Abstract
Free full text

RAB8, RAB10 and RILPL1 contribute to both LRRK2 kinase–mediated centrosomal cohesion and ciliogenesis deficits
Abstract
Mutations in the LRRK2 kinase are the most common cause of familial Parkinson’s disease, and variants increase risk for the sporadic form of the disease. LRRK2 phosphorylates multiple RAB GTPases including RAB8A and RAB10. Phosphorylated RAB10 is recruited to centrosome-localized RILPL1, which may interfere with ciliogenesis in a disease-relevant context. Our previous studies indicate that the centrosomal accumulation of phosphorylated RAB8A causes centrosomal cohesion deficits in dividing cells, including in peripheral patient-derived cells. Here, we show that both RAB8 and RAB10 contribute to the centrosomal cohesion deficits. Pathogenic LRRK2 causes the centrosomal accumulation not only of phosho-RAB8 but also of phospho-RAB10, and the effects on centrosomal cohesion are dependent on RAB8, RAB10 and RILPL1. Conversely, the pathogenic LRRK2-mediated ciliogenesis defects correlate with the centrosomal accumulation of both phospho-RAB8 and phospho-RAB10. LRRK2-mediated centrosomal cohesion and ciliogenesis alterations are observed in patient-derived peripheral cells, as well as in primary astrocytes from mutant LRRK2 mice, and are reverted upon LRRK2 kinase inhibition. These data suggest that the LRRK2-mediated centrosomal cohesion and ciliogenesis defects are distinct cellular readouts of the same underlying phospho-RAB8/RAB10/RILPL1 nexus and highlight the possibility that either centrosomal cohesion and/or ciliogenesis alterations may serve as cellular biomarkers for LRRK2-related PD.
Introduction
Autosomal-dominant point mutations in the leucine-rich repeat kinase 2 (LRRK2) gene are the most common cause of familiar Parkinson’s disease (PD), and protein coding as well as non-coding variants at the LRRK2 locus are major genetic susceptibility factors for sporadic PD (1). These genetic findings point towards a key role for LRRK2 in the entire PD disease spectrum. Recent mass spectroscopy studies have revealed that LRRK2 is able to phosphorylate a small subset of RAB proteins in vitro and in vivo (2–5), with their phosphorylation enhanced by all pathogenic LRRK2 mutants in a cellular context (3). Current data suggest that RAB8 and RAB10 serve as the most prominent LRRK2 kinase substrates, and their phosphorylation is detectable in various cells and tissues including brain (6–8). Therefore, and given the relevance of LRRK2 for PD, it is crucial to determine the cellular processes which are most significantly affected by an increase in LRRK2 kinase activity via RAB8 and/or RAB10 phosphorylation.
RAB GTPases are key regulators of all eukaryotic vesicular membrane–trafficking events (9,10). LRRK2 phosphorylates a crucial region within RAB proteins, whose conformation changes dependent on the nucleotide-bound state. Effectors and regulatory proteins interact with RAB proteins in a nucleotide-dependent manner through this region, and phosphorylation interferes with the ability of the RABs to bind to most of these proteins (3). This is expected to lead to a loss of function in terms of RAB-mediated membrane-trafficking events, as we have recently shown for the LRRK2-mediated phosphorylation of RAB8A (11).
Recent studies also highlight additional, gain-of-function mechanisms for phosphorylated RAB8 and RAB10. Our data show that phosphorylated RAB8A accumulates in a pericentrosomal/centrosomal area to cause deficits in centrosomal positioning with effects on neurite outgrowth, cell polarization and directed migration (12), all cellular phenotypes previously reported to be impaired by pathogenic LRRK2 (13–18). The LRRK2-mediated pericentrosomal/centrosomal accumulation of phospho-RAB8 further causes deficits in centrosomal cohesion in dividing cells, including peripheral LRRK2 PD patient-derived cells, in a manner dependent upon LRRK2 kinase activity (12). Surprisingly, whilst the phosphorylated RABs lose their interactions with their effector and regulatory proteins, they were reported to specifically gain the ability to interact with RILPL1 (2), a poorly characterized protein which localizes to the mother centriole and regulates ciliogenesis (19). The primary cilium emerges from the basal body, which is a special form of the mother centriole, and a recent study reports that pathogenic LRRK2 interferes with ciliogenesis in a manner dependent on both phospho-RAB10 and RILPL1 (20). Ciliogenesis deficits were observed in various cell types, including the somatosensory cortex and the small population of cholinergic neurons present in the striatum of LRRK2 transgenic mice (20). The latter neurons require cilia to sense Sonic hedgehog signalling (21,22), which provides a neuroprotective circuit to support dopaminergic neurons (23). In this manner, ciliogenesis deficits may contribute to PD pathogenesis in a non-cell-autonomous manner. However, it remains unknown whether the reported LRRK2-mediated centrosomal and ciliogenesis deficits are distinct cellular reflections of the same phospho-RAB8/10-RILPL1 pathway.
Here, we show that centrosomal cohesion deficits are mediated by both RAB8A/B and RAB10 and correlate with the centrosomal accumulation of both phospho-RAB proteins. The pathogenic LRRK2-mediated centrosomal cohesion alterations are abolished in RAB8A, RAB10 or RILPL1 knockout cells. In addition, the pathogenic LRRK2-mediated ciliogenesis deficits also correlate with the centrosomal accumulation of phospho-RAB8/10. Both centrosome-related deficits can be observed in LRRK2 PD patient-derived peripheral cells as well as in primary astrocytes from pathogenic LRRK2 knockin mice. These data suggest that these LRRK2-mediated deficits are distinct cellular readouts of the same underlying RAB8/RAB10/RILPL1 nexus and highlight the possibility that either centrosomal cohesion and/or ciliogenesis alterations may serve as cellular biomarkers for LRRK2-related PD.
Results
Both RAB8A/B and RAB10 cause centrosomal cohesion deficits when co-expressed with wild-type LRRK2
We first wondered whether the centrosomal cohesion deficits induced upon co-expression of wild-type LRRK2 with RAB8A (12) are dependent on the RAB8A prenylation status, since prenylation is known to increase the affinity of RAB proteins for membranes and is required for their function (24). We mutated the residue required for prenylation and confirmed that the RAB8A-C204A mutant was diffusely distributed as in contrast to wild-type RAB8A when expressed in HeLa cells, consistent with the intended effect of the mutation in blocking prenylation (Fig. S1A,B). Whilst expression of either RAB8A or LRRK2 on their own was without effect, co-expression of both caused centrosomal cohesion deficits, which was not observed when co-expressing LRRK2 with RAB8A-C204A (Fig. 1A), even though both wild-type and prenylation-deficient RAB8A mutants were expressed to similar degrees (Fig. 1B). Interestingly, and as previously described for RAB10 (25), only wild-type but not RAB8A-C204A was phosphorylated by LRRK2 (Fig. 1B), indicating that RAB8A needs to be prenylated to be phosphorylated, so as to then cause the resultant LRRK2-mediated effects on centrosome cohesion.
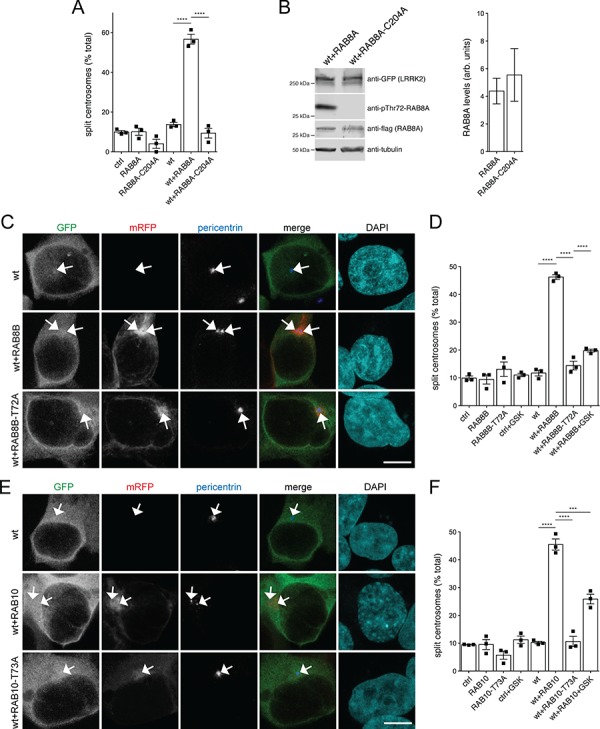
Expression of prenylation-competent RAB8A, or of RAB8B or RAB10 with LRRK2, causes centrosomal cohesion deficits in a manner dependent on LRRK2 kinase activity. (A) Quantification of the percentage of cells with split centrosomes (duplicated centrosomes with a distance between their centres > 1.5 μm) in HEK293T cells transfected with the different constructs as indicated. Bars represent mean ± s.e.m. (n
1.5 μm) in HEK293T cells transfected with the different constructs as indicated. Bars represent mean ± s.e.m. (n =
= 3 experiments); ****, P
3 experiments); ****, P <
< 0.001. (B) Left: HEK293T cells were co-transfected with either wild-type LRRK2 and RAB8A, or RAB8A-C204A as indicated, and extracts blotted for LRRK2 levels, RAB8A levels, phospho-RAB8A levels and tubulin as loading control. Right: no significant differences in the levels of RAB8A or RAB8A-C204A expression. Blots of the type indicated on the left were quantified for RAB8A levels and normalized to tubulin. Bars represent mean ± s.e.m. (n
0.001. (B) Left: HEK293T cells were co-transfected with either wild-type LRRK2 and RAB8A, or RAB8A-C204A as indicated, and extracts blotted for LRRK2 levels, RAB8A levels, phospho-RAB8A levels and tubulin as loading control. Right: no significant differences in the levels of RAB8A or RAB8A-C204A expression. Blots of the type indicated on the left were quantified for RAB8A levels and normalized to tubulin. Bars represent mean ± s.e.m. (n =
= 6 experiments). (C) Example of HEK293T cells transfected with either GFP-tagged wild-type LRRK2 (wt), or wild-type LRRK2 along with either mRFP-tagged RAB8B or phosphorylation-deficient RAB8B-T72A mutant as indicated and stained for pericentrin antibody and DAPI. Scale bar, 5 μm. (D) Quantification of the split centrosome phenotype in cells expressing the indicated constructs, in either the absence or presence of the LRRK2 kinase inhibitor GSK2578215A (500 nm, 2 h) before fixation. Bars represent mean ± s.e.m. (n
6 experiments). (C) Example of HEK293T cells transfected with either GFP-tagged wild-type LRRK2 (wt), or wild-type LRRK2 along with either mRFP-tagged RAB8B or phosphorylation-deficient RAB8B-T72A mutant as indicated and stained for pericentrin antibody and DAPI. Scale bar, 5 μm. (D) Quantification of the split centrosome phenotype in cells expressing the indicated constructs, in either the absence or presence of the LRRK2 kinase inhibitor GSK2578215A (500 nm, 2 h) before fixation. Bars represent mean ± s.e.m. (n =
= 3 experiments); ****, P
3 experiments); ****, P <
< 0.001. (E) Example of HEK293T cells transfected with either GFP-tagged wild-type LRRK2 (wt), or wild-type LRRK2 along with either mRFP-tagged RAB10 or phosphorylation-deficient RAB10-T73A mutant as indicated and stained for pericentrin antibody and DAPI. Scale bar, 5 μm. (F) Quantification of the centrosomal cohesion phenotype in cells expressing the indicated constructs, in either the absence or presence of the LRRK2 kinase inhibitor GSK2578215A (500 nm, 2 h) before fixation. Bars represent mean ± s.e.m. (n
0.001. (E) Example of HEK293T cells transfected with either GFP-tagged wild-type LRRK2 (wt), or wild-type LRRK2 along with either mRFP-tagged RAB10 or phosphorylation-deficient RAB10-T73A mutant as indicated and stained for pericentrin antibody and DAPI. Scale bar, 5 μm. (F) Quantification of the centrosomal cohesion phenotype in cells expressing the indicated constructs, in either the absence or presence of the LRRK2 kinase inhibitor GSK2578215A (500 nm, 2 h) before fixation. Bars represent mean ± s.e.m. (n =
= 3 experiments); ****, P
3 experiments); ****, P <
< 0.001; ***, P
0.001; ***, P <
< 0.005.
