Abstract
Free full text
Gut microbiota in wheezing preschool children and the association with childhood asthma
To the Editor,
Reliable biomarkers to predict asthma in wheezing preschool children are lacking. Recently, the impact of gut microbial perturbations on the development of asthma gained widespread attention. Gut microbial dysbiosis in the first year of life was associated with asthma in multiple birth cohort studies.1, 2, 3, 4, 5 Microbial metabolites might play a crucial role in maintaining an adequate immune balance and preventing asthma through its influence on regulatory T‐cells (Tregs) and the Foxp3 gene.1, 6 However, most data are derived from animal studies, whereas most human studies have focused on the association between infant gut microbiota and asthma‐like symptoms at an age when a reliable diagnosis of asthma cannot yet be made. Furthermore, no studies have been performed that investigated gut microbial composition in wheezing children, and its association with subsequent development of asthma.
In the Asthma DEtection and Monitoring (ADEM) study (clinicaltrial.gov: NCT 00422747), 202 wheezing children and 50 healthy controls aged 2‐4 years were prospectively followed until 6 years of age, when a definitive diagnosis of asthma was made. The study was approved by the Dutch national medical ethical committee, and written informed consent was given by all parents. A detailed study protocol was previously published.7, 8
At inclusion, faecal and blood samples were collected. Faecal microbial composition was analysed by sequencing of the 16S rRNA V3‐V4 gene region. In total, 230 samples (70 true asthmatics, 114 transient wheezers and 46 healthy controls) were successfully analysed (see flow chart, Figure S1). In blood, atopic sensitisation (Phadiatop Infant test), proportion of Tregs by flow cytometry (CD4+CD25highCD127‐), and Foxp3 gene expression were assessed. See Appendix S1 for a detailed description of study methods. The baseline characteristics are displayed in Table S1.
First, we examined whether microbial richness and diversity at preschool age were predictive for future asthma development. Neither the microbial richness (OR 0.99 [95%CI 0.98‐1.01]; P = .46), nor the microbial diversity as assessed by the Shannon index (OR 1.01 [0.98‐1.04]; P = .53) were significantly different between transient wheezing children and true asthmatics while adjusting for potential confounders (sex, breastfeeding, birth season, atopy parents, siblings, parental smoking status, day care attendance; Figure Figure1A‐B).1A‐B). At preschool age, these indices were also not different between wheezers and healthy controls, while adjusting for potential confounders (Figure S2).
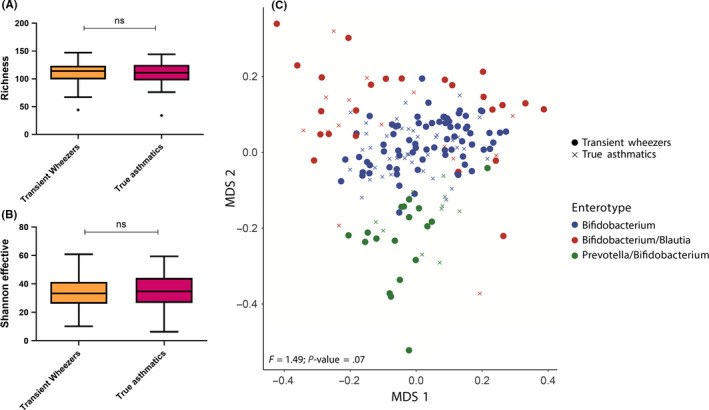
Microbial richness, diversity and community structure among preschool wheezing children who did (true asthmatics) or did not (transient wheezers) subsequently develop asthma. Microbial richness (observed species) (A) and diversity (Shannon effector index) (B) are not significantly different between transient wheezers and true asthmatics (ns: nonsignificant; Kruskal‐Wallis). C, Multidimensional scaling (MDS) based on Bray‐Curtis dissimilarity indicates three different enterotypes driven by Bifidobacterium, Bifidobacterium/Blautia and Prevotella/Bifidobacterium. Overall, microbial community structure is not statistically significantly different between transient wheezers and true asthmatics (permutational analysis of variance [PERMANOVA])
Next, we examined the overall microbial community structure (as assessed by the Bray‐Curtis dissimilarity), which was neither significantly different between transient wheezers and true asthmatics (PERMANOVA P = .07, Figure Figure1C),1C), nor between preschool wheezers and healthy controls (PERMANOVA P = .22, Figure S2). Microbial profiles of all children were clustered using Dirichlet Multinomial Mixture (DMM) modelling. Three distinct clusters (enterotypes) were identified, that is those that were driven by a relatively high abundance of Bifidobacterium, Bifidobacterium combined with Blautia, and Prevotella combined with Bifidobacterium, respectively. Neither the amount of wheezing children who developed asthma, nor the proportion of children with preschool wheeze were significantly different among the three enterotypes, while adjusting for multiple confounders in multivariable logistic regression analyses (Table S2 and S3). Altogether these results indicate that microbial diversity and overall microbial community structure are not predictive for subsequent asthma development among preschool wheezing children.
