SNP, and searching for functional variant sites by FAST SNP. Finally, we investigated the following six SNPs of the ABCB1 gene: SNP1 (rs1045642) in exon 27, SNP2 (rs2032583) in intron 22, SNP3 (rs2032582) in exon 22, SNP4 (rs2235040) in intron boundary exon 21, SNP5 (rs1128503) in exon 13, and SNP6 (rs2235015) in intron 5 of the ABCB1 gene. All genotyping experiments were carried out by Shanghai BioWing Applied Biotechnology Company (http://www.biowing.com.cn). The AxyPrep Blood Genomic DNA Kit was used for extraction, and the ligase detection reaction (LDR) was used to detect the six SNPs. The LDR was performed in 30 cycles at 95°C for 2 min, 94°C for 15
min, 94°C for 15 s, and 50°C for 25
s, and 50°C for 25 s. Target DNA sequences were amplified using a multiplex polymerase chain reaction method. The fluorescent products of the LDR were differentiated using a 3730 ABI sequencer.
s. Target DNA sequences were amplified using a multiplex polymerase chain reaction method. The fluorescent products of the LDR were differentiated using a 3730 ABI sequencer.
Haplotype and Statistical Analysis
Two independent sample t-test and χ2 test were used to examine the clinical and demographic variables between responders and nonresponders. Genotype and allele frequency distributions were compared between the patients and controls and between the responders and nonresponders using the χ2 test for independence. The observed genotype frequencies were compared with the predicted frequencies to investigate the concordance with the Hardy–Weinberg (H-W) equilibrium. Logistic regression analysis was used to estimate the therapeutic effect associated with each genotype; odds ratios with 95% confidence intervals were obtained. A p < 0.05 was considered to be statistically significant. The SHEsis online analysis software was used for linkage disequilibrium and haplotype analysis. Logistic regression analyses were performed using Statistical Package for the Social Sciences version 17.0 for Windows software (SPSS Inc., Chicago, IL). Adjustment for multiple comparisons was performed by Bonferroni correction.
Results
Comparison of General Data Between Control and Case Groups
A total of 208 cases in the control group, 101 males and 107 females, the average age was 31.71 ± 8.25 years. There were 292 patients in the study group, 143 males and 149 females; the average age was 30.89 ± 10.92
years. There were 292 patients in the study group, 143 males and 149 females; the average age was 30.89 ± 10.92 years. There was no significant difference in gender and age composition between the control group and the case group (χ2 = 0.008, p = 0.927; t = 0.954, p = 0.341), which was comparable.
years. There was no significant difference in gender and age composition between the control group and the case group (χ2 = 0.008, p = 0.927; t = 0.954, p = 0.341), which was comparable.
Hardy–Weinberg Balance Analysis of ABCB1 Gene Polymorphisms in the Control and Case Groups
Among the 208 control subjects, the theoretical number of genotypes was 81, the number of genotypes of CT was 98, and the number of genotypes of TT was 29. H-W analysis showed that there was no significant difference in the actual genotype distribution of rsl045642 SNP and the theoretical gene type distribution under H-W equilibrium, χ2 = 1.231, p = 0.267. According to this method, the H-W balance test was performed on the polymorphic loci rsl045642, rs2032583, rs2032582, rs2235040, rsl128503, and rs2235015 between the control group and the case group. As shown in Table 1 , the six SNPs loci in the control and case groups all met the H-W balance. The population selected in this study is representative of the Han population and suitable for genetic analysis.
Table 1
Equilibrium test of six single-nucleotide polymorphisms (SNPs) between the control group and the case group.
| SNP | Genotype | Controls (n = 208) | Cases (n = 292) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Actual number | Theoretical number | χ 2 | p | Actual number | Theoretical number | χ 2 | p | ||
| rs1045642 | CC | 85 | 81 | 1.23 | 0.267 | 103 | 109 | 2.27 | 0.132 |
| CT | 90 | 98 | 151 | 139 | |||||
| TT | 33 | 29 | 38 | 44 | |||||
| rs2032583 | CT | 22 | 21 | 0.65 | 0.42 | 38 | 37 | 1.41 | 0.234 |
| TT | 186 | 187 | 254 | 255 | |||||
| rs2032582 | GG | 78 | 72 | 3.39 | 0.065 | 86 | 78 | 3.44 | 0.06 |
| GT | 88 | 101 | 130 | 146 | |||||
| TT | 42 | 36 | 76 | 68 | |||||
| rs2235040 | AG | 22 | 21 | 0.65 | 0.42 | 40 | 38 | 1.58 | 0.21 |
| GG | 186 | 187 | 252 | 254 | |||||
| rs1128503 | CC | 39 | 34 | 1.8 | 0.18 | 40 | 35 | 1.97 | 0.161 |
| CT | 91 | 100 | 121 | 132 | |||||
| TT | 78 | 73 | 131 | 126 | |||||
| rs2235015 | GG | 187 | 188 | 0.59 | 0.44 | 254 | 255 | 1.414 | 0.23 |
| GT | 21 | 20 | 38 | 37 | |||||
Comparison of Clinical Features Among the Responders and Nonresponders
Among the 292 patients included in this study, 39 patients dropped out because of adverse effects (n = 12), withdrawal of consent (n = 9), contrary to the scheme (n = 2), and lost to follow-up (n = 16); lastly, 253 patients completed the study. Clinical characteristics, use of drugs, and average drug doses between responders and nonresponders are shown in Table 2 . No significant differences were found between the two groups according to the above indicators.
