Abstract
Free full text

Improving risk assessment of the emergence of novel influenza A viruses by incorporating environmental surveillance
Abstract
Reassortment is an evolutionary mechanism by which influenza A viruses (IAV) generate genetic novelty. Reassortment is an important driver of host jumps and is widespread according to retrospective surveillance studies. However, predicting the epidemiological risk of reassortant emergence in novel hosts from surveillance data remains challenging. IAV strains persist and co-occur in the environment, promoting co-infection during environmental transmission. These conditions offer opportunity to understand reassortant emergence in reservoir and spillover hosts. Specifically, environmental RNA could provide rich information for understanding the evolutionary ecology of segmented viruses, and transform our ability to quantify epidemiological risk to spillover hosts. However, significant challenges with recovering and interpreting genomic RNA from the environment have impeded progress towards predicting reassortant emergence from environmental surveillance data. We discuss how the fields of genomics, experimental ecology and epidemiological modelling are well positioned to address these challenges. Coupling quantitative disease models and natural transmission studies with new molecular technologies, such as deep-mutational scanning and single-virus sequencing of environmental samples, should dramatically improve our understanding of viral co-occurrence and reassortment. We define observable risk metrics for emerging molecular technologies and propose a conceptual research framework for improving accuracy and efficiency of risk prediction.
This article is part of the theme issue ‘Dynamic and integrative approaches to understanding pathogen spillover’.
1. Introduction
Reassortment is a prominent mechanism by which segmented viruses produce genetic variation. Reassortment occurs when genetic segments from different co-infecting virions within the same cell are packaged together, generating a novel strain. Some of the most devastating outbreaks caused by influenza A virus (IAV; a segmented RNA virus) are believed to have been driven by reassortants [1,2], yet predicting reassortment remains elusive.
For avian IAVs, new reassortants can be generated in natural reservoir hosts (wild water birds), where multiple strains of virus circulate naturally, or following spillover from a reservoir host to an infected spillover host (e.g. poultry, pigs or humans). Although reassortants with increased fitness may be produced less commonly than deleterious ones [3], reassortants are observed frequently at the population scale among natural virus isolates [4,5] suggesting that reassortment is an important source of adaptation. Both co-infections (a necessary precursor to reassortment) and reassortants are often detected retrospectively during surveys of wild waterfowl [6–11]. A comprehensive analysis of publicly available genomic data from avian and mammalian host species found that a substantial proportion of genomes (3%—646 total) were first time reports of novel reassortants [4]. Yet, emergence of novel IAV reassortants remains unpredictable and depends on host and virus populations [4]. One model estimated a 3-year cycle of emergence and replacement of avian IAV reassortants [9], but the highly pathogenic goose/Guangdong/96 clade 2.3.4.4 H5 appears to be persisting and contributing to the ongoing generation of novel reassortants [12].
In wild birds, IAV transmission via the environment is a major mechanism facilitating co-infection and host contact with pathogens [13]. Environmental reservoirs, such as ponds, can accumulate virus particles shed in faeces by many individuals, increasing the co-occurrence and exposure to different viral strains [13,14], and hence the likelihood of co-infection. By extension, reassortment events that have occurred in hosts using the same water body could be revealed by sequencing individual whole viruses from the environment over time and conducting genomic analyses, thus increasing detection capabilities for the reassortant combinations present. Environmental reservoirs also increase contact between species—i.e. species that may never contact each other directly drink or feed from the same water source. They additionally increase the infectious period (time frame during which infectious particles can be transmitted following infection) and expand the spatial availability of viruses (locations where infectious particles can be acquired), thus increasing the likelihood that novel host species will make contact with a variety of different viruses. Therefore, environmental transmission can overcome barriers of spillover by increasing interspecies contact with viable pathogens, and facilitating large jumps in viral diversity and fitness by allowing co-infection and reassortment.
One of the most important public/livestock health challenges is to predict the emergence of novel strains from surveillance systems. Predicting this epidemiological risk for viruses that reassort frequently, such as IAV, requires an understanding of the viral evolutionary ecology in reservoir and spillover hosts and its significance in driving viral fitness across scales [15]. Factors involved include predicting the rate with which new reassortants are generated in cell tissues and individual hosts (de novo reassortment), estimating the probability that a new reassortant will become predominant in reservoir-host populations locally and regionally, and the probability of spillover and transmission in spillover-host populations (figure 1–1, 1-2).