Furthermore, we examined whether the relative abundance of specific bacterial genera was predictive for future asthma development. Using multivariable logistic regression, we found that the relative abundance of the genera Gemmiger (P = .03) and Escherichia (P = .02) was significantly higher in wheezing children who developed asthma at age 6 years (Figure (Figure2A‐B).2A‐B). The risk of developing asthma was highest in those children who harboured the highest relative abundance of these two bacterial genera (Figure (Figure2C‐D).2C‐D). In particular, a high relative abundance of Escherichia was associated with 4.6‐fold increased odds of asthma (P = .02, Figure Figure2D).2D). When comparing preschool wheezers with healthy controls, the relative abundance of Collinsella (P = .01) and Dorea (P = .02) was significantly lower in wheezing children (Figure S3).
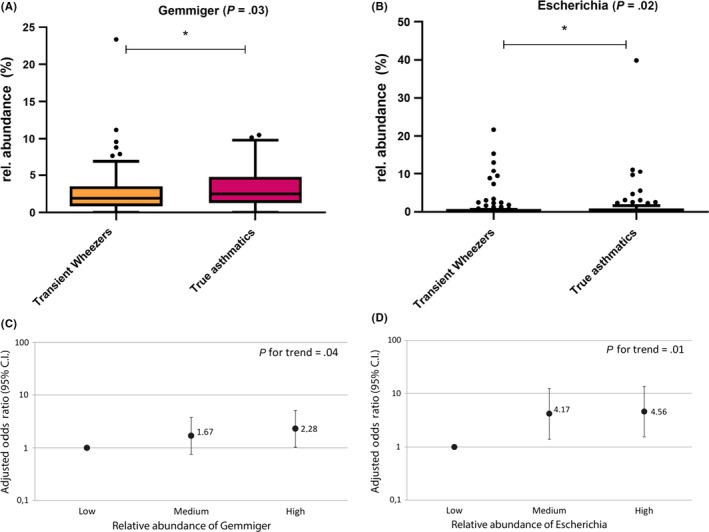
The relative abundance of specific microbial taxa increases the risk of subsequent asthma. The relative abundance of bacterial genera Gemmiger (P = .03) and Escherichia (P = .02) is higher in true asthmatic children compared with transient wheezers (A, B). Multiple logistic regression analyses show that the higher risk to develop asthma among children with a high abundance of Gemmiger and Escherichia remained statistically significant upon adjustment for sex, breastfeeding, birth season, atopy parents, siblings, parental smoking status and day care attendance (C, D)
Finally, we examined whether gut microbial profiles were related to atopic sensitisation, Tregs and Foxp3 gene expression. Besides a weak, yet statistically significant, positive correlation between Foxp3 gene expression and bacterial diversity (Shannon index) within the entire study population (Spearman's rho = 0.16; P = .02), atopic sensitisation, Tregs and Foxp3 gene expression were neither associated with the overall microbial community structure (Bray‐Curtis dissimilarity) nor with the abundance of specific bacterial genera.
To our knowledge, this is the first study to examine the gut microbiota in wheezing preschool children and its association with asthma progression. Our findings suggest that at age 2‐4 years, the microbiota perturbations associated with asthma might only be modest. This is in line with the proposed early window‐of‐opportunity, the first months of life, during which the microbiome is thought to have its strongest impact on immune maturation and tolerance development and stabilises beyond infancy.1, 9 However, this early time‐window might not be a suitable age to identify biomarkers for asthma prediction as early asthma symptoms may not have occurred yet. Moreover, the bacterial genera Gemmiger and in particular Escherichia were significantly associated with asthma, suggesting that some microbial dysbiosis might still exist at preschool age among wheezing children prone for developing asthma. In a recent paediatric study, Escherichia was one of three significantly predominant genera in children with asthma compared with healthy controls.10 Furthermore, in a recent adult study, Escherichia was one of two genera that discriminated asthmatics with fixed airway obstruction from those with no airway obstruction.11 These results are in line with our findings, potentially indicating that the abundance of Escherichia in particular may play a role in the early development of asthma. It has been suggested that an increased abundance of aerotolerant bacteria might be a nonspecific response to inflammatory conditions.12 Recently, an increase in Escherichia appeared to reduce butyrate production, which was associated with asthma.10 Potentially, Escherichia abundance has a wider influence on short‐chain fatty acids production, thereby causing an immunological dysbalance leading to a Th2‐response.