Table 2
Clinical features, dosage, and drug among the responders and the nonresponders.
| Nonresponders (n = 54) | Responders (n = 199) | F | p | ||
|---|---|---|---|---|---|
| Age (years) | 31.19 ± 10.124 | 30.78 ± 11.484 | 1.089 | 0.298 | |
| Weight | 60.778 ± 9.162 | 57.31 ± 9.3811 | 0.033 | 0.856 | |
| Education | 12.17 ± 3.994 | 11.93 ± 3.968 | 0.274 | 0.601 | |
| Sex | Male n (%) | 27(50) | 98(49.2) | 0.012 | 0.912 |
| Female n (%) | 27(50) | 101(50.8) | |||
| Marriage | Spinsterhood n (%) | 29(53.7) | 105(52.8) | 2.441 | 0.119 |
| Married n (%) | 21(38.9) | 86(43.2) | |||
| Divorced n (%) | 3(5.6) | 8(4.0) | |||
| Remarriage n (%) | 1(1.9) | 0(0.0) | |||
| Drug | Escitalopram n (%) | 19(35.2) | 67(33.7) | 5.713 | |
| Paroxetine n (%) | 6(11.1) | 44(22.1) | |||
| Venlafaxine n (%) | 19(35.2) | 46(23.1) | 0.222 | ||
| Duloxetine n (%) | 7(13.0) | 34(17.1) | |||
| Sertraline n (%) | 3(5.6) | 8(4.0) | |||
| Doses | Escitalopram | 10.79 ± 2.507 | 10.67 ± 2.446 | 3.723 | |
| Paroxetine | 21.67 ± 4.082 | 20.91 ± 2.908 | |||
| Venlafaxine | 157.89 ± 23.648 | 158.15 ± 23.602 | 0.055 | ||
| Duloxetine | 60.0 ± 0.00 | 60.0 ± 0.00 | |||
| Sertraline | 58.33 ± 14.434 | 105.63 ± 52.470 |
Genotype and Allele Frequencies in the Responders and Nonresponders
Genotype and allele distributions for the examined ABCB1 gene SNPs in nonresponders and responders are shown in Table 3 . The genotype and allelic distributions of rs1045642, rs2032582, rs2235040, rs1128503, and rs2235015 SNPs were not significantly different between nonresponders and responders. For rs2032583, genotype and allelic distributions significantly differed between the nonresponders and responders. The distribution of TT genotype and T allele frequency was higher in the responders than that in the nonresponders (p = 0.027, p = 0.033, respectively).
Table 3
Genotype and allele frequencies of six SNPs of the ABCB1 gene in the nonresponders and the responders.
| Genotype/allele | Responders (n = 199)(%) | Nonresponders (n = 54)(%) | χ 2 | p | OR (95% CI) | Adjust OR (95% CI) | ||
|---|---|---|---|---|---|---|---|---|
| rs1045642 | ||||||||
| CC | 73 (36.7) | 21 (38.9) | 1 | 1 | ||||
| CT | 97 (48.7) | 26 (48.1) | 0.136 | 0.934 | 1.073 | 0.560–2.056 | 1.159 | 0.575–2.336 |
| TT | 29 (14.6) | 7 (13.0) | 1.192 | 0.457–3.105 | 0.826 | 0.301–2.270 | ||
 C allele C allele | 243 (61.1) | 68 (63.0) | 1 | 1 | ||||
 T allele T allele | 155 (38.9) | 40 (37.0) | 0.131 | 0.718 | 1.084 | 0.699–1.683 | 0.762 | 0.299–1.945 |
| rs2032583 | ||||||||
| TT | 175 (87.9) | 41 (75.9) | 1 | 1 | ||||
| CT | 24 (12.1) | 13 (24.1) | 4.91 | 0.027* | 0.433* | 0.203–0.921 | 0.4* | 0.179–0.896 |
 T allele T allele | 374 (94.0) | 95 (88.0) | 1 | 1 | ||||
 C allele C allele | 24 (6.0) | 13 (12.0) | 4.52 | 0.033* | 0.469* | 0.23–0.955 | 0.4* | 0.179–0.8966 |
| rs2032582 | ||||||||
| GG | 63 (31.7) | 16 (29.6) | 1 | 1 | ||||
| GT | 81 (40.7) | 25 (46.3) | 0.574 | 0.751 | 0.823 | 0.405–1.671 | 0.878 | 0.41–1.877 |
| TT | 55 (27.6) | 13 (24.1) | 1.074 | 0.475–2.431 | 0.917 | 0.386–2.178 | ||
 G allele G allele | 207 (52.0) | 57 (52.8) | 1 | 1 | ||||
 T allele T allele | 191 (48.0) | 51 (47.2) | 0.02 | 0.887 | 1.031 | 0.674–1.579 | 0.989 | 0.473–2.07 |
| rs2235040 | ||||||||
| AG | 27 (13.6) | 12 (22.2) | 1 | 1 | ||||
| GG | 172 (86.4) | 42 (77.8) | 2.44 | 0.118 | 1.82 | 0.852–3.888 | 2.031 | 0.905–4.558 |
 A allele A allele | 27 (6.8) | 12 (11.1) | 1 | 1 | ||||
 G allele G allele | 371 (93.2) | 96 (88.9) | 2.236 | 0.135 | 1.718 | 0.839–3.515 | 2.047 | 0.919–4.56 |
| rs1128503 | ||||||||
| CC | 26 (13.1) | 8 (14.8) | 1 | 1 | ||||
| CT | 79 (39.7) | 27 (50.0) | 2.554 | 0.279 | 0.9 | 0.364–2.225 | 0.905 | 0.348–2.352 |
 C allele C allele | 131 (32.9) | 43 (39.8) | 1 | 1 | ||||
 T allele T allele | 267 (67.1) | 65 (60.2) | 1.793 | 0.181 | 1.348 | 0.87–2.09 | 1.41 | 0.727–2.734 |
| rs2235015 | ||||||||
| GG | 174 (87.4) | 42 (77.8) | 1 | 1 | ||||
| GT | 25 (12.6) | 12 (22.2) | 3.174 | 0.075 | 0.503 | 0.234–1.082 | 0.416 | 0.204–1.04 |
 G allele G allele | 373 (93.7) | 96 (88.9) | 1 | 1 | ||||
 T allele T allele | 25 (6.3) | 12 (11.1) | 2.924 | 0.087 | 0.536 | 0.26–1.108 | 0.530 | 0.21–1.01 |
*Statistically significant difference between the groups (p < 0.05).