0.005.
LRRK2 phosphorylates both the A and B isoforms of RAB8 (2,3). These isoforms are encoded by different genes, are highly similar in sequence, but display differential tissue-specific expression patterns (26), and their functions are not redundant in all cases (27,28). We therefore next analyzed whether RAB8B may play similar roles in modulating the LRRK2-mediated centrosomal cohesion deficits. Indeed, when co-expressed with wild-type LRRK2, RAB8B caused cohesion deficits identical to those observed for RAB8A, which were not observed with a phosphorylation-deficient RAB8B-T72A mutant (Fig. 1C and D). Both wild-type and mutant RAB8B were expressed to similar degrees and localized to a perinuclear tubular-like compartment like RAB8A, but additional cytosolic localization was observed with the phosphorylation-deficient RAB8B mutant (Fig. S1A,B). As an additional means to assure that the effects were mediated by the LRRK2 kinase activity, we employed the LRRK2 kinase inhibitor GSK2578215A, which largely reverted the centrosomal cohesion deficits in the presence of LRRK2 and RAB8B (Fig. 1D). Thus, both RAB8A and RAB8B cause centrosomal cohesion deficits in a manner dependent on the LRRK2 kinase activity.
RAB10 has been identified as another prominent LRRK2 kinase substrate (2,3), and both RAB8 and RAB10 have been reported to be involved in centrosome-related events (27). These findings prompted us to analyze for the effects of RAB10 expression on centrosomal alterations in the presence of wild-type LRRK2. Co-expression of wild-type LRRK2 with RAB10, but not a phosphorylation-deficient RAB10-T73A mutant, caused centrosomal cohesion deficits identical to those observed with RAB8A or RAB8B, respectively (Fig. 1E and F). Both wild-type and mutant RAB10 were expressed to similar degrees and localized to a perinuclear tubular-like compartment identical to RAB8A, with additional cytosolic localization observed with the phosphorylation-deficient RAB10 mutant (Fig. S1A–C). Application of the LRRK2 kinase inhibitor GSK2578215A largely reverted the centrosomal cohesion deficits observed in the presence of LRRK2 and RAB10 (Fig. 1F). Therefore, co-expression of wild-type LRRK2 with either RAB8A, RAB8B or RAB10 causes centrosomal cohesion deficits in a manner dependent on the LRRK2 kinase activity.
Pathogenic LRRK2-induced centrosomal cohesion deficits correlate with aberrant pericentrosomal/centrosomal accumulation of phospho-RAB8 and phospho-RAB10
We previously showed that the pathogenic LRRK2-induced centrosomal cohesion deficits correlate with an aberrant centrosomal/pericentrosomal accumulation of endogenous phosphorylated RAB8 (12). Given that increasing RAB10 levels also caused centrosomal cohesion deficits in the presence of wild-type LRRK2, we wondered whether pathogenic LRRK2 may also trigger the pericentrosomal/centrosomal accumulation of endogenous phosphorylated RAB10.
We first analyzed for the specificity of distinct RAB8, RAB10, phospho-RAB8 and phospho-RAB10 antibodies by western blotting and immunocytochemistry techniques. As assessed by western blotting, both knockout-validated sheep and mouse total anti-RAB10 antibodies were specific for RAB10, and both knockout-validated sheep and mouse total anti-RAB8 antibodies were specific for RAB8, respectively (Fig. S2) (12). By immunocytochemistry, the sheep knockout-validated total anti-RAB10 and anti-RAB8 antibodies were also specific for the respective RAB proteins (Fig. S3A,B). In contrast, the sheep phospho-RAB10 antibody detected accumulation of phosphorylated RAB species in cells expressing wild-type LRRK2 with either RAB10 or with RAB8, indicating that it was detecting both phosphorylated RAB species, and similar results were obtained for the sheep phospho-RAB8 antibody (Fig. S3C,D). Since the high-affinity knockout-validated commercial rabbit monoclonal anti-phospho-RAB10 and anti-phospho-RAB8 antibodies (6) were not suitable for immunocytochemistry purposes under the conditions employed (Fig. S3E,F), we used the sheep phospho-RAB10 antibody, which detects both phosphorylated RAB10/8 (Fig. S3D), as well as the knockout-validated mouse monoclonal anti-RAB10 antibody highly specific for total RAB10 (Fig. S3G), for further cell biological analysis.
Staining of HEK293T cells expressing pathogenic Y1699C LRRK2 revealed pericentrosomal/centrosomal phospho-RAB10/8 accumulation (Fig. 2A and B). The signal was specific for detecting the phosphorylated RAB species, as not observed upon preincubation of the antibody with phospho-peptide or upon treatment of cells with MLi2, another distinct and highly specific LRRK2 kinase inhibitor (Fig. 2A and B). We further used a knockout-validated total anti-RAB10 antibody highly specific for RAB10 (Fig. S3G). Endogenous RAB10 was localized to a pericentrosomal/centrosomal area in cells expressing either pathogenic Y1699C, R1441C or G2019S mutant LRRK2 (Fig. 2C and D). Such localization was reverted upon application of MLi2 and was not observed in non-transfected, wild-type LRRK2-transfected or kinase-inactive G2019S-K1906M LRRK2-transfected cells, respectively, indicating that it reflects a phosphorylated species of RAB10 (Fig. 2C and D).
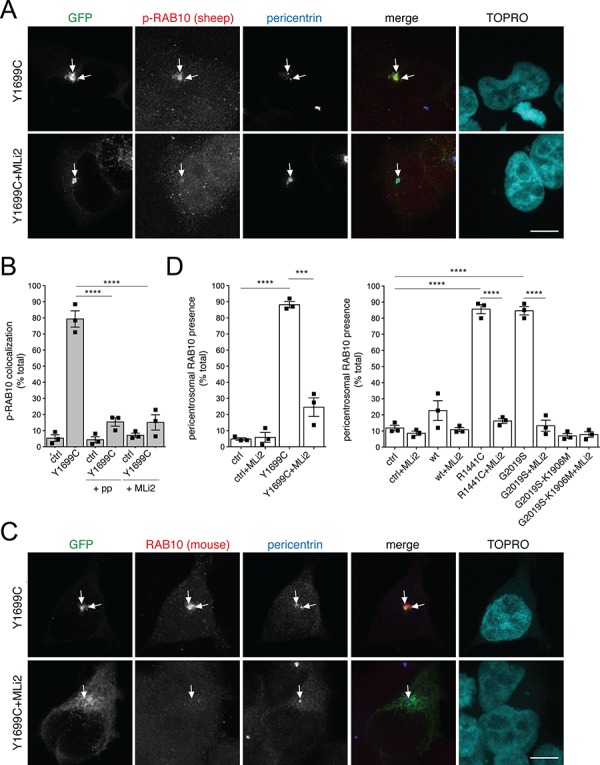
Pathogenic LRRK2 causes kinase-dependent pericentrosomal/centrosomal accumulation of endogenous phospho-RAB10/8. (A) Example of HEK293T cells transfected with Y1699C mutant GFP-tagged LRRK2, in either the absence or presence of 100 nm of the LRRK2 kinase inhibitor MLi2 for 60 min prior to immunocytochemistry as indicated. Cells were stained using a sheep anti-T73-phospho-RAB10 antibody preabsorbed with dephospho-peptide, an anti-pericentrin antibody and TOPRO. Scale bar, 5 μm. (B) Quantification of the percentage of cells displaying pericentrosomal/centrosomal phospho-RAB10/8 staining colocalizing with centrosomes within a 3 μm diameter circle in either non-transfected cells (ctrl), or in pathogenic LRRK2-transfected cells as indicated, in either the absence or presence of antibody preabsorption with dephospho-peptide or phospho-peptide (+pp), or pretreatment of cells with MLi2 as described above. Around 100–150 cells were quantified per condition per experiment. Bars represent mean ± s.e.m. (n =
= 3 independent experiments); ****, P
3 independent experiments); ****, P <
< 0.001. (C) Cells were transfected with pathogenic GFP-tagged LRRK2 and subjected to immunocytochemistry using a knockout-validated anti-RAB10 antibody, pericentrin and TOPRO upon incubation of cells with 100 nm MLi2 for 60 min prior to immunocytochemistry as indicated. Scale bar, 5 μm. (D) Quantification of the percentage of cells displaying pericentrosomal RAB10 accumulation in either non-transfected cells (ctrl), wild-type, pathogenic LRRK2, or kinase-inactive pathogenic LRRK2 (G2019S-K1906M)-transfected cells as indicated, either in the absence or presence of MLi2 (100 nm, 1 h). Bars represent mean ± s.e.m. (n
0.001. (C) Cells were transfected with pathogenic GFP-tagged LRRK2 and subjected to immunocytochemistry using a knockout-validated anti-RAB10 antibody, pericentrin and TOPRO upon incubation of cells with 100 nm MLi2 for 60 min prior to immunocytochemistry as indicated. Scale bar, 5 μm. (D) Quantification of the percentage of cells displaying pericentrosomal RAB10 accumulation in either non-transfected cells (ctrl), wild-type, pathogenic LRRK2, or kinase-inactive pathogenic LRRK2 (G2019S-K1906M)-transfected cells as indicated, either in the absence or presence of MLi2 (100 nm, 1 h). Bars represent mean ± s.e.m. (n =
= 3 independent experiments); ****, P
3 independent experiments); ****, P <
< 0.001; ***, P
0.001; ***, P <
< 0.005.
0.005.