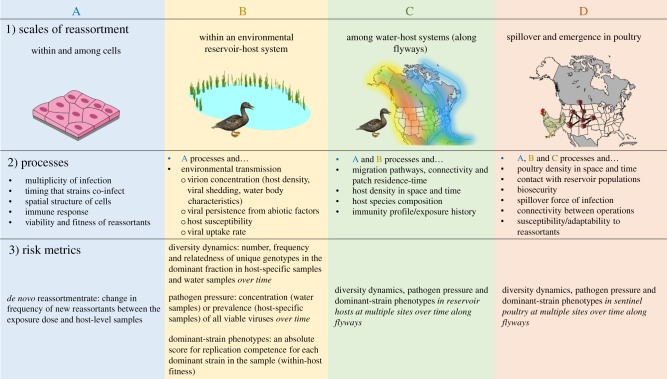
Scales at which reassortment occurs (1). In order to predict the emergence of reassortants in poultry (D), reassortment and transmission processes (2) need to be understood at each scale. We propose risk metrics that can be measured at each scale (3) for development of predictive models of epidemiological risk (figure 2).
We first review what is known from experimental studies of within-host reassortment rates, environmental transmission of IAV and assaying strain-specific fitness. We then outline emerging technologies in environmental sampling, phenotyping, genomics and bioinformatics that could improve detection and assessment of reassortants in environmental surveillance samples. We describe how emerging technology could allow efficient observation of reassortant dynamics in host populations, by incorporating environmental surveillance. Lastly, we define risk metrics (e.g. figure 1-3) that can be measured using these molecular technologies and describe conceptual strategies for using those metrics to inform quantitative frameworks designed for prediction of epidemiological risk from environmental surveillance samples.
2. Knowledge from experimentation
Experiments have provided foundational knowledge on de novo reassortment rates and combinatorial constraints within hosts and cell tissues, yet predicting within-host rates of reassortment from specific factors has just begun [16,17]. Similarly, quantifying reassortment rates within an environmental reservoir-host system (e.g. figure 1b) has only rarely been addressed [11]. Below, we describe experimental approaches that could be used to measure de novo reassortment rates within hosts, and transmission of reassortants in an environmental reservoir-host system, allowing improved prediction of epidemiological risk from surveillance data.
(a) Co-infection and reassortment processes
Significant progress has been made in understanding within-host reassortment processes in spillover host models (i.e. poultry and mammals) using experimental inoculation techniques [16,17]. Rates of de novo reassortment within hosts can be determined by co-infecting single hosts with known parental genotypes and measuring the proportion of shed virions that are reassortants. Such experiments have helped to develop assays for studying reassortment rates within hosts, and demonstrated substantial and highly variable rates of reassortment within-host individuals ([18–22]; e.g. 8.7% of viruses recovered from ferrets co-infected with human and avian strains were reassortants [18], to approximately 50% of viruses recovered from co-infections of chickens with other chicken-derived subtypes [19], to 86% in swine and guinea pigs co-infected with swine or human strains [20,21]). The differences are thought to be owing to functional incompatibilities between viral genome segments or differing receptor binding specificities or other host restrictions, leading to low levels of co-infection in the same cells.
Within-host studies have determined that transmission route, timing of exposures, dose, strains and strain competition all can influence co-infection and reassortment rates [16,17,21]. Higher doses lead to higher rates of co-infected cells [21]. When exposure with two different strains is offset by a short period of time (12 h), co-infection and reassortment rates are higher than when the host is exposed to the two strains simultaneously [21]. Delayed introduction of the second strain likely increases co-infection rates because it allows the first strain time to infect many cells, thus maximizing co-infection potential by the second strain [21]. By contrast, exposures with longer time lags can reduce co-infection because of super-infection exclusion mechanisms that occur later post-infection (i.e. host innate immune function, and viral processes such as receptor interference) [17] and because death of infected cells occurs. As expected, effects of co-infection time lags on reassortment rates can differ depending on host-strain combinations and experimental design [19]. Importantly, these advances in our understanding of within-host reassortment provide a foundation for predictive models to account for co-infection and reassortment processes mechanistically in epidemiological models. An important gap for experimentation is that little work has focused on understanding reassortment in reservoir hosts under natural transmission conditions (figure 1A); how the lessons learned from animal models apply to wild birds infected by environmental transmission remains unknown.