CI, confidence interval; OR, odds ratio.
ABCB1 Gene Polymorphism Loci and Clinical Response to Selective Serotonin Reuptake Inhibitors
For SSRIs (sertraline, paroxetine, and escitalopram), no significant difference in genotype and allele frequency distribution was observed between the responders and nonresponders (p > 0.05) ( Table 4 ).
Table 4
Genotype, allelic distribution of all genotyped single nucleotide polymorphisms among the treatment responders and nonresponders for selective serotonin reuptake inhibitor (SSRI) drugs group.
| Responders (n = 119) (%) | Nonresponders (n = 28) (%) | χ2 | p | OR (95% CI) | Adjust OR (95% CI) | |
|---|---|---|---|---|---|---|
| rs1045642 | ||||||
| CC | 42 (35.3) | 9 (32.1) | 1 | 1 | ||
| CT | 60 (50.4) | 14 (50.0) | 0.259 | 0.879 | 0.918 (0.364–2.317) | 0.717 (0.255–2.02) |
| TT | 17 (14.3) | 5 (17.9) | 0.729 (0.213–2.492) | 0.389 (0.098–1.551) | ||
 C allele C allele | 144 (60.5) | 32 (57.1) | 1 | 1 | ||
 T allele T allele | 94 (39.5) | 24 (42.9) | 0.213 | 0.644 | 0.87 (0.483–1.57) | 0.479 (0.143–1.601) |
| rs2032583 | ||||||
| TT | 106 (89.1) | 24 (85.7) | 1 | 1 | ||
| CT | 13 (10.9) | 4 (14.3) | 0.25 | 0.617 | 0.736 (0.221–2.455) | 0.751 (0.205–2.747) |
 T allele T allele | 225 (94.5) | 52 (92.9) | 1 | 1 | ||
 C allele C allele | 13 (5.5) | 4 (7.1) | 0.235 | 0.628 | 0.751 (0.235–2.397) | 0.751 (0.205–2.747) |
| rs2032582 | ||||||
| GG | 38 (31.9) | 5 (17.9) | 1 | 1 | ||
| GT | 53 (44.5) | 14 (50.0) | 2.352 | 0.309 | 0.498 (0.165–1.501) | 0.336 (0.09–1.256) |
| TT | 28 (23.5) | 9 (32.1) | 0.409 (0.124–1.355) | 0.237 (0.057–0.984) | ||
 G allele G allele | 129 (54.2) | 24 (42.9) | 1 | 1 | ||
 T allele T allele | 109 (45.8) | 32 (57.1) | 2.338 | 0.126 | 0.634 (0.352–1.14) | 0.509 (0.189–1.37) |
| rs2235040 | ||||||
| AG | 15 (12.6) | 4 (14.3) | 1 | 1 | ||
| GG | 104 (87.4) | 24 (85.7) | 0.056 | 0.813 | 1.156 (0.352–3.794) | 1.129 (0.316–4.032) |
 A allele A allele | 15 (6.3) | 4 (7.1) | 1 | 1 | ||
 G allele G allele | 223 (93.7) | 52 (92.9) | 0.052 | 0.82 | 1.144 (0.364–3.588) | 1.129 (0.316–4.032) |
| rs1128503 | ||||||
| CC | 16 (13.4) | 2 (7.1) | 1 | 1 | ||
| CT | 39 (32.8) | 14 (50.0) | 3.124 | 0.21 | 0.348 (0.071–1.711) | 0.303 (0.057–1.627) |
| TT | 64 (53.8) | 12 (42.9) | 0.667 (0.135–3.282) | 0.483 (0.091–2.57) | ||
 C allele C allele | 71 (29.8) | 18 (32.1) | 1 | 1 | ||
 T allele T allele | 167 (70.2) | 38 (67.9) | 0.115 | 0.735 | 1.114 (0.596–2.083) | 1.224 (0.494–3.033) |
| rs2235015 | ||||||
| GG | 106 (89.1) | 24 (85.7) | 1 | 1 | ||
| GT | 13 (10.9) | 4 (14.3) | 0.25 | 0.617 | 0.736 (0.221–2.455) | 0.696 (0.191–2.543) |
 G allele G allele | 225 (94.6) | 52 (92.9) | 1 | 1 | ||
 T allele T allele | 13 (5.5) | 4 (7.1) | 0.235 | 0.628 | 0.751 (0.235–2.397) | 0.751 (0.235–2.397) |
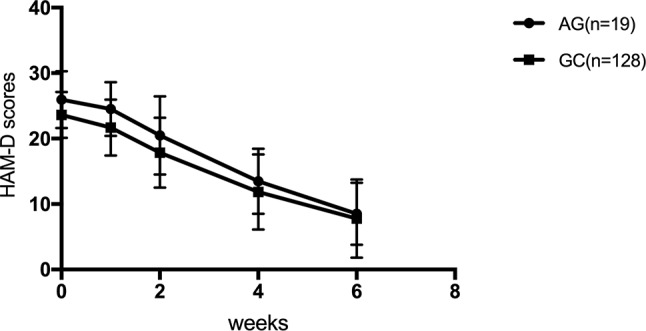
The efficacy of SSRIs was examined for genotypes in the SSRI treatment group according to the HAM-D17 scores, decreased scores, and reducing score rate during the first, second, fourth, and sixth weeks. The HAM-D17 scores, decreased scores, and reducing score rate at the first, second, fourth, and sixth weeks were not significantly different among rs1045642, rs2032583, rs2032582, rs1128503, and rs2235015 (p > 0.05). HAM-D17 decreased scores and reducing score rate did not reveal any significant difference with rs2235040 ( Table 5 ); however, a significant difference was observed for the genotype of rs2235040 in the HAM-D17 scores (F = 4.349, p = 0.039) ( Figure 1 ).