The accumulation of phospho-RAB10 was next evaluated in another cell system. The phospho-RAB10/8 antibodies were not of sufficient affinity to detect the centrosomal accumulation of endogenous phospho-RABs in G2019S LRRK2 mutant SH-SY5Y cells but did detect centrosomal accumulation of phospho-RAB10/8 when expressing wild-type RAB10 in either wild-type or G2019S LRRK2-mutant SH-SY5Y cells to increase the phosphorylated RAB10 species (Fig. S4A,B). The increase in the phospho-RAB10 signal disappeared when pretreating cells with MLi2 and was not detected when expressing the non-phosphorylatable RAB10-T73A mutant (Fig. S4A,B), indicating that it was specifically detecting a LRRK2-phosphorylated version of RAB10. Expression of wild-type RAB10, but not phosphorylation-deficient RAB10 mutant, in wild-type or G2019S LRRK2-mutant SH-SY5Y cells caused a centrosomal cohesion deficit which was reversed by MLi2 (Fig. S4C,D). Therefore, as analyzed in two distinct cell systems, increasing the pericentrosomal/centrosomal amount of phosphorylated RAB10 also correlates with the observed centrosomal cohesion deficits.
Pathogenic LRRK2-mediated centrosomal cohesion deficits depend on RAB8A, RAB10 and RILPL1
To further analyze how pathogenic LRRK2 causes centrosomal cohesion deficits, we expressed wild-type or distinct LRRK2 mutants in either wild-type A549 cells, or in cells deficient in RAB8A, RAB10 or RILPL1. In wild-type cells, expression of pathogenic G2019S, R1441C or Y1699C LRRK2 mutant caused a centrosomal cohesion phenotype which was abolished upon treatment with the LRRK2 kinase inhibitor MLi2 and was not observed when expressing wild-type or kinase-dead K1906M LRRK2 (Fig. 3A and B), with all constructs expressed to similar degrees (Fig. 3C and D). We next attempted to correlate the LRRK2-mediated centrosomal cohesion phenotype with alterations in the levels of phospho-RAB8 or phospho-RAB10 as assessed by western blotting techniques. In non-transfected A549 RAB8A knockout cells, the total anti-RAB8 antibody, previously reported to be specific for RAB8(A+B) as assessed by western blotting of RAB8A/RAB8B double-knockout mouse extracts (27), revealed the remaining presence of RAB8B. The rabbit monoclonal phospho-RAB8A antibody revealed the presence of a phospho-RAB band in the A549 RAB8A knockout cells, likely based on the reported cross-reactivity of this antibody with other LRRK2-phosphorylated RAB substrates including RAB8B and/or RAB10 (6) (Fig. 3C). In contrast, the mouse monoclonal anti-RAB10 and the rabbit monoclonal anti-phospho-RAB10 antibodies were highly specific, as no signal was detected in the A549 RAB10 knockout cells (Fig. 3D). Importantly, the centrosomal cohesion deficits mediated by pathogenic LRRK2 correlated with increases in phospho-RAB8/phospho-RAB10 levels in a manner modulated by the LRRK2 kinase inhibitor MLi2 (Fig. 3C and D).
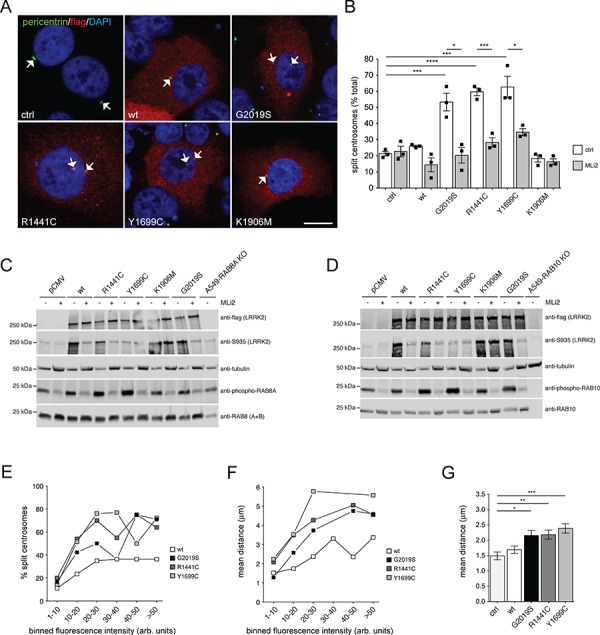
Pathogenic LRRK2 causes kinase-dependent and dose-dependent centrosomal cohesion deficits. (A) Example of wild-type A549 cells transfected with either pCMV (ctrl), or with the indicated flag-tagged LRRK2 constructs and stained with an antibody against flag, pericentrin and DAPI. Scale bar, 10 μm. (B) Quantification of the percentage of wild-type A549 cells with split centrosomes (duplicated centrosomes with a distance between their centres > 2.5 μm) transfected with the different LRRK2 constructs, and either left untreated or incubated with 500 nm MLi2 for 2 h before immunocytochemistry as indicated. Bars represent mean ± s.e.m. (n
2.5 μm) transfected with the different LRRK2 constructs, and either left untreated or incubated with 500 nm MLi2 for 2 h before immunocytochemistry as indicated. Bars represent mean ± s.e.m. (n =
= 3 experiments); ****, P
3 experiments); ****, P <
< 0.001; ***, P
0.001; ***, P <
< 0.005; *, P
0.005; *, P <
< 0.05. (C) Wild-type A549 cells were transfected with the indicated LRRK2 constructs, either left untreated or incubated with 500 nm MLi2 for 2 h as indicated, and extracts blotted for flag-tagged LRRK2, phosphorylated LRRK2 (phospho-S935), endogenous RAB8 (A+B), phospho-T72-RAB8A (rabbit monoclonal antibody) and tubulin as loading control. (D) Same as in (C), but extracts blotted for flag-tagged LRRK2, phosphorylated LRRK2 (phospho-S935), endogenous RAB10, phospho-T73-RAB10 (rabbit monoclonal antibody) and tubulin as loading control. (E) Wild-type A549 cells were transfected with the indicated flag-tagged constructs, and the percentage of cells with split centrosomes, along with the flag staining intensity of each cell were quantified from around 100 cells with duplicated centrosomes per condition, with fluorescence intensities grouped into distinct bins as indicated. (F) Same as in (E), but depicting the mean distance of duplicated centrosomes against the distinct binned fluorescence intensities. (G) Same as in (F), but depicting the mean distance of duplicated centrosomes in cells transfected with the indicated constructs irrespective of flag staining intensity. ***, P
0.05. (C) Wild-type A549 cells were transfected with the indicated LRRK2 constructs, either left untreated or incubated with 500 nm MLi2 for 2 h as indicated, and extracts blotted for flag-tagged LRRK2, phosphorylated LRRK2 (phospho-S935), endogenous RAB8 (A+B), phospho-T72-RAB8A (rabbit monoclonal antibody) and tubulin as loading control. (D) Same as in (C), but extracts blotted for flag-tagged LRRK2, phosphorylated LRRK2 (phospho-S935), endogenous RAB10, phospho-T73-RAB10 (rabbit monoclonal antibody) and tubulin as loading control. (E) Wild-type A549 cells were transfected with the indicated flag-tagged constructs, and the percentage of cells with split centrosomes, along with the flag staining intensity of each cell were quantified from around 100 cells with duplicated centrosomes per condition, with fluorescence intensities grouped into distinct bins as indicated. (F) Same as in (E), but depicting the mean distance of duplicated centrosomes against the distinct binned fluorescence intensities. (G) Same as in (F), but depicting the mean distance of duplicated centrosomes in cells transfected with the indicated constructs irrespective of flag staining intensity. ***, P <
< 0.005; **, P
0.005; **, P <
< 0.01; *, P
0.01; *, P <
< 0.05.
0.05.
When analyzing the relationship between centrosomal cohesion deficits and LRRK2 expression levels as measured by quantitative light microscopy, the percentage of split centrosomes and the overall mean distance between duplicated centrosomes increased with increasing LRRK2 expression levels until reaching a plateau, suggesting that pathogenic LRRK2 expression causes a concentration-dependent centrosomal cohesion deficit which becomes saturated at higher expression levels (Fig. 3E and F). Measuring the distance between duplicated centrosomes in all cells irrespective of protein expression levels also revealed a significant increase in the mean distance between duplicated centrosomes in cells expressing pathogenic LRRK2 as in contrast to control cells, with the mean centrosomal distance in control cells similar to what has been previously described in this cell type (20) (Fig. 3G).
A recent report suggested that RILPL1 expression in A549 cells causes a cohesion deficit in a manner independent on expression levels (20). When expressing either C-terminally flag-tagged, or C-terminally or N-terminally eGFP-tagged human RILPL1, no differences in centrosomal cohesion were observed when quantifying the percentage of split centrosomes or the mean distance between duplicated centrosomes, including in cells expressing very high levels of RILPL1 (Fig. 4A, B and C). Thus, our data indicate that pathogenic LRRK2 causes centrosomal cohesion deficits in a concentration-dependent manner, which is not observed upon human RILPL1 expression.
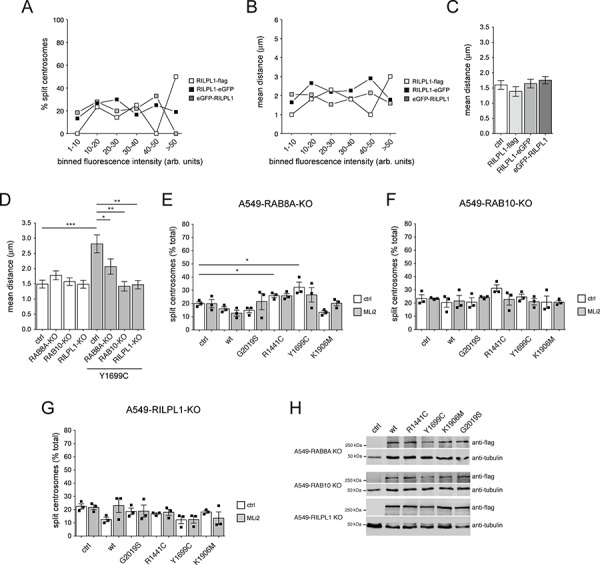
Pathogenic LRRK2-mediated centrosomal cohesion deficits depend on RAB8A, RAB10 and RILPL1. (A) Wild-type A549 cells were transfected with either C-terminally flag-tagged or eGFP-tagged human RILPL1, or N-terminally eGFP-tagged human RILPL1, and the percentage of cells with split centrosomes, along with the flag staining intensity (or eGFP fluorescence intensity) of each cell quantified from around 100 cells with duplicated centrosomes per condition, with fluorescence intensities grouped into distinct bins as indicated. (B) Same as in (A), but depicting the mean distance of duplicated centrosomes against the distinct binned fluorescence intensities. (C) Same as in (B), but depicting the mean distance of duplicated centrosomes in cells transfected with the indicated constructs irrespective of flag staining/eGFP fluorescence intensity. (D) Quantification of the mean distance of duplicated centrosomes from either wild-type A549 cells (ctrl), or A549 cells in which the different proteins were knocked out using CRISPR-Cas9 (RAB8A-KO, RAB10-KO, RILPL1-KO), in the absence or presence of Y1699C pathogenic LRRK2 expression as indicated. Between 50 and 100 cells with duplicated centrosomes per condition were analyzed. ***, P <
< 0.005; *, P
0.005; *, P <
< 0.05. (E) Quantification of the percentage of RAB8A A549 knockout cells transfected with the different LRRK2 constructs displaying duplicated split centrosomes, in either the presence or absence of MLi2 (500 nm, 2 h) as indicated. Bars represent mean ± s.e.m. (n
0.05. (E) Quantification of the percentage of RAB8A A549 knockout cells transfected with the different LRRK2 constructs displaying duplicated split centrosomes, in either the presence or absence of MLi2 (500 nm, 2 h) as indicated. Bars represent mean ± s.e.m. (n =
= 3 experiments); *, P
3 experiments); *, P <
< 0.05. (F) Quantification of the percentage of RAB10 A549 knockout cells transfected with the different LRRK2 constructs displaying duplicated split centrosomes, in either the presence or absence of MLI2 (500 nm, 2 h). Bars represent mean ± s.e.m. (n
0.05. (F) Quantification of the percentage of RAB10 A549 knockout cells transfected with the different LRRK2 constructs displaying duplicated split centrosomes, in either the presence or absence of MLI2 (500 nm, 2 h). Bars represent mean ± s.e.m. (n =
= 3 experiments). (G) Quantification of the percentage of RILPL1 A549 knockout cells transfected with the different LRRK2 constructs displaying duplicated split centrosomes, in either the presence or absence of MLI2 (500 nm, 2 h). Bars represent mean ± s.e.m. (n
3 experiments). (G) Quantification of the percentage of RILPL1 A549 knockout cells transfected with the different LRRK2 constructs displaying duplicated split centrosomes, in either the presence or absence of MLI2 (500 nm, 2 h). Bars represent mean ± s.e.m. (n =
= 3 experiments). (H) The distinct A549 knockout cells were transfected with the indicated flag-tagged LRRK2 constructs, and extracts blotted for flag and tubulin as loading control.