(b) Environmental transmission
Models suggest that environmental reservoirs play an important role in the ecology of IAVs because they allow infection between hosts that infrequently come into direct contact, such as migratory wild waterfowl on breeding grounds [13,14,23,24]. Experimental systems have shown the occurrence of environmental transmission of IAVs among reservoir hosts and from reservoir hosts to spillover hosts [25–28]. However, the role of indirect transmission though the environment relative to other transmission mechanisms is not well understood empirically, which is important for informing models aimed at predicting epidemiological risk. One study identified a correlation between host population density and environmental levels of IAV [29]. Environmental transmission experiments combined with molecular characterization and disease-dynamic models would provide powerful tools for improving our quantitative knowledge of environmental transmission, co-infection and reassortment processes.
One key driver of environmental transmission is persistence of viral viability after shedding into the environment. There is substantial variation in environmental durability of IAVs (between 1 and 600 days) depending on abiotic factors, especially temperature, salinity, pH and mineral content of the medium [30–35] and viral genotype [33,35], including under natural conditions [36]. While some information is available for water and bird faeces, studies with soil [37], sediments [29,33,38] and plant material are few. Variation in viral environmental persistence could be an important component of fitness [39] and should be included in models predicting the ecological and evolutionary dynamics of IAVs. A final important knowledge gap is understanding the relationship between environmental virus concentrations and infectivity.
(c) Strain-specific fitness
Experimental studies are also helpful for evaluating phenotypes (e.g. measures of within-host replication fitness) of parental and reassortant viruses, a predictor of co-infection and reassortment rates within hosts (figure 2a). Fitness of reassortant strains is a crucial filter on emergence risk. Fitness studies on human IAV strains provide some insights where deleterious reassortment events were characterized by fewer reassortant progeny and more limited replication efficiency in spillover host cells [3,40]. Also poorly understood is the connection between within-host fitness and transmissibility (e.g. [41]).

Approaches to predicting epidemiological risk in an environmental reservoir-host system. (a) Bottom-up approach. Develop a mechanistic disease transmission model of the evolutionary and ecological processes (bolded numbers) involved in reassortment. Design controlled environmental transmission studies with multiple strains and hosts. Then, iteratively fit and validate the mechanistic model and refine models and experiments based on learning. Top part of the plot shows processes that need to be considered in the mechanistic model. Note that there are additional within-host processes (not shown here) such as super-infection exclusion, within cell compartmentalization, spatial heterogeneity of target cells and others that can also affect the likelihood of reassortment. Numbers represent the following processes: (1) some viruses degrade owing to environmental conditions, (2) selection for persistence, (3) viruses that persist can be transmitted, (4) more than one virus strain can infect a single host either by co- or super-infection, (5) viral particles propagate within-hosts, as determined by the initial dose and fitness of the inoculating strain(s), (6) co- or super-infection of individual cells and reassortment may occur as a function of multiplicity of infection in target cells, (7) mutations with fitness effects occur during within-host replication, (8) selection for within-host replication influences the bottleneck size at shedding into the environment (9). (b) Top-down approach. Develop a statistical model for predicting diversity-pressure-phenotypes. Collect surveillance data from water, wild birds and sentinel chickens in multiple areas. Fit the model to the data to predict true reassortant diversity-pressure-phenotypes metrics. For both bottom-up and top-down approaches, the first step is to develop predictive tools. Once the predictive tools are validated through research studies, they can be applied to surveillance data (e.g. figure 1D) to predict epidemiological risk in poultry.
Comparative genomics of IAV genomes from wild bird and poultry samples identify putative genetic constellations that can readily produce reassortants with high fitness in reservoir and spillover host species (‘high risk’ markers). These could be tested for potential fitness effects in vitro with deep-mutational scanning approaches [42] to efficiently phenotype a large number of strains for which the genotype is known. Then putative ‘high risk’ markers can be identified by statistically analysing the relationship between genotype and phenotype. Using synthetically engineered or naturally isolated genotypes in in vivo experimental infection and transmission experiments could produce estimates of fitness for particular genotypes or molecular markers in a given host system (e.g. see techniques in [36]). At present, deep-mutational scanning and in vivo techniques are too time-intensive and costly to incorporate in large-scale surveillance systems, but they are feasible for experimental settings and thus hold promise for improving our understanding of viral evolutionary dynamics and emergence.
3. Prospects from genomic technology
A fundamental hurdle with studying reassortment using genetic data is knowing how different segments are distributed among virions. In this section, we describe challenges at each step, from acquiring genome sequences to detecting reassortants in a collection of genomes. We further describe cutting-edge advances that may make it possible to detect and understand reassortment from an environmental reservoir-host system (figure 1B). In particular, the rapidly growing field of environmental metagenomics provides a framework for capturing IAV diversity from the environment ([43,44]; figure 1B). When applied in tandem with population genomics analyses, environmental metagenomics could be a useful tool for inferring reassortment events and for understanding the role of the environment in co-infection risk and reassortant emergence (figure 1).