Table 5
Rs2235040 and response to antidepressants.
| 1 week | 2 weeks | 4 weeks | 6 weeks | F | p | ||
|---|---|---|---|---|---|---|---|
| Decreased score | AG (n = 19) | 1.42 ± 1.71 | 5.47 ± 5.28 | 12.47 ± 5.78 | 17.42 ± 5.27 | 0.147 | 0.702 |
| GG (n = 128) | 1.93 ± 2.49 | 5.74 ± 4.57 | 11.75 ± 5.99 | 15.82 ± 6.37 | |||
| Reducing score rate (%) | AG (n = 19) | 5.31 ± 6.43 | 20.74 ± 18.96 | 47.36 ± 19.63 | 67.3 ± 17.42 | 0.323 | 0.571 |
| GG (n = 128) | 8.38 ± 11.31 | 24.50 ± 19.09 | 49.64 ± 22.97 | 67.04 ± 23.77 |
ABCB1 Gene Polymorphism Loci and Clinical Response to Serotonin–Norepinephrine Reuptake Inhibitors
For SNRIs (venlafaxine and duloxetine), no significant difference was observed in the distribution of genotype and allele frequency of the rs1045642, rs2032582, rs2235040, rs1128503, and rs2235015 SNPs between the responders and nonresponders (p > 0.05). For rs2032583, the T allele frequency and TT genotype were significantly increased in the responders compared with those in the nonresponders (p = 0.025 and p = 0.018, respectively) ( Table 6 ).
Table 6
Genotype, allelic distribution of all genotyped SNPs among the treatment responders and nonresponders for serotonin-norepinephrine reuptake inhibitors (SNRIs).
| Responders (n = 80) (%) | Nonresponders (n = 26) (%) | X2 | p | OR (95% CI) | Adjust OR (95% CI) | |
|---|---|---|---|---|---|---|
| rs1045642 | ||||||
| CC | 31 (38.8) | 12 (46.2) | 1 | 1 | ||
| CT | 37 (46.3) | 12 (46.2) | 1.059 | 0.589 | 1.194 (0.47–3.03) | 1.898 (0.644–5.593) |
| TT | 12 (15.0) | 2 (7.7) | 2.323 (0.451–11.96) | 1.499 (0.263–8.537) | ||
 C allele C allele | 99 (61.9) | 36 (69.2) | 1 | 1 | ||
 T allele T allele | 61 (38.1) | 16 (30.8) | 0.918 | 0.338 | 1.386 (0.71–2.709) | 1.11 (0.208–5.932) |
| rS2032583 | ||||||
| TT | 69 (86.3) | 17 (65.4) | 1 | 1 | ||
| CT | 11 (13.8) | 9 (34.6) | 5.581 | 0.018* | 0.301* (0.108–0.842) | 0.261* (0.085–0.807) |
 T allele T allele | 149 (93.1) | 43 (82.7) | 1 | |||
 C allele C allele | 11 (6.9) | 9 (17.3) | 4.999 | 0.025* | 0.353* (0.137–0.907) | 0.261* (0.085–0.807) |
| rs2032582 | ||||||
| GG | 25 (31.3) | 11 (42.3) | 1 | 1 | ||
| GT | 28 (35) | 11 (42.3) | 3.254 | 0.196 | 1.12 (0.414–3.028) | 1.466 (0.473–4.541) |
| TT | 27 (33.8) | 4 (15.4) | 2.97 (0.836–10.55) | 2.502 (0.644–9.72) | ||
 G allele G allele | 78 (48.75) | 33 (63.5) | 1 | 1 | ||
 T allele T allele | 82 (51.25) | 19 (36.5) | 3.405 | 0.065 | 1.826 (0.959–3.477) | 2.07 (0.598–7.170) |
| rs2235040 | ||||||
| AG | 12 (15) | 8 (30.8) | 1 | 1 | ||
| GG | 68 (85) | 18 (69.2) | 3.188 | 0.074 | 2.519 (0.895–7.086) | 2.98 (0.96–9.245) |
 A allele A allele | 12 (7.5) | 8 (15.4) | 1 | 1 | ||
 G allele G allele | 148 (92.5) | 44 (84.6) | 2.856 | 0.091 | 2.242 (0.862–5.832) | 2.98 (0.96–9.245) |
| rs1128503 | ||||||
| CC | 10 (12.5) | 6 (23.1) | 1 | 1 | ||
| CT | 40 (50.0) | 13 (50.0) | 2.083 | 0.353 | 1.846 (0.562–6.068) | 1.838 (0.495–6.818) |
| TT | 30 (37.5) | 7 (26.9) | 2.571 (0.698–9.476) | 1.819 (0.422–7.839) | ||
 C allele C allele | 60 (37.5) | 25 (48.1) | 1 | 1 | ||
 T allele T allele | 100 (62.5) | 27 (51.9) | 1.828 | 0.176 | 1.543 (0.821–2.901) | 1.149 (0.385–3.43) |
| rs2235015 | ||||||
| GG | 68 (85.0) | 18 (69.2) | 1 | 1 | ||
| GT | 12 (15.0) | 8 (30.8) | 3.188 | 0.074 | 0.397 (0.141–1.117) | 0.983 (0.944–1.024) |
 G allele G allele | 148 (92.5) | 44 (84.6) | 1 | 1 | ||
 T allele T allele | 12 (7.5) | 8 (15.4) | 2.856 | 0.091 | 0.446 (0.171–1.16) | 0.446 (0.171–1.16) |
*Statistically significant difference between the groups (p < 0.05).