3 experiments). (H) The distinct A549 knockout cells were transfected with the indicated flag-tagged LRRK2 constructs, and extracts blotted for flag and tubulin as loading control.
To address whether the LRRK2-mediated centrosomal cohesion deficits are dependent on the presence of RAB8A, RAB10 or RILPL1, we employed A549 cells where the proteins were knocked out using CRISPR-Cas9 (20). Non-transfected A549 cells lacking RAB8A, RAB10 or RILPL1 did not display centrosomal cohesion deficits (Fig. 4D), with the lack of the respective proteins confirmed by western blotting techniques (Fig. S5). In addition, no alterations were found in the levels of RAB8B in either the RAB8A or RAB10 knockout cells, in the levels of RAB8A in the RAB10 or RILPL1 knockout cells, or in the levels of RAB10 in the RAB8A or RILPL1 knockout cells, respectively (Fig. S5). Importantly though, and as in contrast to A549 wild-type cells, the pathogenic LRRK2-mediated centrosomal cohesion deficits were drastically reduced in cells deficient for RAB8A (Fig. 4D and E) and abolished in cells deficient for RAB10 or RILPL1 (Fig. 4D, F and G), respectively, even though the various LRRK2 variants were expressed to similar degrees (Fig. 4H). Therefore, the LRRK2-mediated centrosomal cohesion deficit is crucially dependent on RAB8A, RAB10 and RILPL1.
Pathogenic LRRK2 causes ciliogenesis deficits associated with the pericentrosomal/centrosomal accumulation of phospho-RAB10/8
We next aimed to determine whether the pericentrosomal/centrosomal accumulation of phospho-RAB10/8, shown to interfere with centrosomal cohesion in dividing cells, may also be responsible for the reported LRRK2-mediated ciliogenesis deficits (20). HEK293T cells were transfected with either wild-type, kinase-inactive or pathogenic mutant LRRK2 constructs, and the percentage of ciliated cells quantified in both serum-fed as well as serum-starved conditions (Fig. 5A, B and C). Serum starvation did not increase the amount of ciliated cells (Fig. 5B and C), likely based on the limited time of starvation possible without compromising viability in this cell type. Expression of all pathogenic LRRK2 mutants, but not of wild-type or kinase-dead LRRK2, caused a decrease in the percentage of ciliated cells in both serum-fed and serum-starved conditions (Fig. 5B and C). This was associated with a significant increase in the accumulation of phospho-RAB10/8 in transfected cells (Fig. 5D and E), with all constructs expressed to similar degrees (Fig. 5F). The phospho-RAB accumulation was reverted upon MLi2 treatment (Fig. 5D and E), with only a slight rescue of the ciliogenesis phenotype observed (Fig. 5B and C), suggesting that the reformation of cilia upon phospho-RAB10/8 removal may take additional time.
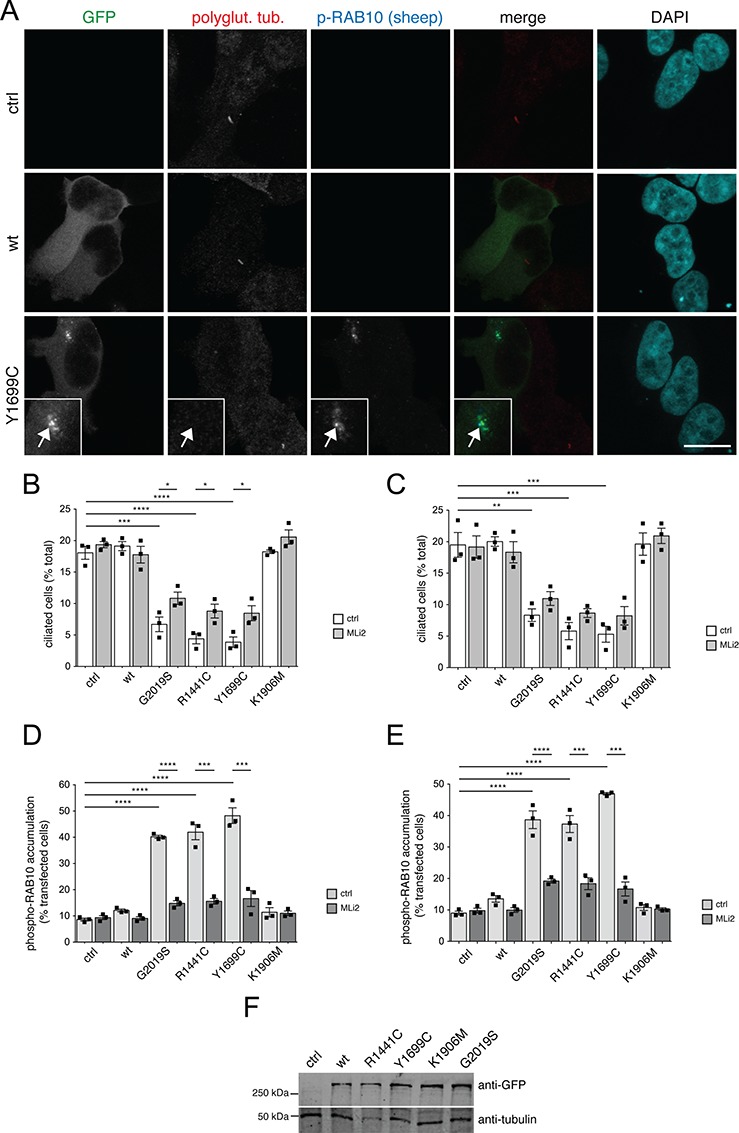
Pathogenic LRRK2 causes ciliogenesis deficits in a kinase-dependent manner. (A) Example of HEK293T cells transfected with either pCMV (ctrl), or with GFP-tagged wild-type or pathogenic Y1699C constructs, and stained with an antibody against polyglutamylated tubulin, sheep anti-phospho-T73-RAB10 and DAPI. Scale bar, 10 μm. (B) HEK293T cells were transfected with the indicated LRRK2 constructs, and either left untreated or treated with MLi2 (200 nm, 12 h) before immunocytochemistry using an antibody against polyglutamylated tubulin and against phospho-T73-RAB10 as described in (A). The percentage of ciliated cells expressing the distinct constructs was quantified from around 200 cells per condition per experiment. Bars represent mean ± s.e.m. (n =
= 3 experiments); ****, P
3 experiments); ****, P <
< 0.001; ***, P
0.001; ***, P <
< 0.005; *, P
0.005; *, P <
< 0.05. (C) Same as in (B), but cells were serum-starved for 12 h, either in the absence or presence of 200 nm MLi2 as indicated, and the percentage of ciliated transfected cells quantified from around 200 cells per condition per experiment. Bars represent mean ± s.e.m. (n
0.05. (C) Same as in (B), but cells were serum-starved for 12 h, either in the absence or presence of 200 nm MLi2 as indicated, and the percentage of ciliated transfected cells quantified from around 200 cells per condition per experiment. Bars represent mean ± s.e.m. (n =
= 3 experiments); ***, P
3 experiments); ***, P <
< 0.005; **, P
0.005; **, P <
< 0.01. (D) Same as (B), with cells either left untreated or treated with MLi2 (200 nm, 12 h) before immunocytochemistry, and the perinuclear accumulation of phospho-T73-RAB10 quantified from around 200 transfected cells per condition and experiment. Bars represent mean ± s.e.m. (n
0.01. (D) Same as (B), with cells either left untreated or treated with MLi2 (200 nm, 12 h) before immunocytochemistry, and the perinuclear accumulation of phospho-T73-RAB10 quantified from around 200 transfected cells per condition and experiment. Bars represent mean ± s.e.m. (n =
= 3 experiments); ****, P
3 experiments); ****, P <
< 0.001; ***, P
0.001; ***, P <
< 0.005. (E) Same as (D), with cells serum-starved for 12 h either in the absence or presence of 200 nm MLi2 as indicated before immunocytochemistry, and the perinuclear accumulation of phospho-T73-RAB10 quantified from around 200 transfected cells per condition and experiment. Bars represent mean ± s.e.m. (n
0.005. (E) Same as (D), with cells serum-starved for 12 h either in the absence or presence of 200 nm MLi2 as indicated before immunocytochemistry, and the perinuclear accumulation of phospho-T73-RAB10 quantified from around 200 transfected cells per condition and experiment. Bars represent mean ± s.e.m. (n =
= 3 experiments); ****, P
3 experiments); ****, P <
< 0.001; ***, P
0.001; ***, P <
< 0.005. (F) HEK293T cells were transfected with the indicated GFP-tagged LRRK2 constructs, extracts analyzed by western blotting for GFP and tubulin as loading control.
0.005. (F) HEK293T cells were transfected with the indicated GFP-tagged LRRK2 constructs, extracts analyzed by western blotting for GFP and tubulin as loading control.
To assure that the observed phospho-RAB accumulation occurred at the ciliary base, cells were co-transfected with pathogenic LRRK2 and Smoothened-EGFP (smo-EGFP) to label cilia (Fig. 6A and B). When selectively quantifying the pathogenic LRRK2-expressing cells still displaying cilia, phospho-RAB10/8 accumulation at/around the ciliary base could be detected, but quantification of the fluorescence intensity of the phospho-RAB10/8 signal indicated that there was significantly less overall accumulation in ciliated as in contrast to non-ciliated cells, or as in contrast to only smo-EGFP-transfected ciliated or non-ciliated cells, respectively (Fig. 6B). Finally, when staining cells for both ciliary and centrosomal markers, a significant percentage of the pathogenic LRRK2-expressing cells which still contained cilia was found to display a duplicated split centrosome phenotype (Fig. 6C and D). Since cilia are usually reabsorbed in G2 phase of the cell cycle (29), these data suggest that pathogenic LRRK2 may additionally interfere with proper ciliary resorption.