(a) Environmental sampling
A major challenge with environmental surveillance is capturing the strain diversity of IAVs in the environment [38]. Questions include: do you sample water or soils or both? How many samples per site? How deep do you sample? How many technical replicates? A gold standard for capturing, isolating and identifying true IAV diversity from the environment is still not established [45]. Similarly, the effects of common environmental variables such as water turbidity, rates of dispersion and diffusion and spatial heterogeneity across water or other environments have not been examined, although they affect environmental DNA capture from aquatic organisms [46]. Because knowledge of how to sample the environment for target RNA is still in its infancy, pilot studies structured to optimize sampling protocols are critical (e.g. [47]).
Another major sampling challenge is RNA preservation [48]. Filtration to capture virions/RNA requires hours at a single site (e.g. 800 L) to achieve adequate probability of virus detection [49]. Decontamination of pumping equipment at each site is necessary, further increasing sampling time. While these methods provide a useful means of collecting environmental genetic sequences [46], methodological advancements in RNA preservation/collection techniques are needed to improve the efficiency of collecting high-quality RNA viromes from environmental samples.
(b) Generating sequence data from environmental samples
An initial field-based screen can rapidly prioritize localities for further sampling efforts. Technological advances will soon allow diagnostic PCR assays in the field, with results within hours to minutes. Companies such as Biomeme (Philadelphia, Pennsylvania, USA) have developed field-based nucleic acid extraction kits (M1 Sample Prep Kits) and a hand held real-time PCR thermocycler that attaches to a smart phone (two3™ Real-Time PCR Thermocycler). Both of these have been deployed in the field to identify mosquito pools positive for RNA viruses in less than 2 h to target samples for more thorough sequencing [50].
New sequencing platforms (e.g. MinION from Oxford Nanopore Technologies) hold promise for field-based surveillance of IAV owing to their rapid library prep and run times, and elimination of PCR amplification. The MinION generates long reads that allow sequencing of entire gene segments in a single read, single-molecule sequencing and direct RNA sequencing without conversion to cDNA and PCR amplification [51,52]. For field-based pathogen surveillance, it is possible to run a sequencer from a smartphone (e.g. SmidgION from Oxford Nanopore Technology). All of these technologies currently lack high-throughput capacities, have cost-prohibitive library prep, low sensitivity and high error rates for application to broad-scale surveillance. Despite these hurdles, there is a continual push to improve mobile diagnostic capabilities [53,54], which will inevitably improve efficiency, accuracy and precision of obtaining IAV genomic data from environmental samples.
All contemporary sequencing platforms require a minimum concentration of input nucleic acids, and water samples tend to fall below this threshold, requiring enrichment of target IAV RNA [55]. Centrifugation following filtration has been effective at concentrating viral particles [56]. The gold standard for IAV enrichment is virus culture in either embryonated chicken eggs or Madin–Darby canine kidney cells. These methods work well for clinical samples, but there can be high costs and challenges with isolating the virus from environmental samples [57–59]. Additionally, using virus isolates to study reassortment can be counter-productive because of subtype and genomic selection bias in media used to propagate isolates. PCR is another method for enriching viral genomes, however, PCR can preferentially amplify specific strains and introduce nucleotide errors that can be difficult to distinguish from true, low-frequency virus variants. The ideal approach is direct sequencing of viral genomes without PCR (see below).
Another major challenge with viromic data from environmental samples is delineation of individual genomes—i.e. how do we distinguish potential reassortants from multiple parental genotypes or viral mixtures? A common method is to isolate individual genotypes using limiting dilution plaque assays and subsequent whole-genome sequencing. However, plaque assays can cause biased replication levels between multiple strains because of tissue tropism [60], in vitro strain mutation [61] and overlapping plaques [62], all which can obscure the delineation of genotypes. Serial dilution can reduce plaque overlap but is very time consuming.
The advent of single-virus sequencing techniques [63,64], similar to single-cell genome sequencing techniques of eukaryotic and bacterial genomes [65], holds promise for circumventing several potential biases from viral isolation techniques while providing sequence data from individual virons. Using flow cytometry to isolate virions has improved sequencing of single-virus genomes [63,66], but RNA input requirements for library prep are still too high to allow the sequencing of a single, non-enriched virus genome from environmental samples. However, methods for conducting single-virus RNA sequencing on a Nanopore or similar single-molecule sequencing platform will undoubtedly be realized in the near future, providing a valuable method of delineating IAV genotypes that have not been subject to selective media.