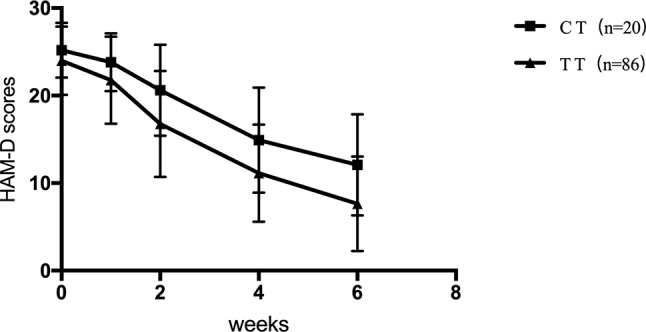
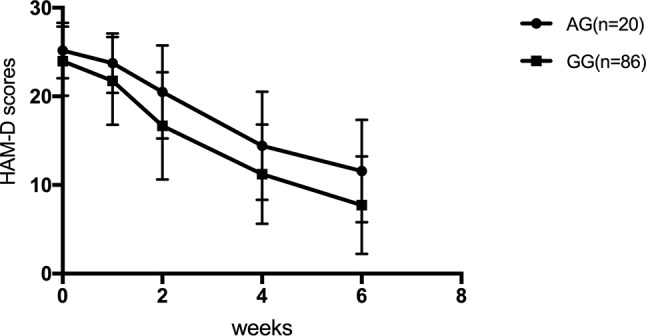
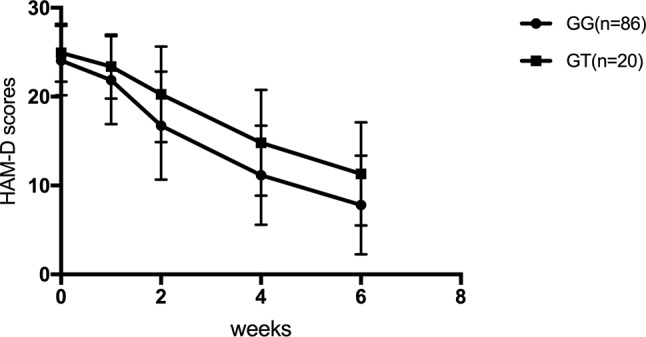
For SNRIs (venlafaxine and duloxetine), no significant difference was observed for the rs1045642, rs2032582, and rs1128503 SNPs in any of the HAM-D17 scores, decreased scores, and reducing score rate during the first, second, fourth, and sixth weeks (p > 0.05). For rs2032583, the HAM-D17 scores of TT genotype are lower than those of the CT genotype, whereas the decreased scores and reducing score rate are higher than those of the CT genotype ( Table 7 and Figure 2 ). The GG genotypes of rs2235040 have lower HAM-D17 scores than AG genotypes and higher decreased score and reducing score rate than AG genotypes ( Table 8 and Figure 3 ). For rs2235015, the GG genotypes have lower HAM-D17 scores than those of the GT genotypes and higher in decreased score and reducing score rate than those of the GT genotypes ( Table 9 and Figure 4 ).
Table 7
Rs2032583 and response to antidepressants.
| 1 week | 2 weeks | 4 weeks | 6 weeks | F | p | ||
|---|---|---|---|---|---|---|---|
| Decreased score | CT (n = 17) | 1.40 ± 1.79 | 4.60 ± 4.11 | 10.3 ± 6.33 | 13.1 ± 6.51 | 5.949 | 0.016* |
| TT (n = 130) | 2.23 ± 2.67 | 7.33 ± 4.62 | 12.86 ± 5.28 | 16.36 ± 5.29 | |||
| Reducing score rate (%) | CT (n = 17) | 5.49 ± 7.33 | 18.69 ± 17.19 | 40.50 ± 24.09 | 51.4 ± 24.0 | 9.241 | 0.003* |
| TT (n = 130) | 9.71 ± 11.76 | 31.29 ± 19.91 | 53.91 ± 20.80 | 68.8 ± 20.77 |
*Statistically significant difference between the groups (p < 0.05).
Table 8
Rs2235040 and response to antidepressants.
| 1 week | 2 weeks | 4 weeks | 6 weeks | F | p | ||
|---|---|---|---|---|---|---|---|
| Decreased score | AG (n = 20) | 1.45 ± 1.76 | 4.70 ± 4.07 | 10.75 ± 6.02 | 13.60 ± 6.06 | 4.288 | 0.041* |
| GG (n = 86) | 2.22 ± 2.68 | 7.30 ± 4.64 | 12.76 ± 5.41 | 16.24 ± 5.47 | |||
| Reducing score rate (%) | AG (n = 20) | 5.74 ± 7.21 | 19.19 ± 17.07 | 42.75 ± 23.17 | 53.9 ± 22.57 | 6.64 | 0.011* |
| GG (n = 86) | 9.65 ± 11.80 | 31.18 ± 20.00 | 53.39 ± 21.33 | 68.22 ± 21.56 |
*Statistically significant difference between the groups (p < 0.05).