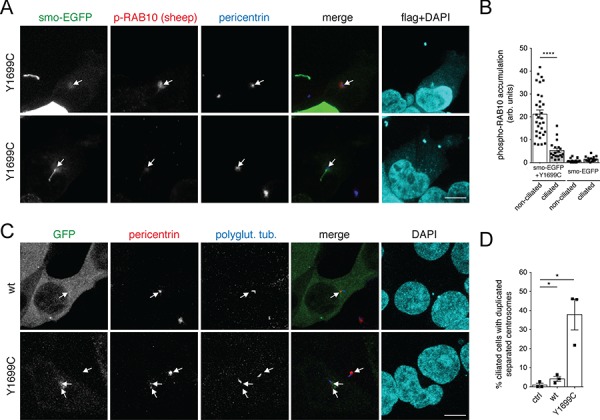
Pathogenic LRRK2 causes phospho-RAB accumulation preferentially in non-ciliated cells and may cause ciliary resorption deficits. (A) HEK293T cells were co-transfected with pathogenic flag-tagged LRRK2 and smo-EGFP and stained for phospho-T73-RAB10 (secondary Alexa594 antibody, red), pericentrin (secondary Alexa647 antibody, pseudo-coloured in blue), anti-flag (secondary Alexa405 antibody, pseudo-coloured in cyan) and DAPI (pseudo-coloured in cyan). Example of either non-ciliated (top) or ciliated (bottom) cell co-expressing smo-EGFP and flag-tagged Y1699C LRRK2. Scale bar, 5 μm. (B) Quantification of the fluorescence intensity of the phospho-T73-RAB10 accumulation at the ciliary base in cells only expressing smo-EGFP, or co-expressing pathogenic Y1699C LRRK2, according to the absence or presence of cilia from experiments of the type described in (A). Bars represent mean ± s.e.m. from around 20–30 cells analyzed for each condition; ****, P <
< 0.001. (C) Example of HEK293T cells transfected with GFP-tagged wild-type or Y1699C mutant LRRK2 as indicated and stained for pericentrin, polyglutamylated tubulin and DAPI. Scale bar, 5 μm. (D) Quantification of the percentage of ciliated cells in either the absence of transfection (ctrl), or upon expression of wild-type or Y1699C mutant LRRK2 as indicated, which display a duplicated split centrosome phenotype. The centrosomal phenotype was quantified from around 50 ciliated cells per condition per experiment. Bars represent mean ± s.e.m. (n
0.001. (C) Example of HEK293T cells transfected with GFP-tagged wild-type or Y1699C mutant LRRK2 as indicated and stained for pericentrin, polyglutamylated tubulin and DAPI. Scale bar, 5 μm. (D) Quantification of the percentage of ciliated cells in either the absence of transfection (ctrl), or upon expression of wild-type or Y1699C mutant LRRK2 as indicated, which display a duplicated split centrosome phenotype. The centrosomal phenotype was quantified from around 50 ciliated cells per condition per experiment. Bars represent mean ± s.e.m. (n =
= 3 independent experiments); *, P
3 independent experiments); *, P <
< 0.05.
0.05.
As another means to determine the effect of phospho-RAB accumulation on ciliogenesis, we co-expressed wild-type LRRK2 with either RAB8A or RAB10, respectively (Fig. S6). Whilst expression of either wild-type RAB8A or wild-type RAB10 was without effect on ciliogenesis, co-expression with LRRK2 caused a deficit in ciliogenesis (Fig. S6A–C) associated with the detection of phospho-RAB8/10 (Fig. S6D), which is analogous to what we observed regarding centrosomal cohesion deficits (Figs 1 and and2).2). Altogether, these data indicate that pathogenic LRRK2 interferes with both ciliogenesis and centrosome cohesion events via the increased pericentrosomal/centrosomal accumulation of phospho-RAB10/8.
Ciliogenesis and centrosomal cohesion deficits in human dermal fibroblasts from G2019S LRRK2 patients and murine G2019S knockin astrocytes
Our previous studies reported centrosomal cohesion deficits in human dermal fibroblasts from G2019S LRRK2 patients as in contrast to healthy controls, which were reverted upon addition of LRRK2 kinase inhibitor (12). We thus wondered whether these patient-derived cells also display a ciliogenesis phenotype. Under serum-fed conditions, G2019S LRRK2-PD fibroblasts displayed a ciliogenesis deficit as in contrast to healthy control fibroblasts (Fig. 7A and B). In this cell type, starvation prominently induced ciliogenesis (Fig. 7B), and the deficit in ciliogenesis in the pathogenic LRRK2-expressing cells as in contrast to control cells was also observed under starvation conditions (Fig. 7B). Application of LRRK2 kinase inhibitor MLi2 had a minor effect in rescuing such deficit, even though some fibroblast cells responded better than others. Further, quantification of cilia length did not show drastic differences between control and G2019S LRRK2-PD fibroblasts (Fig. 7C and D), in agreement with what has been previously described for cilia in iPSCs from LRRK2-PD patients, or in the cortex of pathogenic LRRK2-expressing mice (20).
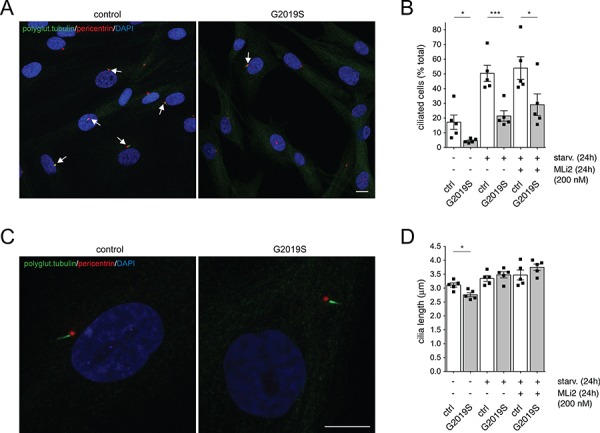
Ciliogenesis deficits in human dermal fibroblasts from G2019S mutant LRRK2 PD patients in contrast to healthy controls. (A) Example of control and G2019S mutant LRRK2 PD patient fibroblasts stained with polyglutamylated tubulin antibody, pericentrin antibody and DAPI. Scale bar, 10 μm. (B) The percentage of ciliated cells was quantified from around 100 cells per line, and from 5 control and 5 G2019S mutant LRRK2 fibroblast lines. Control or G2019S mutant LRRK2 fibroblasts were treated with or without MLi2 (200 nm, 24 h), either in the absence or presence of serum starvation (starv., 48 h) as indicated. Bars represent mean ± s.e.m. (between five independent lines); ***, P <
< 0.005; *, P
0.005; *, P <
< 0.05. (C) High-resolution deconvolved image of a control and G2019S mutant LRRK2 PD patient fibroblast cell stained with polyglutamylated tubulin antibody, pericentrin antibody and DAPI. Scale bar, 10 μm. (D) Pictures of the type acquired in (C) were analyzed for cilia length as described in Materials and Methods, with 30–60 ciliated cells analyzed for each cell line. Bars represent mean ± s.e.m. (between five independent lines); *, P
0.05. (C) High-resolution deconvolved image of a control and G2019S mutant LRRK2 PD patient fibroblast cell stained with polyglutamylated tubulin antibody, pericentrin antibody and DAPI. Scale bar, 10 μm. (D) Pictures of the type acquired in (C) were analyzed for cilia length as described in Materials and Methods, with 30–60 ciliated cells analyzed for each cell line. Bars represent mean ± s.e.m. (between five independent lines); *, P <
< 0.05.
0.05.
Finally, we analyzed for both centrosomal cohesion and ciliogenesis deficits in primary mouse astrocytes cultured from either control or G2019S knockin mice (30). Cells were stained with GFAP, and centrosomal cohesion deficits quantified in both the absence and presence of starvation (Fig. 8A, B and C). G2019S knockin astrocytes displayed a centrosomal cohesion deficit as assessed by quantifying either the percentage of split centrosomes or the mean distance between duplicated centrosomes, and such cohesion deficit was reverted upon application of LRRK2 kinase inhibitor MLi2 (Fig. 8B and C). Similarly, G2019S knockin astrocytes displayed a significant deficit in ciliogenesis as in contrast to control cells under serum-fed conditions, which was reverted by MLi2 (Fig. 8D and E). Thus, pathogenic LRRK2 causes both centrosomal cohesion and ciliogenesis deficits in a disease-relevant cell type (31,32), and in a manner mediated by the LRRK2 kinase activity.
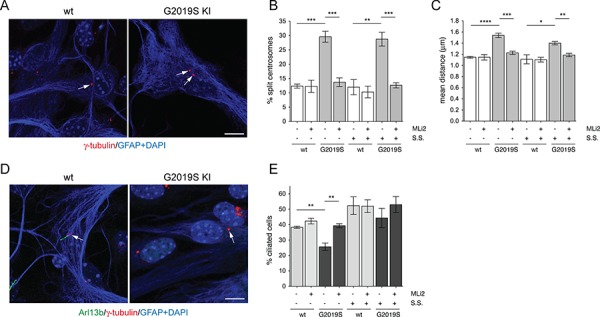
Primary astrocytes from G2019S knockin mice display both centrosomal cohesion and ciliogenesis deficits as in contrast to control astrocytes. (A) Example of wild-type and G2019S knockin primary astrocytes stained with anti-gamma-tubulin, anti-GFAP and DAPI. Scale bar, 10 μm. (B) Quantification of the percentage of wild-type or G2019S knockin primary astrocytes with split centrosomes (duplicated centrosomes with a distance between their centres > 1.5 μm), in the presence or absence of 48 h of serum starvation (S.S.), and in the presence or absence of 200 nm MLi2 for 24 h before immunocytochemistry as indicated. Around 250–350 cells were quantified per condition per experiment. Bars represent mean ± s.e.m. (n =
= 3 independent experiments); ***, P
3 independent experiments); ***, P <
< 0.005; **, P
0.005; **, P <
< 0.01. (C) Same as in (B), but depicting the mean distance of duplicated centrosomes. Between 50 and 100 cells with duplicated centrosomes per condition per experiment were analyzed. Bars represent mean ± s.e.m. (n
0.01. (C) Same as in (B), but depicting the mean distance of duplicated centrosomes. Between 50 and 100 cells with duplicated centrosomes per condition per experiment were analyzed. Bars represent mean ± s.e.m. (n =
= 3 independent experiments); ****, P
3 independent experiments); ****, P <
< 0.001; ***, P
0.001; ***, P <
< 0.005; **, P
0.005; **, P <
< 0.01; *, P
0.01; *, P <
< 0.05. (D) Example of wild-type and G2019S knockin primary astrocytes stained with anti-Arl13b, anti-gamma-tubulin, anti-GFAP and DAPI. Scale bar, 10 μm. (E) The percentage of ciliated cells was quantified from around 200 wild-type or G2019S knockin cells, in either the absence or presence of serum starvation (48 h, S.S.), and absence or presence of 200 nm MLi2 (24 h) as indicated. Bars represent mean ± s.e.m. (n
0.05. (D) Example of wild-type and G2019S knockin primary astrocytes stained with anti-Arl13b, anti-gamma-tubulin, anti-GFAP and DAPI. Scale bar, 10 μm. (E) The percentage of ciliated cells was quantified from around 200 wild-type or G2019S knockin cells, in either the absence or presence of serum starvation (48 h, S.S.), and absence or presence of 200 nm MLi2 (24 h) as indicated. Bars represent mean ± s.e.m. (n =
= 3 independent experiments); **, P
3 independent experiments); **, P <
< 0.01.