(c) Assembling genes and genomes for reassortment analysis
The most widely used sequencing platforms generate short reads, which makes genome assembly challenging because there may not be enough overlap between reads to determine alignments with high enough certainty. A common first step is to remove non-target sequence reads. Environmental samples include nucleic acids from non-target organisms, for which sequenced genomes are not available. The effectiveness of filtering is thus reduced, which increases computing time for genome assembly. The two main approaches for viral genome assembly are reference-based mapping and de novo assembly. Mapping reads to a reference genome can simplify assembly, but reference genome choice is not trivial and IAV genomic diversity in wild birds is very high, which can bias results when trying to recover viral population diversity [67]. De novo assembly of segmented viral genomes is complicated because of incomplete and uneven coverage of different segments, which can lead to skewed results or failed assemblies [68]. A useful approach could be de novo assembly to build reference genomes directly from the sample reads then map all the reads back to this genome to recover the viral population diversity, infer haplotypes and estimate their frequencies [67,69,70]. The ultimate goal is to accurately assemble all genome variants in systems with high levels of diversity and determine dominant and low-frequency strains, which can greatly influence epidemiologic outcomes.
(d) Detecting reassortants
Classically reassortants are identified by sequence alignment, and if necessary by generating phylogenetic trees of each gene segment to look for incongruent clades. An isolate can be considered to be a reassortant if one or more gene segments have a significantly different position in a fully resolved phylogenetic tree relative to other gene segments [71]. However, in wild birds, IAV diversities are frequently very high with multiple strains co-circulating, such that both mixtures of parental sequences and reassortants may be quite prevalent, resulting in phylogenetic trees that do not differ significantly from trees generated at random [6]. To detect such reassortants, we must define a minimum degree of difference in the topologies (e.g. [4]). The chosen thresholds will impact inferred rates of reassortment in surveillance samples.
Another challenge of detecting reassortants is that some assumptions of phylogenetic methods are violated, as ecological and evolutionary processes occur simultaneously in viral populations. For example, phylogenetic trees assume ancestral alleles are extinct, evolutionary patterns are bifurcating and no reticulation events have occurred [72]. Even creative approaches to estimating the frequency of reassortants in phylogenetic trees, such as discrete trait mapping and Bayesian ancestral state reconstruction [73], do not circumvent assumptions. Furthermore all eight gene segments need to be analysed separately, which is computationally burdensome. To better detect viral reassortants, we need to develop or apply approaches that align with viral evolutionary processes rather than applying theories and concepts developed for organisms with highly disparate biology [74].
Population genetic approaches may be more appropriate for identifying the frequency of reassortants in a set of genomes. Phylogenetic networks are methods that account for reticulate evolution and can identify reassortment events [11,72,75,76]. There is promise in using coalescent theory [77] and maximum-likelihood or Bayesian estimates of migration [78]. An additional approach could be from population genetic clustering algorithms (e.g. [79,80]), using hemagglutinin (HA) and neuraminidase (NA) subtypes as population delimiters and then using those as a framework for estimating the replacement and ‘gene flow’ of the internal segments. One final method could take advantage of recombination metrics by using concatenated IAV gene segments and treating the genomes as chromosomes. The exchange of gene segments will thus lead to estimates of recombination rates that can serve as a proxy for reassortment frequencies [76].
4. Approaches to predicting epidemiological risk: the way forward
(a) Observing reassortment across scales
Within individual hosts (figure 1A), the important risk metric is the de novo reassortment rate. Reassortment rates ideally would be measured using the genomic diversity (number of unique genotypes, frequency of each unique genotype, and relatedness between genotypes) in samples from individual hosts over time during an infection, where the first time point is the exposure dose of co-infecting viruses [22]. The frequency of reassortants in each sample relative to parental viruses at the first time point would give an empirical index of the host-level rate of reassortment. From the distribution of frequencies for all samples from an individual host over time, one could calculate the mean and variation of host-level reassortment rates. However, except in controlled field experiments the genomic diversity of the exposure dose will be unknown. As a proxy, genomic diversity of samples from the environment could be used.