Table 9
Rs2235015 and response to antidepressants.
| 1 week | 2 weeks | 4 weeks | 6 weeks | F | p | ||
|---|---|---|---|---|---|---|---|
| Decreased score | GG (n = 130) | 2.20 ± 2.67 | 7.30 ± 4.64 | 12.90 ± 5.28 | 16.23 ± 5.42 | 4.93 | 0.029* |
| GT (n = 17) | 1.55 ± 1.85 | 4.70 ± 4.08 | 10.15 ± 6.25 | 13.65 ± 6.29 | |||
| Reducing score rate (%) | GG (n = 130) | 9.53 ± 11.75 | 31.12 ± 20.0 | 53.96 ± 20.86 | 68.1 ± 21.32 | 7.151 | 0.009* |
| GT (n = 17) | 6.27 ± 7.82 | 19.44 ± 17.28 | 40.31 ± 23.73 | 54.4 ± 23.90 |
*Statistically significant difference between the groups (p < 0.05).
Discussion
We investigated the association among the six SNPs of the ABCB1 gene and therapeutic response in the local Chinese Han population. Among the five SNPs, only one SNP (rs2032583) differed in genotype and allele frequencies between the responders and nonresponders. In particular, the TT genotype of this SNP was significantly more common in the responders than that in the nonresponders. This finding suggests that the C allele of rs2032583 may be a risk factor for MDD. In the SSRI therapy group, no correlation was found between the six SNPs of the ABCB1gene and therapeutic response to SSRIs (p > 0.05). However, for the SNRI therapy group, only rs2032583 is associated with a therapeutic response to SNRIs in patients with MDD. The C allele of the ABCB 1 gene SNP rs2032583 was negatively correlated with therapeutic response according to the logistic regression analyses. The genotype of ABCB1 gene SNPs rs2235015 and rs2235040 were associated with decreased score and reducing score rate in the SNRIs therapy group, but because of the small sample size, there was no significant difference in the final effective and ineffective grouping. This finding indicated that ABCB1 gene polymorphisms may not be associated with the treatment response to SSRIs, but with SNRIs. The TT genotype of the ABCB1 gene SNP rs2032583 could be a predictive factor of improved treatment response to SNRIs. To our knowledge, this study is the first to report the genetic association of ABCB1 gene polymorphism with therapeutic responses in a case-control design in Chinese Han people in Mainland China.
P-gp can restrict the entry of various substrates, such as antidepressants, from the bloodstream into the brain. Studies on ABCB1 knockout animals have shown that ABCB1−/− mice possess higher intracerebral concentrations of escitalopram, trimipramine, amitriptyline, doxepin, venlafaxine, and paroxetine compared with those in wild-type mice (Uhr et al., 2000; Uhr et al., 2002; Uhr et al., 2003). Kato et al. (2008) reported a significant association of the nonsynonymous SNP G2677T/A (rs2032582) with treatment response to paroxetine in depressed patients. Moreover, the wild variant haplotype 3435C–2677G–1236T is associated with poor response (Kato et al., 2008). In contrast, Nikisch et al. (2008) found that depressed patients harboring the 2677 GG/GT genotype responded to escitalopram treatment significantly better than patients with the 2677TT genotype and suggested that this polymorphism could be used as genetic markers for predicting treatment response to escitalopram treatment in MDD (n = 15). Uhr et al. (2008) showed that polymorphisms in the ABCB1 gene predict the response to antidepressant treatment in depressed patients (n = 133) receiving drugs that have been identified as substrates of P-gp (amitriptyline, paroxetine, venlafaxine, and escitalopram). However, Mihaljevic et al. (2008) found that MDR1 variants G2677T (rs2032582) and C3435T (rs1045642) are not associated with therapeutic response to paroxetine in patients with MDD (n = 127). Furthermore, Peters et al. (2008) did not find an association between ABCB1C3435T, G2677T, and C1236T polymorphisms and response and tolerance to citalopram in a large study sample (n = 831).
ABCB1 gene polymorphism loci mutation might cause the variation of P-gp, thereby increasing drug concentrations in the brain to improve therapeutic effects. The specific function of rs2032583 (intron 22) is not clear. Clinical research has found the association between rs2032583 and antidepressant effects. The wild-type allele of rs2032583 is T and the mutant allele is C. The study by Sarginson et al. (2010) showed that carriers of the C allele remitted faster than those with the TT genotype in the rs2032583 among paroxetine-treated patients and support the findings of Uhr et al. (2002) who found that rs2032583 genetic variants affected efficacy in patients of various ages treated with paroxetine and other ABCB1 substrates (Uhr et al., 2008; Sarginson et al., 2010). By contrast, the two previous results are inconsistent with our present results. For rs2032583, we found that TT genotype has lower HAM-D17 scores than those of CT genotype and higher in decreased score and reducing score rate than those of CT genotype. This finding suggested that the TT genotype of rs2032583 is likely to be a predictive factor of better treatment response to SNRIs.
Whether the locus mutation manipulates the coding region of P-protein conformation remains to be further basic research. The rs2032583 locus mutation might change the dimensional conformation of P-gp by allowing entry of drugs and toxic substances into the brain simultaneously to exacerbate the symptoms. In addition, the locus mutation changes P-gp conformation and then stops the SNRIs into the brain but does not affect SSRI substrate. Furthermore, locus mutation may not change the conformation of P-gp. The positive results of SNRIs may be attributed to the small sample size, thereby causing false-positive results in the study. Foreign studies may have different results because of various factors such as race, region, environment, and diet.