The strength of our study is that we assessed the gut microbial profiles of a large group of preschool wheezing children, using modern sequencing techniques. Another strength is a reliable asthma diagnosis at age 6 years based on symptoms, medication use and lung function measurements.8
The current study has several limitations. First of all, microbiome data are high dimensional, and it cannot be excluded that the observed associations are the result of multiple comparisons. Replication in future studies is therefore needed. Additionally, albeit being the first study to examine the gut microbiota in wheezing preschool children and its association with asthma progression, the sample size limits the power of stratified analyses. The number of sensitised preschool children might for example have been too low to detect significant associations when stratifying for atopic and nonatopic asthma. It is also plausible that microbial perturbations especially impact the risk of asthma in children with certain genetic asthma risk loci, which requires stratified analyses for genotype. Finally, the number of children with eczema was substantial in our population, which might have been accompanied by dietary adaptations. Unfortunately, we had no additional information on the children's diets or restrictions.
In conclusion, gut microbial diversity and overall gut microbial community structure at age 2‐4 years were not associated with preschool wheezing or future asthma development at age 6. When compared to microbial perturbations during infancy, microbial perturbations at preschool age appear to be only modestly associated with asthma. On a genus level, some bacterial genera were associated with wheezing (Collinsella and Dorea) or subsequent development of asthma (Gemmiger and Escherichia), suggesting some microbial dysbiosis in children prone for developing asthma. The role of these genera in the development of asthma warrants further investigation.
AUTHORS' CONTRIBUTIONS
MB and N.v.B. drafted the manuscript; N.v.B., LB and JP were responsible for gut microbial analysis; MB, N.v.B., LB, K.v.d.K, QJ, ED and JP contributed to the design of the study, data collection and interpretation of data. All authors read, revised and approved the final manuscript.
FUNDING INFORMATION
This study was funded by the Dutch Foundation for Asthma Prevention (project number SAB 2017/006).
Notes
Bannier and Best are first authors contributed equally to this manuscript.
Dompeling and Penders are last authors contributed equally to this manuscript.
Contributor Information
Michiel A. G. E. Bannier, Email: [email protected].
Niels van Best, Email: [email protected].
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1111/all.14156
Read article for free, from open access legal sources, via Unpaywall:
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1111/all.14156
Citations & impact
Impact metrics
Article citations
Gut microbiome impact on childhood allergic rhinitis and house dust mite IgE responses.
Pediatr Res, 21 Oct 2024
Cited by: 0 articles | PMID: 39433961
The Role of Biodiversity in the Development of Asthma and Allergic Sensitization: A State-of-the-Science Review.
Environ Health Perspect, 132(6):66001, 27 Jun 2024
Cited by: 1 article | PMID: 38935403 | PMCID: PMC11218706
Review Free full text in Europe PMC
The Footprint of Microbiome in Pediatric Asthma-A Complex Puzzle for a Balanced Development.
Nutrients, 15(14):3278, 24 Jul 2023
Cited by: 11 articles | PMID: 37513696 | PMCID: PMC10384859
Review Free full text in Europe PMC
Risk Factors Affecting Development and Persistence of Preschool Wheezing: Consensus Document of the Emilia-Romagna Asthma (ERA) Study Group.
J Clin Med, 11(21):6558, 04 Nov 2022
Cited by: 6 articles | PMID: 36362786 | PMCID: PMC9655250
Heterogeneous Condition of Asthmatic Children Patients: A Narrative Review.
Children (Basel), 9(3):332, 01 Mar 2022
Cited by: 1 article | PMID: 35327702 | PMCID: PMC8947522
Review Free full text in Europe PMC
Go to all (11) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
[Wheezing in infants and young children. Signs of asthma risk or remission with age?].
MMW Fortschr Med, 153(18):12-13, 01 May 2011
Cited by: 0 articles | PMID: 21604585
Epidemiology of asthma and recurrent wheeze in childhood.
Clin Rev Allergy Immunol, 22(1):33-44, 01 Feb 2002
Cited by: 63 articles | PMID: 11803801
Review
Infections and their role in childhood asthma inception.
Pediatr Allergy Immunol, 25(2):122-128, 17 Nov 2013
Cited by: 22 articles | PMID: 24236893 | PMCID: PMC3977202
Review Free full text in Europe PMC
Life Cycle of Childhood Asthma: Prenatal, Infancy and Preschool, Childhood, and Adolescence.
Clin Chest Med, 40(1):125-147, 20 Dec 2018
Cited by: 3 articles | PMID: 30691707
Review
Funding
Funders who supported this work.
Stichting Astma Bestrijding (1)
Grant ID: SAB 2017/006

 1
1