Extending these ideas, the next scale is reassortment in an individual environmental reservoir-wild bird system (figure 1B). We propose three complementary risk metrics at this scale: the ‘diversity dynamics’ are captured by measuring the genomic diversity in the water and faecal samples from individual hosts over time; the ‘pathogen pressure’ [81] is the concentration of virions in water and their prevalence in the host population; and the ‘dominant-strain phenotypes' is a rapid assessment of replicative fitness for strains that are most frequent in the surveillance samples. We refer to these three important risk metrics together as ‘diversity-pressure-phenotyping’: data that can be observed to understand and predict epidemiological risk. An important condition is that these data are collected over time at the same site (longitudinal sampling) in order to monitor the process of reassortants being generated and selected within hosts, and then shed and selected for persistence in the environment (e.g. figure 2a). Linking the molecular markers that are identified in the ‘diversity’ data to phenotypes will provide a mechanistic foundation for diversity dynamics.
At the next level (figure 1C), diversity-pressure-phenotyping in water and hosts could be measured across space and time to surveil for reassortant emergence along flyways. Lastly, concurrent surveillance of diversity-pressure-phenotyping in sentinel poultry at sites where water-host systems are being surveilled would provide an ideal measure of predicted epidemiological risk for spillover and emergence of reassortants in poultry (figure 1D).
(b) Bottom-up approaches: disease-dynamic modelling
Within-host viral dynamics of IAV have now been reasonably well studied using experimental data [82,83], providing a strong foundation for developing predictive models of reassortment dynamics. Important factors determining within-host viral kinetics for IAV include target-cell availability [84], immune system factors [85], spatial distribution of target cells [86] and initial dose [87]. While these viral kinetic models have been developed using data from human infections [82,83], their insights and methods have begun to be applied to avian hosts [88]. The most important factor for reassortment is co-infection [16,17]. Thus, initial dose, target-cell availability and the distribution of target cells will play a primary role in predicting de novo reassortment rates. Once reassortment has occurred within a host, selection within the host and the size of the transmission bottleneck will determine whether a reassortant is shed into the environment [16] (figure 2a). The recent advances in understanding of intracellular and within-host processes that govern reassortment [16,17] provide conceptual and quantitative knowledge for models of within-host viral and reassortment dynamics. In the larger context, the reassortment processes and within-host viral dynamic models can be nested in between-host models that account for selection, transmission bottlenecks and environmental transmission processes (e.g. [39]) to develop a predictive framework that can be validated with experimental data (figure 2a).
Controlled experiments that allow transmission to occur naturally coupled with data-driven modelling frameworks (as above, figure 2a) together form a valuable stepping-stone toward inferring epidemiological risk from genetic samples. Controlled experiments are important for restricting the range of potential genotypes and environmental factors so that models can be designed to capture epidemiological behaviour based on a limited and known set of factors. In high-dimensional systems such as IAV, model-guided fieldwork [89]—i.e. iterative model development, prediction and experimentation (figure 2a)—can be applied for efficient experimental design and knowledge building. By this approach, models could be used to predict which processes are the most important to assess. Then experiments would test our knowledge of how ill-understood processes such as environmental transmission contribute to the observed outcome. For example, a poor-fitting model of diversity-pressure-phenotyping would suggest that there are ill-defined processes occurring in the system that are not well captured or are absent in the model. Then the model and experimental design could be adjusted to elucidate these ill-understood mechanisms. This concept is not new [89] but it is surprisingly underused for designing empirical studies of complex ecological and epidemiological systems with ample process uncertainty. The iterative approach of hypothesis generation → model building and prediction → data collection → model evaluation aims to reduce process uncertainty by targeted learning. Thus, it is a powerful approach for identifying important gaps in our ability to predict reassortment in environmental reservoir-host systems (figure 1B), and to design effective experiments that will best inform the identified gaps. Once this type of mechanistic epidemiological model is developed and validated using experimental data from controlled studies with natural transmission, it could be used to predict epidemiological risk from surveillance of diversity-pressure-phenotypes data in environmental reservoir-host systems.
As an illustration of how these cross-scale processes could be explored empirically, we describe a possible experimental study to measure diversity-pressure-phenotypes and resulting infection risks in an environmental reservoir-host system. A potential study would simulate a scenario where multiple hosts infected with different virus strains would be allowed to contaminate a quasi-natural environment. Infected hosts would then be removed and susceptible individuals would be exposed to the contaminated environment to allow for infection. Hosts and the environment would be sampled longitudinally at regular intervals throughout the experiment. All samples would be sequenced to determine de novo reassortment rates and diversity-pressure-phenotypes (figure 1; which measures the frequency dynamics of particular genotypes and their associated phenotypes, i.e. reassortant emergence). Testing both reservoir and spillover host species (e.g. mallards and chickens) would allow for determination of potential host differences in susceptibility and the impacts of diversity-pressure-phenotypes on epidemiological dynamics. A similar design could be applied to conditions that only allow direct contact between hosts. Comparing results from both experiments would help quantify transmission mechanisms and determine which mechanisms lead to higher reassortment risk.