Several limitations of the present study should be acknowledged. First, we did not distinguish the first and recurrence of depressive patients joining the patients’ group. Regression analysis determines the total course of the disease and whether they accepted treatment; however, previous treatment had some influence on the overall response rate. Hence, the present study can only obtain a trend about differences. Second, the present study only selected the drugs that have been identified as substrates of P-gp. Future studies should include no-P-gp substrates such as mirtazapine. Third, genetic effect is slight because of the small sample size. A large sample size is needed to improve the power of the test. Fourth, many receptors are involved to determine the effect of antidepressant drugs; hence, a combination of gene should be considered. Larger and more homogenous samples will be included in our future research to improve the statistical power. Because there may be a delayed response to symptom relief, 6 weeks of treatment may not be sufficient to fully demonstrate the relationship between ABCB1 gene and clinical response, and we consider extending the duration of efficacy observation in future studies.
Ethics Statement
The study protocol was approved by the Medical Ethics Committee of Second Xiangya Hospital, Central South University. Written informed consent was obtained from each patient after the study was explained.
Author Contributions
X-XS and YQ contributed equally to this work, mainly responsible for essay writing and conducting the study. W-WX mainly participated in conducting the study. YY assisted the study. R-RW, H-SW and L-HL contributed to guide of conducting the study and essay writing.
Funding
This research was supported by National Natural Science Foundation of China (grant no. 81501163), National Natural Science Foundation of China (grant no. 81270019), National R&D Special Fund for Health Profession (grant no. 201002003), and National Science and Technology Major Projects for “Major New Drugs Innovation and Development” (2012ZX09303014-001).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Ameyaw M., Regateiro F., Li T., Liu X., Tariq M., Mobarek A., et al. (2001). MDR1 pharmacogenetics: frequency of the C3435T mutation in exon 26 is significantly influenced by ethnicity. Pharmacogenetics 11 (3), 217–221. 10.1097/00008571-200104000-00005 [Abstract] [CrossRef] [Google Scholar]
- de Klerk O. L., Nolte I. M., Bet P. M., Bosker F. J., Snieder H., Den Boer J. A., et al. (2013). ABCB1 gene variants influence tolerance to selective serotonin reuptake inhibitors in a large sample of Dutch cases with major depressive disorder. Pharmacogenomics J. 13 (4), 349–353. 10.1038/tpj.2012.16 [Abstract] [CrossRef] [Google Scholar]
- Dong C., Wong M. L., Licinio J. (2009). Sequence variations of ABCB1, SLC6A2, SLC6A3, SLC6A4, CREB1, CRHR1 and NTRK2: association with major depression and antidepressant response in Mexican-Americans. Mol. Psychiatry 14 (12), 1105–1118. 10.1038/mp.2009.92 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kato M., Fukuda T., Serretti A., Wakeno M., Okugawa G., Ikenaga Y., et al. (2008). ABCB1 (MDR1)gene polymorphisms are associated with the clinical response to paroxetine in patients with major depressive disorder. Prog. Neuropsychopharmacol. Biol. Psychiatry 15 (32), 398–404. 10.1016/j.pnpbp.2007.09.003 [Abstract] [CrossRef] [Google Scholar]
- Lin K. M., Chiu Y. F., Tsai I. J., Chen C. H., Shen W. W., Liu S. C., et al. (2011). ABCB1 gene polymorphisms are associated with the severity of major depressive disorder and its response to escitalopram treatment. Pharmacogenet. Genomics 21 (4), 163–170. 10.1097/FPC.0b013e32833db216 [Abstract] [CrossRef] [Google Scholar]
- Menu P., Gressier F., Verstuyft C., Hardy P., Becquemont L., Corruble E. (2010). Antidepressants and ABCB1 gene C3435T functional polymorphism: a naturalistic study. Neuropsychobiology 62 (3), 193–197. 10.1159/000319361 [Abstract] [CrossRef] [Google Scholar]
- Mihaljevic Peles A., Bozina N., Saqud M., Roinic Kuzman M. (2008). Lovric M. Prog. Neuropsychopharmacol. Biol. Psychiatry 32 (6), 1439–1444. 10.1016/j.pnpbp.2008.03.018 [Abstract] [CrossRef] [Google Scholar]
- Nikisch G., Eap C. B., Baumann P. (2008). Citalopram enantiomers in plasma and cerebrospinal fluid of ABCB1 genotyped depressive patients and clinical response: A pilot study. Pharmacol, Res. 58 (5–6), 344–347. 10.1016/j.phrs.2008.09.010 [Abstract] [CrossRef] [Google Scholar]
- Peters E. J., Slager S. L., Kraft J. B., Jenkins G. D., Reinalda M. S., McGrath P. J., et al. (2008). Pharmacokinetic genes do not influence response or tolerance to citalopram in the STAR*D sample. PLoS One 3 (4), e1872. 10.1371/journal.pone.0001872 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Rosenhagen M. C., Uhr M. (2011). The clinical impact of ABCB1 polymorphisms on the treatment of psychiatric diseases. Curr. Pharm. Des. 17 (26), 2843–2851. 10.2174/138161211797440140 [Abstract] [CrossRef] [Google Scholar]