0.01.
Discussion
We have shown here that pathogenic LRRK2 causes the pericentrosomal/centrosomal accumulation of phospho-RAB8 as well as of phospho-RAB10, and centrosomal cohesion deficits in a manner dependent on RAB8, RAB10 and RILPL1. RAB8 and RAB10 share various effector proteins, and the phosphorylation by pathogenic LRRK2 kinase interferes with the ability of most of these effector proteins to interact with the phosphorylated RABs (3). This is expected to lead to a loss of function in the membrane-trafficking pathways these specific RAB-effector complexes are involved in. However, a very small amount of effector proteins including RILPL1 behave in the opposite way by binding to only the phosphorylated forms of RAB8 and RAB10 (2). These unusual phospho-RAB-specific effectors are therefore of crucial importance to understand the downstream physiological consequences of hyperactivated LRRK2 kinase underlying PD in a possibly neomorphic, gain-of-function manner.
RILPL1 localizes to a pericentriolar compartment adjacent to the mother centriole and has been reported to recruit phospho-RAB10 to this location (19,20). Under conditions of endogenous protein expression, pathogenic LRRK2-mediated ciliogenesis defects were rescued upon knockdown of either RAB10 or RILPL1 (20). Similarly, we here describe that the pathogenic LRRK2-mediated centrosomal cohesion deficits are largely and/or completely abolished in cells lacking RAB8A, RAB10 or RILPL1. These data highlight the importance of these proteins in pathogenic LRRK2 signalling with respect to both ciliogenesis and centrosomal cohesion (Fig. 9). In the future, it will be important to determine how pericentrosomal/centrosomal protein complexes of phospho-RAB8/10 bound to RILPL1 block cilia formation and interfere with centrosome cohesion.
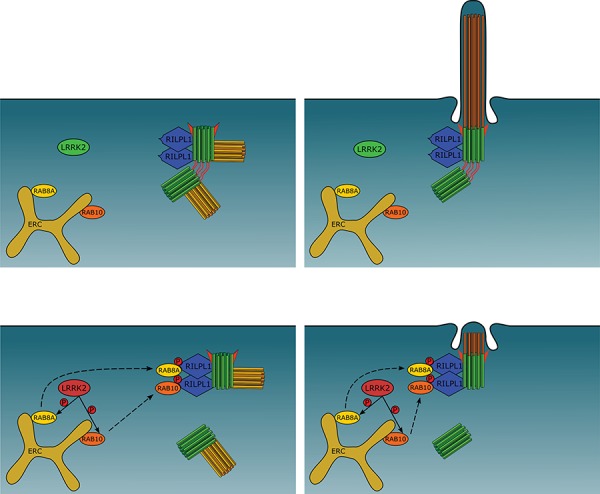
Cartoon summarizing pathogenic LRRK2-mediated effects on centrosome cohesion and ciliogenesis. Top row: under normal conditions, RAB8A and RAB10 are localized to the early recycling compartment (ERC), and RILPL1 is localized to the mother centriole (green, with appendages depicted in red). In dividing cells, upon centriole duplication (daughter centrioles in yellow), the duplicated centrosomes are properly held together in G2 phase by centrosomal linker proteins (red) (left). Ciliogenesis in interphase or non-dividing cells, nucleated by the mother centriole-derived basal body, occurs normally as well (right). Bottom row: pathogenic LRRK2 phosphorylates RAB8A/RAB10, which causes their relocalization to the centrosome via binding to RILPL1, followed by a deficit in centrosome cohesion in dividing cells (left), or a deficit in primary cilia formation in interphase or non-dividing cells (right).
RAB8 plays an important role in ciliary vesicle trafficking required for ciliogenesis in a variety of cell types (28,33–37), whilst the role of RAB10 in primary ciliogenesis remains less clear (38). Ciliogenesis has been reported to be unimpaired in mouse embryonic fibroblasts from RAB8A/RAB8B double knockout mice but is impaired when additionally silencing RAB10 (27). These data suggest that RAB10 may act synergistically with RAB8 in ciliogenesis, such that most ciliary transport processes occur normally in the absence of RAB8A/B as long as RAB10 remains available. In contrast, ciliogenesis was reported to be impaired in A549 RAB8A knockout cells but enhanced in A549 RAB10 knockout cells, suggesting that RAB10 may act as a suppressor of cilia formation (20). Further work is required to understand the relative contributions of RAB8A/B and RAB10 to ciliogenesis, which may involve cell type-specific differences in the precise vesicular trafficking steps mediated by these RABs which directly and/or indirectly impinge upon cilia formation. Importantly, our data indicate that the pericentrosomal/centrosomal accumulation of phospho-RAB8/10 upon pathogenic LRRK2 expression is observed in non-ciliated cells but is much reduced and/or absent in ciliated cells. In addition, increasing the levels of phospho-RAB8/10 by co-expressing the RABs with wild-type LRRK2 caused a pronounced deficit in ciliogenesis, which was not observed when expressing either RAB8/10 or wild-type LRRK2 on their own. Therefore, and apart from a role for endogenous RAB8 and/or RAB10 in regulating ciliary vesicular trafficking events, our data indicate that the pericentrosomal/centrosomal accumulation of phospho-RAB8/10 interferes with ciliogenesis.
Some proteins implicated in vesicular trafficking events have been reported to display additional moonlighting functions at different stages of the cell cycle, including effects on centrosome cohesion (39). We did not detect centrosomal cohesion deficits in A549 cells deficient for either RAB8A, RAB10 or RILPL1, indicating that these three proteins do not modulate centrosome cohesion under normal conditions. However, these three proteins are crucial for the centrosomal cohesion deficits induced by pathogenic LRRK2, since LRRK2-mediated deficits were largely abolished in either A549 RAB8A, RAB10 or RILPL1 knockout cells. Centrosomal cohesion deficits were also observed when increasing the levels of phospho-RAB8/10 by co-expressing the RABs with wild-type LRRK2, but not upon co-expression of phospho-deficient versions of these RAB proteins. Since phospho-site RAB8/10 mutants have been reported to be non-functional in various cellular contexts (11,20), we additionally used LRRK2 kinase inhibitors to demonstrate that the effects were mediated by the LRRK2 kinase activity. Thus, the pericentrosomal/centrosomal recruitment of phospho-RAB8/10 is crucial to mediate the centrosomal cohesion deficits mediated by pathogenic LRRK2 in a kinase activity-dependent manner.
Overexpression of RILPL1 has been suggested to cause a deficit in ciliogenesis as well as centrosomal cohesion (20). This is in contrast to previous studies showing no effect of RILPL1 overexpression on ciliogenesis (19), and no effect of RILPL1 overexpression on centrosomal cohesion as determined in the present study. Further work is needed to corroborate or refute the effects of increasing cellular RILPL1 levels on ciliogenesis and/or centrosomal cohesion events. In either case, loss of RILPL1 abolished the pathogenic LRRK2-mediated deficits in ciliogenesis (20), and our data show that loss of RILPL1 also abolishes the LRRK2-mediated deficits in centrosome cohesion. Therefore, this unique effector protein for only the phosphorylated versions of RAB8/10 plays an essential role in mediating both centrosome-related cellular readouts.
The observed ciliogenesis deficits in cholinergic neurons in the striatum of pathogenic LRRK2 mutant mice (20) is expected to impair a described neuroprotective circuit, whereby Sonic hedgehog signalling from dopaminergic neurons is sensed by cilia in the cholinergic neurons, which in turn triggers the secretion of neuroprotective glial cell line–derived neurotrophic factor from these cholinergic neurons back towards the dopaminergic neurons, in this manner maintaining their health (23). Whilst formal proof for a deficit in this neuroprotective circuit in the context of pathogenic LRRK2 is currently lacking, LRRK2-mediated ciliogenesis deficits have also been described in distinct brain areas of pathogenic LRRK2 knockin mice, and in various cell types including iPS cells from PD patients based on LRRK2 mutations, or murine embryonic fibroblasts from pathogenic LRRK2 knockin mice (20). Additionally, our data indicate the presence of LRRK2-mediated centrosomal (12) as well as ciliogenesis deficits in primary dermal fibroblasts from LRRK2-PD patients, and centrosomal as well as ciliogenesis defects in astrocytes from pathogenic LRRK2 knockin mice. Therefore, these pathogenic LRRK2-mediated deficits seem to be a prominent cellular phenotype observed in a variety of neuronal and non-neuronal cell types and modulated by LRRK2 kinase inhibitors. Astrocytes comprise a disease-relevant cell type which supports the health of dopaminergic neurons (31,32,40), and aberrant ciliary signalling can affect astrocyte survival under certain conditions (41). In the future, it will be important to probe for astrocytic ciliary deficits in the intact rodent and human brain and evaluate the possible consequences of such deficits for dopaminergic cell survival.
Finally, some cells such as microglia or lymphocytes which express high endogenous LRRK2 levels are largely devoid of a primary cilium (42,43). However, we have observed centrosomal cohesion deficits in lymphoblastoid cell lines derived from LRRK2-PD patients (12). Since our data indicate that the centrosomal cohesion readout is a reflection of the same phospho-RAB8/10-RILPL1 mechanism as the one underlying the LRRK2-mediated ciliogenesis deficits, it will be important to test in the future whether centrosomal cohesion deficits can also be observed in primary blood-derived cells from LRRK2-PD patients, and whether such readout may aid in the stratification of PD patients who may benefit from LRRK2-related treatment strategies.
Materials and Methods
DNA constructs and site-directed mutagenesis
GFP-tagged human LRRK2 and RAB8A constructs have been previously described (12). Myc-DDK-tagged human RAB8B and human RAB10 were obtained from OriGene, human RILPL1-DDK was purchased from GenScript, human RILPL1-eGFP construct was kindly provided by Dr Dario Alessi (University of Dundee, United Kingdom), and 3×FLAG-tagged human LRRK2 constructs were kindly provided by Dr Elisa Greggio (University of Padova, Italy). pEGFP-mSmo was a gift from Philip Beachy (Addgene plasmid 25395) (44). N-terminally GFP-tagged, mRFP-tagged or 3×FLAG-tagged human RAB8A, RAB8B and RAB10, and eGFP-tagged human RILPL1 constructs were generated using Gibson Assembly Master Mix (New England Biolabs). The RAB8A-C204A, RAB8B-T72A and RAB10-T73A mutations were generated by site-directed mutagenesis (QuikChange, Stratagene), and the identity of all constructs verified by sequencing of the entire coding region. DNA constructs were prepared from bacterial cultures at 37°C using PureYield™ Plasmid Midiprep System (Promega) following manufacturer’s instructions.