(c) Top-down approaches: statistical modelling of sentinel surveillance (functional surveillance)
A second important approach to understanding and predicting epidemiological risk is to apply statistical methods to surveillance data from the environment and from reservoir and sentinel hosts. While bottom-up approaches based on experimental data have the power to improve inference by reducing complexity, the inference is specific to the conditions that are tested. For this reason, approaches that measure diversity-pressure-phenotypes in natural populations are equally important. Phylogenetic or phylodynamic methods have mostly been applied to estimate reassortment occurrence, rather than to quantify its frequency or dynamics (i.e. pattern-based surveillance as opposed to process-based, which is ‘functional surveillance’). Additionally, because these methods rely on historical genomic diversity for inference of current genomic diversity, they are retrospective not predictive. We propose that longitudinal surveillance of environmental reservoir-host systems, paired with systematic deployment of sentinel host individuals (e.g. [11]), is an important research direction that could improve the power of top-down statistical tools for prediction of epidemiological risk from environmental surveillance data.
Sentinel host sampling can allow for direct measurement of spillover rates between reservoir and poultry host populations when surveillance occurs at the same locations over time [90]. An example approach would include enclosures of sentinel poultry adjacent to environmental reservoirs used by waterfowl. This type of sentinel host design has allowed for documentation of seasonal dynamics of spillover [91] and evaluation of vaccination effectiveness [92]. The sentinel host design would be coupled with frequent measures of diversity-pressure-phenotypes in the environmental reservoir-host system, and in the sentinel hosts themselves, by applying single-virus sequencing and phenotyping assays to the samples (figure 2b). Analysis of genotype frequencies, linked to measured phenotypes, could identify molecular markers that are most likely to lead to outbreaks in poultry. Another useful dimension within this design would be to collect longitudinal data on the abiotic conditions of environmental samples, densities of hosts and host-species composition, concurrently with the longitudinal diversity-pressure-phenotypes data. These data would be used as covariate data for predicting emergence risk in sentinel hosts (figure 2b). Once validated with out-of-sample data from multiple sites, this type of statistical framework could be used to predict epidemiological risk in spillover hosts using diversity-pressure-phenotypes and abiotic data collected longitudinally from environmental reservoir-host systems. Although these systems might not capture all of the real-life complexity of contact structure between hosts, they would include natural complexity from strain diversity and abiotic factors, which is important for optimizing environmental surveillance technologies and inference from environmentally sampled data.
5. Conclusion
Understanding the emergence of novel reassortments across ecological scales that include environmental transmission, could transform risk assessment capabilities and add to current approaches that use non-mechanistic or retrospective genetic analyses. For example, shifting our focus from inferring which strains have spilled over to predicting which strains will successfully spill over from wild birds to poultry in the near future. Advancements in molecular techniques have brought the goal of predicting reassortment and spillover risk of IAVs onto the near-term horizon. Coupling risk-metric data, such as diversity-pressure-phenotyping, with bottom-up and top-down quantitative models is possible and can yield frameworks for both understanding and predicting reassortant emergence.
Although it is still not possible to collect diversity-pressure-phenotyping data routinely as part of large-scale surveillance systems, technological refinements that dramatically improve efficiency can occur rapidly. Once more efficient molecular technologies are available, scaling up their application to surveillance programmes will require a strong research foundation that has addressed fundamental gaps in the evolutionary ecology of IAVs; a knowledge base that is inextricably linked to the natural life cycle and complexity of this host–pathogen system. Thus, addressing the basic science knowledge gaps with current technology, as we have outlined, is an important stepping-stone towards developing more accurate and efficient risk assessment and surveillance programmes. However, even at a fine scale, the research approaches we outlined will require substantial investment because they require wide-ranging interdisciplinary collaboration and specialized facilities (i.e. animal pathobiology, cutting-edge sequencing technology, bioinformatic resources and skills, and expertise in mathematical and statistical modelling). Collaborations of this breadth are expensive but are feasible. The potential payoffs from improvements to risk assessment and outbreak prevention through evidence-based biosecurity could greatly outweigh the costs in the long term.