- Sarginson J. E., Lazzeroni L. C., Ryan H. S., Ershoff B. D., Schatzberg A. F., Murphy G. M., Jr. (2010). ABCB1 (MDR1) polymorphisms and antidepressant response in geriatric depression. Pharmacogenet. Genomics 20 (8), 467–775. 10.1097/FPC.0b013e32833b593a [Abstract] [CrossRef] [Google Scholar]
- Singh A. B., Bousman C. A., Ng C. H., Byron K., Berk M. (2012). ABCB1 polymorphism predicts escitalopram dose needed for remission in major depression. Transl. Psychiatry 2, e198. 10.1038/tp.2012.115 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Uhr M., Grauer M. T., Yassouridis A., Ebinqer M. (2007). Blood-brain barrier penetration and pharmacokinetics of amitriptyline and its metabolites in p-glycoprotein (abcb1ab) knock-out mice and controls. J. Psychiatr. Res. 41 (1–2), 179–188. 10.1016/j.jpsychires.2005.10.005 [Abstract] [CrossRef] [Google Scholar]
- Uhr M., Grauer M. T., Holsboer F. (2003). Differential enhancement of antidepressantpenetration into the brain in mice with abcb1ab (mdr1ab) P-glycoproteingene disruption. Biol. Psychiatry 54 (8), 840–846. 10.1016/S0006-3223(03)00074-X [Abstract] [CrossRef] [Google Scholar]
- Uhr M., Holsboer F., Müller M. B. (2002). Penetration of endogenous steroid hormones corticosterone, cortisol, aldosterone and progesterone into the brain is enhanced in mice deficient for both mdr1a and mdr1b P-glycoproteins. J. Neuroendocrinol. 14, 753–759. 10.1046/j.1365-2826.2002.00836.x [Abstract] [CrossRef] [Google Scholar]
- Uhr M., Steckler T., Yassouridis A., Holsboer F. (2000). Penetration of amitriptyline, but not of fluoxetine, into brain is enhanced in mice with blood-brain barrier deficiency due to mdr1a P-glycoprotein gene disruption. Neuropsychopharmacology 22, 380–387. 10.1016/S0893-133X(99)00095-0 [Abstract] [CrossRef] [Google Scholar]
- Uhr M., Tontsch A., Namendorf C., Ripke S., Lucae S., Ising M., et al. (2008). Polymorphisms in the drug transporter gene ABCB1 predictantidepressant treatment response in depression. Neuron 57 (2), 203–209. 10.1016/j.neuron.2007.11.017 [Abstract] [CrossRef] [Google Scholar]
- Weizman S., Gonda X., Dome P., Faludi G. (2012). Pharmacogenetics of antidepressive drugs: a way towards personalized treatment of major depressive disorder. Neuropsychopharmacol. Hung. 14 (2), 87–101. 10.5706/nph201206002 [Abstract] [CrossRef] [Google Scholar]
Articles from Frontiers in Pharmacology are provided here courtesy of Frontiers Media SA
Full text links
Read article at publisher's site: https://doi.org/10.3389/fphar.2019.00761
Read article for free, from open access legal sources, via Unpaywall:
https://www.frontiersin.org/articles/10.3389/fphar.2019.00761/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.3389/fphar.2019.00761
Article citations
Omega-3 Polyunsaturated Fatty Acids and Blood-Brain Barrier Integrity in Major Depressive Disorder: Restoring Balance for Neuroinflammation and Neuroprotection.
Yale J Biol Med, 97(3):349-363, 30 Sep 2024
Cited by: 0 articles | PMID: 39351324 | PMCID: PMC11426295
Review Free full text in Europe PMC
The Role of Pharmacogenetics in Personalizing the Antidepressant and Anxiolytic Therapy.
Genes (Basel), 14(5):1095, 16 May 2023
Cited by: 12 articles | PMID: 37239455 | PMCID: PMC10218654
Review Free full text in Europe PMC
Influence of eight ABCB1 polymorphisms on antidepressant response in a prospective cohort of treatment-free Russian patients with moderate or severe depression: An explorative psychopharmacological study with naturalistic design.
Hum Psychopharmacol, 37(3):e2826, 17 Nov 2021
Cited by: 3 articles | PMID: 34788473 | PMCID: PMC9285790
How Can Drug Metabolism and Transporter Genetics Inform Psychotropic Prescribing?
Front Genet, 11:491895, 08 Dec 2020
Cited by: 16 articles | PMID: 33363564 | PMCID: PMC7753050
Review Free full text in Europe PMC
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
SNPs (Showing 10 of 10)
- (19 citations) dbSNP - rs2032583
- (12 citations) dbSNP - rs2032582
- (12 citations) dbSNP - rs2235040
- (11 citations) dbSNP - rs1045642
- (10 citations) dbSNP - rs2235015
- (7 citations) dbSNP - rs1128503
- (1 citation) dbSNP - rs58898486
- (1 citation) dbSNP - rs17064
- (1 citation) dbSNP - rs4728697
- (1 citation) dbSNP - rs2232583
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Case-control association study of ABCB1 gene and major depressive disorder in a local Chinese Han population.
Neuropsychiatr Dis Treat, 11:1967-1971, 04 Aug 2015
Cited by: 10 articles | PMID: 26347319 | PMCID: PMC4531014
Modification of the association between paroxetine serum concentration and SERT-occupancy by ABCB1 (P-glycoprotein) polymorphisms in major depressive disorder.
Psychiatr Genet, 30(1):19-29, 01 Feb 2020
Cited by: 2 articles | PMID: 31634334
ABCB1 Gene Variants and Antidepressant Treatment Outcomes: A Systematic Review and Meta-Analysis Including Results from the CAN-BIND-1 Study.
Clin Pharmacol Ther, 114(1):88-117, 06 Mar 2023
Cited by: 4 articles | PMID: 36681895
Review
ABCB1 polymorphism predicts escitalopram dose needed for remission in major depression.
Transl Psychiatry, 2:e198, 27 Nov 2012
Cited by: 45 articles | PMID: 23188198 | PMCID: PMC3565756