Cell culture and transfections
HEK293T/17 cells were cultured as previously described (12,45). Briefly, cells were grown in full medium [Dulbecco’s modified Eagle’s medium (DMEM) containing low glucose and 10% fetal bovine serum, non-essential amino acids, 100 U/ml penicillin and 100 μg/ml streptomycin] and transfected at 80% confluence with 1 μg of LRRK2 constructs (and 100 ng of RAB constructs where indicated) with 3 μl of LipoD293™ Transfection Reagent (SignaGen Laboratories) per well of a 12-well plate for 5 h in full medium. The next day, cells were split to 25% confluence onto poly-L-lysine-coated coverslips and subjected to immunocytochemistry or western blot analysis 48 h after transfection. For ciliogenesis experiments, cells were processed as described above, and serum starved for 12 h in the absence or presence of MLi2 (200 nm, 12 h).
HeLa cells were cultured as described (11,46). Briefly, cells were grown in full medium (DMEM containing high glucose and 10% fetal bovine serum, non-essential amino acids, 100 U/ml penicillin and 100 μg/ml streptomycin) and transfected at 80% confluence with 200 ng of RAB constructs and 1 μl of Lipofectamine® 2000 Transfection Reagent (Invitrogen) per well of a 12-well plate following manufacturer’s instructions for 5 h in DMEM. The following day, cells were split 1:3 onto coverslips and processed for immunocytochemistry 48 h after transfection.
SH-SY5Y cells stably expressing GFP, flag-tagged wild-type LRRK2, or flag-tagged G2019S-mutant LRRK2 were cultured as previously described (12,47,48). Briefly, cells were grown in full medium (DMEM containing high glucose and GlutaMAX (Life Technologies, 31966-021), supplemented with 15% fetal bovine serum, non-essential amino acids, 50 μg/ml gentamicin (Life Technologies) and 200 μg/ml hygromycin B Gold™ (InvivoGen) and subcultured at a ratio of 1:6 twice a week. Cells were transfected at 80% confluence in DMEM containing high glucose and GlutaMAX with 400 ng of RAB constructs and 2 μl of Lipofectamine® 2000 Transfection Reagent per well of a 24-well plate, and 5 h later, media was replaced with full medium. Cells were split 1:3 the following day onto coverslips and subjected to immunocytochemistry 48 h after transfection.
A549 wild-type cells, as well as CRISPR/Cas9 A549 RAB8A knockout, RAB10 knockout and RILPL1 knockout cells were generously provided by D. Alessi (2,20,49). Cells were cultured in DMEM containing high glucose without glutamine and supplemented with 10% fetal bovine serum, 2 mm L-glutamine, 100 U/ml of penicillin and 100 μg/ml of streptomycin and subcultured at a ratio of 1:6–1:10 twice a week. Cells were transfected at 90% confluence with 1 μg of LRRK2 constructs and 4 μl of LipoD293™ Transfection Reagent (SignaGen Laboratories), and 5 h later, media was replaced with full medium. Cells were split 1:4 onto coverslips the following day and processed for immunocytochemistry or western blotting 48 h after transfection.
Primary human skin fibroblasts established from skin biopsies taken from five age- and sex-matched healthy controls and five PD patients with the G2019S mutation (12,46), with informed consent and ethical approval, were grown in Iscove’s modified Dulbecco’s media (IMDM) and 10% fetal bovine serum, with media exchanged every 2 days. Cells were subcultured at a ratio of 1:4 and seeded at equal densities onto coverslips. For ciliogenesis experiments, cells were serum-starved for 48 h in either the presence or absence of 200 nm MLi2 for the last 24 h. Analyses were carried out on passages 5–10, with no passage-dependent differences observed.
Heterozygous breeding pairs of mice were crossed to yield wild-type and homozygous G2019S progeny, which were then bred to obtain homozygous embryonic wild-type or G2019S knockin pups (30). Mouse cortical astrocytes from wild-type or homozygous G2019S knockin mice were cultured as previously described (50). Briefly, 6–7 brains were obtained from P1 to P4 mouse pups; isolated cortices were mechanically dissociated in BME, and dissociated cells cultured in growth medium [DMEM with pyruvate, high glucose (Life Technologies, 41966-029) and 10% fetal bovine serum (HyClone Fetal Bovine Serum, Fisher, 10309433)]. Cells were seeded in two 175 cm2 cell culture flasks, and the following day, old growth medium was removed and replaced with fresh ice-cold growth medium. Four days later, this procedure was repeated, and subsequently, 50% of growth medium was replaced with warm growth medium every 3–4 days. Once confluent, cells were split at high density onto poly-L-lysine (Sigma, P9155)-coated coverslips and processed for immunocytochemistry. For ciliogenesis, cells were serum-starved for 48 h, in the presence or absence of 200 nm MLi2 for the last 24 h.
Where indicated, cells were treated with MLi2 (MRC PPU, Dundee, UK) or GSK2578215A (Tocris) before fixation.
Immunocytochemistry
For centrosome staining, HEK293T/17 and A549 cells were fixed with 2% paraformaldehyde (PFA) in PBS for 20 min at room temperature, whilst SH-SY5Y cells were fixed with 2% PFA in PBS containing 4% sucrose for 20 min at room temperature. For detection of primary cilia in HEK293T/17, and primary cilia and centrosomes in primary dermal human fibroblasts and astrocytes, cells were fixed with 4% PFA in PBS for 15 min at 37°C, followed by ice-cold MeOH for 5 min. In all cases, cells were permeabilized with 0.2% Triton-X100/PBS for 12 min at room temperature, followed by incubation in blocking solution (0.5% BSA (w/v) in 0.2% Triton-X100/PBS) for 1 h at room temperature. Primary antibodies were diluted in blocking solution and incubated overnight at 4°C. Primary antibodies included rabbit polyclonal anti-pericentrin (Abcam, ab4448, 1:1000), mouse monoclonal anti-pericentrin (Abcam, ab28144, 1:1000), mouse monoclonal anti-γ-tubulin (Abcam, ab11316, 1:1000), mouse monoclonal anti-flag (Sigma, clone M2, F1804, 1:500), rabbit monoclonal knockout-validated anti-RAB8A (Abcam, ab188574, 1:1000), mouse monoclonal knockout-validated anti-RAB10 (Sigma, SAB5300028, 1:1000), mouse monoclonal anti-polyglutamylated tubulin (AdipoGen, AG-20B-0020-C100, 1:1000), rabbit polyclonal anti-Arl13b (ProteinTECH, 17711-1-AP, 1:250), chicken polyclonal anti-GFAP (Abcam, ab4674, 1:500) and mouse monoclonal p230/Golgin245 (BD, 611280, 1:400). The sheep anti-RAB8A (MRC PPU, S969D, 1:250), anti-phospho-T72-RAB8A (MRC PPU, S874D, 1:50), anti-RAB10 (MRC PPU, S945D, 1:250) and anti-phospho-T73-RAB10 (MRC PPU, S873D, 1:50) have been previously described (3) and were generously provided by D. Alessi . The sheep phospho-T72-RAB8A and phospho-T73-RAB10 antibodies were used in the presence of a 10-fold molar excess of the respective dephospho-peptides, or of the respective phospho-peptides where indicated. Importantly, all double- and triple-immunocytochemistry involving sheep antibodies were performed sequentially, with the sheep antibody employed first, and omitting the MeOH fixation step. The rabbit polyclonal anti-phospho-T72-RAB8A and rabbit polyclonal anti-phospho-T73-RAB10 antibodies were kindly provided by D. Alessi and employed at a dilution of 1:250. The rabbit monoclonal knockout-validated anti-phospho-T72-RAB8A (Abcam, ab230260) was used at a dilution of 1:1000. For the rabbit monoclonal knockout-validated anti-phospho-T73-RAB10 (Abcam, ab230261, 1:1000) antibody, 0.2% Triton-X100/PBS was replaced by 0.1% saponin/PBS throughout. The following day, coverslips were washed two times 10 min in 0.2% Triton-X100/PBS (or 0.1% saponin/PBS) (wash buffer) and were incubated with secondary antibodies diluted in wash buffer for 1 h at room temperature.
Note that the sheep total anti-RAB8A and total anti-RAB10 antibodies have been previously validated against extracts from RAB8A or RAB10 A549 knockout cell lines (MRC PPU, Dundee, UK). The commercial mouse total anti-RAB8A antibody has been previously validated against extracts from a RAB8A HAP1 knockout cell line (Abcam, ab188574), and the commercial mouse total anti-RAB10 antibody has been previously validated against HELA cell extracts in the presence or absence of siRNA of RAB10 (Sigma, SAB5300028/Thermo Fisher, MA5-15670, clone 4E2). The sheep phospho-RAB8A and phospho-RAB10 antibodies have been previously validated in HEK293 cells overexpressing the respective RABs in the presence or absence of a LRRK2 kinase inhibitor, as not sensitive enough to detect endogenous phospho-RAB levels by western blotting techniques (6). The rabbit monoclonal phospho-RAB8A and phospho-RAB10 antibodies are able to detect endogenous phospho-RABs by western blotting techniques and have been previously validated by western blotting using both LRRK2 knockout mice extracts, as well as A549 RAB8A or RAB10 knockout cell extracts (6).
Secondary antibodies included Alexa405-conjugated goat anti-mouse, Alexa488-conjugated goat anti-mouse, goat anti-rabbit or donkey anti-sheep, Alexa555-conjugated goat anti-mouse or goat anti-rabbit, Alexa594-conjugated goat anti-mouse, goat anti-rabbit or donkey anti-sheep, Alexa647-conjugated goat anti-mouse, goat anti-rabbit or donkey anti-sheep (all from Invitrogen, 1:1000). Alexa405-conjugated goat anti-chicken was from Abcam (ab175675, 1:250). Coverslips were washed two times in wash buffer, rinsed in PBS and mounted in mounting medium with DAPI (Vector Laboratories).
For the determination of the subcellular localization of GFP-tagged RAB proteins in HeLa cells, cells were transfected and cultured as described above and fixed 48 h after transfection with 4% PFA/PBS for 20 min at room temperature. Coverslips were washed with PBS and mounted with mounting medium with DAPI.