The improved surveillance system we envision can be characterized as ‘functional surveillance’: collecting and integrating appropriate surveillance data into analytical frameworks that describe emergence processes (as opposed to patterns). Functional surveillance would complement current approaches of using pattern-based surveillance by improving our understanding of emergence processes (e.g. [93]). The mechanistic underpinnings of functional surveillance will allow for better prediction as conditions change, a current weakness of retrospective risk assessments.
Acknowledgements
The findings and conclusions in this preliminary publication have not been formally disseminated by the U.S. Department of Agriculture and should not be construed to represent any agency determination or policy. The authors thank three anonymous reviewers for helpful comments and the organizers of a workshop on pathogen spillover in MT 20–23 February, which helped with idea development, and DARPA for funding the workshop.
Authors' contributions
K.M.P., M.W.H., S.A.S., E.S. and A.J.P. wrote the first draft. K.M.P. designed the figures. All authors contributed to idea development and manuscript/figure editing.
Funding
K.M.P., A.J.P., M.W.H., E.S. and S.A.S. were supported by the U.S. Department of Agriculture, APHIS.
References
Articles from Philosophical Transactions of the Royal Society B: Biological Sciences are provided here courtesy of The Royal Society
Full text links
Read article at publisher's site: https://doi.org/10.1098/rstb.2018.0346
Read article for free, from open access legal sources, via Unpaywall:
https://royalsocietypublishing.org/doi/pdf/10.1098/rstb.2018.0346
Citations & impact
Impact metrics
Article citations
Environmental transmission of influenza A virus in mallards.
mBio, 14(5):e0086223, 28 Sep 2023
Cited by: 2 articles | PMID: 37768062 | PMCID: PMC10653830
Avian influenza A virus susceptibility, infection, transmission, and antibody kinetics in European starlings.
PLoS Pathog, 17(8):e1009879, 30 Aug 2021
Cited by: 6 articles | PMID: 34460868 | PMCID: PMC8432794
Experimental virus evolution in cancer cell monolayers, spheroids, and tissue explants.
Virus Evol, 7(1):veab045, 01 Jan 2021
Cited by: 0 articles | PMID: 34040797 | PMCID: PMC8134955
Highly Pathogenic Avian Influenza Viruses at the Wild-Domestic Bird Interface in Europe: Future Directions for Research and Surveillance.
Viruses, 13(2):212, 30 Jan 2021
Cited by: 100 articles | PMID: 33573231 | PMCID: PMC7912471
Review Free full text in Europe PMC
Mammalian pathogenicity and transmissibility of low pathogenic avian influenza H7N1 and H7N3 viruses isolated from North America in 2018.
Emerg Microbes Infect, 9(1):1037-1045, 01 Dec 2020
Cited by: 7 articles | PMID: 32449503 | PMCID: PMC8284977
Go to all (8) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Enhancement of influenza virus transmission by gene reassortment.
Curr Top Microbiol Immunol, 385:185-204, 01 Jan 2014
Cited by: 23 articles | PMID: 25048543
Review
Onward transmission of viruses: how do viruses emerge to cause epidemics after spillover?
Philos Trans R Soc Lond B Biol Sci, 374(1782):20190017, 12 Aug 2019
Cited by: 29 articles | PMID: 31401954 | PMCID: PMC6711314
Review Free full text in Europe PMC
Vaccination decreases the risk of influenza A virus reassortment but not genetic variation in pigs.
Elife, 11:e78618, 02 Sep 2022
Cited by: 4 articles | PMID: 36052992 | PMCID: PMC9439680
Emergence and Evolution of Novel Reassortant Influenza A Viruses in Canines in Southern China.
mBio, 9(3):e00909-18, 05 Jun 2018
Cited by: 30 articles | PMID: 29871917 | PMCID: PMC5989073
Funding
Funders who supported this work.
Animal and Plant Health Inspection Service
Medical Research Council (2)
Understanding herd immunity for influenza using archived sera from the UK and mathematical models
Professor Steve Riley, Imperial College London
Grant ID: MR/J008761/1
MRC Centre for Global Infectious Disease Analysis
Professor Neil Ferguson, Imperial College London
Grant ID: MR/R015600/1
National Institute for Health Research (NIHR) (1)
Grant ID: HPRU-2012-10080
Wellcome Trust (1)
The life course of human immune responses to influenza infection and vaccination
Professor Steve Riley, Imperial College London
Grant ID: 200861/Z/16/Z