Image acquisition and quantification
Images were acquired on a Leica TCS-SP5 confocal microscope using a 63× 1.4 NA oil UV objective (HCX PLAPO CS). Images were collected using single excitation for each wavelength separately and dependent on secondary antibodies [488 nm argon laser line and a 510–540 nm emission band pass, 543 HeNe laser line and a 555–575 nm emission band pass (for Alexa555) or a 590–620 nm emission band pass (for Alexa594), 633 nm HeNe Laser line and a 640–670 nm emission band pass]. GFP-tagged proteins were excited with a 488 nm argon laser line and a 500–530 nm emission band pass, mRFP-tagged proteins were excited with a 543 nm HeNe laser line and a 560–590 nm emission band pass, and DAPI with a 405 nm UV diode and a 430–470 nm emission band pass, respectively.
For centrosome cohesion determination, 10–15 image sections of selected areas were acquired with a step size of 0.5 μm, and z-stack images analyzed and processed using Leica Applied Systems (LAS AF6000) image acquisition software. The same laser intensity settings and exposure times were used for image acquisition of individual experiments to be quantified. For HEK293T/17 cells and SH-SY5Y cells, centrosomes were scored as being separated when the distance between their centres was > 1.5 μm (12). For A549 cells, the mean distance between duplicated centrosomes was 1.49
1.5 μm (12). For A549 cells, the mean distance between duplicated centrosomes was 1.49 ±
± 0.13 μm (mean ± s.e.m., n
0.13 μm (mean ± s.e.m., n =
= 82 cells), and duplicated centrosomes were scored as separated when the distance was >
82 cells), and duplicated centrosomes were scored as separated when the distance was > 2.5 μm. For mouse cortical astrocytes, the mean distance between duplicated centrosomes was 1.18
2.5 μm. For mouse cortical astrocytes, the mean distance between duplicated centrosomes was 1.18 ±
± 0.06 μm (mean ± s.e.m., n
0.06 μm (mean ± s.e.m., n =
= 96 cells), and duplicated centrosomes were scored as separated when the distance was >
96 cells), and duplicated centrosomes were scored as separated when the distance was > 1.5 μm. In all cases, mitotic cells were excluded from the analysis.
1.5 μm. In all cases, mitotic cells were excluded from the analysis.
Quantification of phospho-RAB10 signal in SH-SY5Y or HEK293T/17 cells was performed over non-processed and non-saturated images acquired on the same day and with the same laser intensities with Leica Applied Systems (LAS AF6000) Image Acquisition Software. Circular ROIs of 3 μm diameter were set on top of the centrosome signal as detected by anti-pericentrin staining, and the mean intensity from the phospho-RAB10 signal obtained from 50–70 cells per condition and experiment. Background signal was subtracted in all cases by placing the ROI in a different aleatory place within the same cell.
For determination of the percentage of ciliated cells, cells were visualized on an inverted microscope (Zeiss) using a 100× 1.4 NA Plan APO oil objective. For each experiment, around 200 random cells were scored for either the presence or absence of primary cilia as assessed by either polyglutamylated tubulin or Arl13b staining, with identical results obtained in both cases. Quantification of the percentage of ciliated cells was performed by two additional observers blind to condition, with identical results obtained in all cases. As an additional means of primary cilia quantification, 20–25 image sections were acquired with a step size of 0.25 μm, and images deconvolved using Huygens Essential Deconvolution Software. For measuring cilia length, a method based on the Pythagorean theorem (PyT) was employed, and around 40 cells per condition were quantified (51,52). Cilia length was also quantified based on maximum intensity projection (MIP), with the determined cilia length smaller as expected (52), but with no difference between genotypes observed.
Western blotting
HEK293T/17, A549 and HeLa cells were collected 48 h after transfection from a well of a 6-well plate, washed in PBS, resuspended in 75 μl of PBS, lysed with 25 μl of 4× Nu PAGE LDS sample buffer (Novex, Life Technologies, NP00008) supplemented with β-mercaptoethanol to a final volume of 2.5% (v/v) and sonicated and boiled at 70°C for 10 min. Around 10–15 μl (around 20 μg of protein) were resolved by SDS-PAGE polyacrylamide gel electrophoresis using 4–20% precast gradient gels (Bio-Rad, 456-1096), and proteins electrophoretically transferred onto nitrocellulose membranes (GE Healthcare). Membranes were blocked in blocking buffer (Li-COR Biosciences, Li-COR Odyssey PBS blocking buffer, 927-40000) for 1 h at room temperature and incubated with primary antibodies in blocking buffer overnight at 4°C. Primary antibodies included rabbit polyclonal anti-GFP (Abcam, ab6556, 1:1000), mouse monoclonal anti-flag (Sigma, clone M2, F1894, 1:500), rabbit monoclonal knockout-validated anti-RAB8A (Abcam, ab188574, 1:1000), mouse monoclonal anti-RAB8(A+B) (BD, 610844, 1:500), rabbit polyclonal anti-RAB8B (Invitrogen, PA5-67354, 1:1000), mouse monoclonal knockout-validated anti-RAB10 (Sigma, SAB5300028, 1:1000), rabbit monoclonal anti-phospho-T72-RAB8A (Abcam, ab230260, 1:1000), rabbit monoclonal anti-phospho-T73-RAB10 (Abcam, ab230261, 1:1000), rabbit monoclonal anti-phospho-S935-LRRK2 (Abcam, ab133450, 1:500), mouse monoclonal anti-α-tubulin (Sigma, clone DM1A, T6199, 1:25 000) and mouse monoclonal anti-GAPDH (Santa Cruz, sc-32
000) and mouse monoclonal anti-GAPDH (Santa Cruz, sc-32 233, 1:2000). Secondary antibodies included goat anti-rabbit or anti-mouse IRDye 800CW, and goat anti-rabbit or anti-mouse IRDye 680CW (1:14
233, 1:2000). Secondary antibodies included goat anti-rabbit or anti-mouse IRDye 800CW, and goat anti-rabbit or anti-mouse IRDye 680CW (1:14 000). Blots were imaged via near infrared fluorescent detection using Odyssey CLx Imaging System, and quantification was done using the instrument’s Image Studio Software.
000). Blots were imaged via near infrared fluorescent detection using Odyssey CLx Imaging System, and quantification was done using the instrument’s Image Studio Software.
For sheep antibodies, HEK293T/17 and A549 cells were collected 48 h after transfection, washed in PBS, resuspended in 80 μl of PBS and lysed with 20 μl of 5× Laemmli SDS sample buffer supplemented with β-mercaptoethanol (final 2.5% (v/v)). Extracts were sonicated and boiled at 95°C for 5 min. Proteins were resolved by SDS-PAGE polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes as described above. Membranes were blocked in 5% milk in 0.1% Tween-20/TBS for 8 h, and primary antibodies diluted in 5% milk in 0.1% Tween-20/TBS and incubated overnight at 4°C. Antibodies included sheep anti-RAB8A and anti-phospho-T72-RAB8A (MRC PPU, S969D and S874D, 1:500 and 1:250, respectively), sheep anti-RAB10 and anti-phospho-T73-RAB10 (MRC PPU, S945D and S873D, 1:500 and 1:250, respectively) and sheep anti-RILPL1 (generously provided by D. Alessi). Membranes were washed three times in 0.1% Tween-20/TBS, followed by incubation with HRP-conjugated rabbit anti-sheep secondary antibodies diluted in 5% milk in 0.1% Tween-20/TBS for 1 h. Western blotting of sheep antibodies was performed with ECL Prime Western Blotting Detection Reagent (GE Healthcare).
Statistical analysis
One-way ANOVA with Tukey’s post hoc test was employed, with significance set at P <
< 0.05. Significance values for all data are indicated in figure legends. All statistical analysis and graphs were performed using Prism software version 7.0 (GraphPad, San Diego, CA).
0.05. Significance values for all data are indicated in figure legends. All statistical analysis and graphs were performed using Prism software version 7.0 (GraphPad, San Diego, CA).
Acknowledgements
We thank Laura Montosa for technical assistance with microscopy, and Rafael Fernández-Chacón for help with G2019S knockin mice. We are especially grateful to Dario Alessi for providing a large variety of reagents.
Funding
Michael J. Fox Foundation for Parkinson’s research (to S.H.); European Regional Development Fund; Spanish Ministry of Economy and Competitiveness (SAF2017-89402-R to S.H.); Spanish Ministry of Education, Culture and Sport (FPU15/05233 to A.J.L.O.).
Conflict of Interest statement. None declared.
Ethics approval and consent to participate
The experiments reported in this study followed the ethical guidelines for investigations of experimental animals approved by the Ethical Committee of Spanish National Research Council (CSIC) and performed in accordance with the guidelines from Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes.
References
Articles from Human Molecular Genetics are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/hmg/ddz201
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/hmg/article-pdf/28/21/3552/31606394/ddz201.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/hmg/ddz201
Article citations
CK and LRRK2 Involvement in Neurodegenerative Diseases.
Int J Mol Sci, 25(21):11661, 30 Oct 2024
Cited by: 0 articles | PMID: 39519213 | PMCID: PMC11546471
Review Free full text in Europe PMC
PAK6 rescues pathogenic LRRK2-mediated ciliogenesis and centrosomal cohesion defects in a mutation-specific manner.
Cell Death Dis, 15(10):752, 17 Oct 2024
Cited by: 0 articles | PMID: 39419978 | PMCID: PMC11487180
RAB12-LRRK2 complex suppresses primary ciliogenesis and regulates centrosome homeostasis in astrocytes.
Nat Commun, 15(1):8434, 29 Sep 2024
Cited by: 0 articles | PMID: 39343966 | PMCID: PMC11439917
Loss of primary cilia and dopaminergic neuroprotection in pathogenic LRRK2-driven and idiopathic Parkinson's disease.
Proc Natl Acad Sci U S A, 121(32):e2402206121, 01 Aug 2024
Cited by: 4 articles | PMID: 39088390 | PMCID: PMC11317616
LRRK2 kinase activity is necessary for development and regeneration in Nematostella vectensis.
Neural Dev, 19(1):16, 08 Aug 2024
Cited by: 0 articles | PMID: 39118162 | PMCID: PMC11308222
Go to all (50) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The LRRK2 signaling network converges on a centriolar phospho-Rab10/RILPL1 complex to cause deficits in centrosome cohesion and cell polarization.
Biol Open, 11(8):bio059468, 29 Jul 2022
Cited by: 11 articles | PMID: 35776681 | PMCID: PMC9346292
Parkinson disease-associated mutations in LRRK2 cause centrosomal defects via Rab8a phosphorylation.
Mol Neurodegener, 13(1):3, 23 Jan 2018
Cited by: 57 articles | PMID: 29357897 | PMCID: PMC5778812
Pathogenic LRRK2 regulates centrosome cohesion via Rab10/RILPL1-mediated CDK5RAP2 displacement.
iScience, 25(6):104476, 30 May 2022
Cited by: 10 articles | PMID: 35721463 | PMCID: PMC9198432
LRRK2 and Rab GTPases.
Biochem Soc Trans, 46(6):1707-1712, 22 Nov 2018
Cited by: 26 articles | PMID: 30467121
Review
Funding
Funders who supported this work.
European Regional Development Fund
Spanish Ministry of Economy and Competitiveness (1)
Grant ID: SAF2017-89402-R
Spanish Ministry of Education, Culture and Sport (1)
Grant ID: FPU15/05233





