Abstract
Free full text

GBM-Targeted oHSV Armed with Matrix Metalloproteinase 9 Enhances Anti-tumor Activity and Animal Survival
Abstract
The use of mutant strains of oncolytic herpes simplex virus (oHSV) in early-phase human clinical trials for the treatment of glioblastoma multiforme (GBM) has proven safe, but limited efficacy suggests that more potent vector designs are required for effective GBM therapy. Inadequate vector performance may derive from poor intratumoral vector replication and limited spread to uninfected cells. Vector replication may be impaired by mutagenesis strategies to achieve vector safety, and intratumoral virus spread may be hampered by vector entrapment in the tumor-specific extracellular matrix (ECM) that in GBM is composed primarily of type IV collagen. In this report, we armed our previously described epidermal growth factor receptor (EGFR)vIII-targeted, neuronal microRNA-sensitive oHSV with a matrix metalloproteinase (MMP9) to improve intratumoral vector distribution. We show that vector-expressed MMP9 enhanced therapeutic efficacy and long-term animal survival in a GBM xenograft model.
Introduction
Glioblastoma multiforme (GBM) is an incurable brain malignancy.1 Oncolytic vectors (OVs) are under development as a potential alternative therapy to the standard of care consisting of surgery, radiation, and temozolomide chemotherapy.2 OVs derived from herpes simplex virus (oHSV) have been designed for virotherapy of nervous system tumors, but early-phase clinical trials have resulted in only limited success.3 Analyses of oncolytic virotherapy of human glioma have revealed several barriers that impede effective lytic infection and tumor cell killing in patients, including inadequate virus replication as well as poor virus distribution within the tumor mass and to tumor cells migrating along white matter tracks.4 To address these problems, we designed our oHSV backbone to express the full complement of viral functions, thus maximizing virus growth in tumors. Tumor-selective infection was achieved by vector retargeting to cancer cells based on recognition of the highly expressed tumor-specific antigens epidermal growth factor receptor (EGFR)/EGFRvIII and elimination of HSV glycoprotein D (gD)’s ability to interact with the cognate HSV entry receptors.5 To further limit oHSV replication to cancer cells, we exploited a highly expressed, endogenous neuronal microRNA, miR-124, that is absent in glioma cells. Introduction of four miR-124 recognition sites into the 3′ UTR of the essential IE gene ICP4 compromised the synthesis of ICP4 protein, preventing escalation in normal brain to high-dose vector toxicity.6 Treatment of an orthotopic model of primary human GBM in nude mice demonstrated that the combination of vector retargeting to EGFR and miR-124 control of virus replication achieved long-term survival of at least 50% of the animals, a level comparable to that obtained with the EGFR-retargeted vector alone.6 However, the complex GBM extracellular matrix (ECM) likely restricts vector distribution and widespread tumor cell virolysis. We previously demonstrated that increased expression of matrix metalloproteinase 9 (MMP9) in neuroblastoma spheroids in culture and in intracranial tumors resulted in enhanced virus dissemination, offering a rationale for expression of MMP9 from our oHSV.7 In this study, we show that this new-generation oHSV provides greatly improved virus penetration in GBM stem-like spheroids in vitro and exhibits remarkable therapeutic efficacy in an orthotopic, xenogeneic model of human GBM.
Results
Arming oHSV with MMP9
By limiting intercellular contact in GBM, the ECM is thought to minimize intratumoral virus spread via direct cell-to-cell contact (lateral spread).8 MMP9 is a membrane-associated and secreted metalloprotease with specificity for type IV amorphous collagen, a core component of the GBM ECM.8 We armed our oHSV with MMP9 in an attempt to reduce the extracellular barrier to virus spread. The genome structures of our MMP9-armed oHSV (KMMP9) and its unarmed precursor (KGW) are shown in Figure 1A. Both vectors contain repeated recognition sites for miR-124 in the 3′ UTR of the ICP4 gene and both are retargeted to EGFR/EGFRvIII.6 KGW contains a Gateway (GW) recombination cassette in the HSV UL3-UL4 intergenic region of the viral genome, which was replaced with MMP9 cDNA under control of the CAG promoter in KMMP9. Both vectors retain the bacterial artificial chromosome (BAC) sequences used for vector propagation in Escherichia coli and express EGFP under control of the glycoprotein C (gC) (UL44) promoter from a bicistronic transcript containing a T2A peptide sequence for ribosome reentry. Since the gC promoter becomes active after viral genome replication, GFP expression is indicative of oHSV replication.
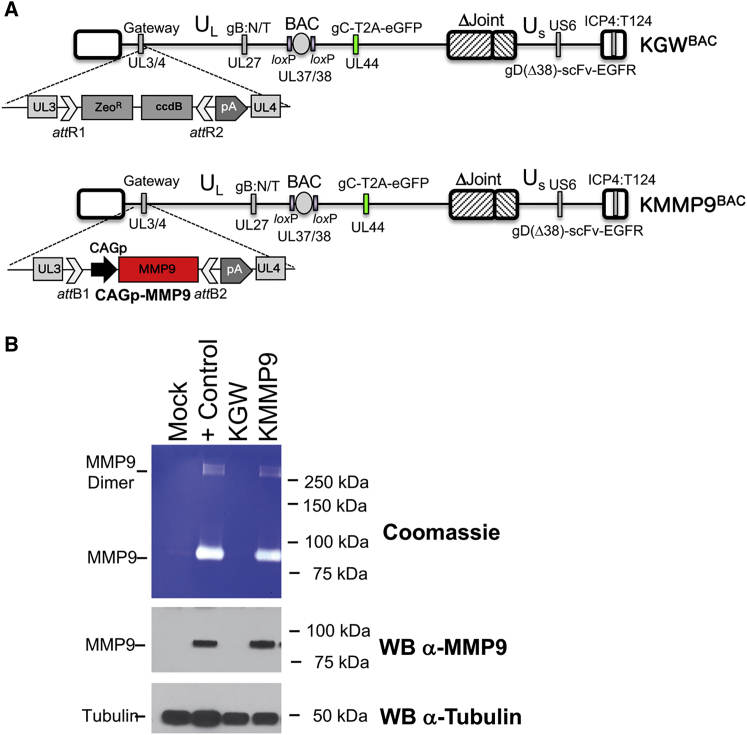
KMMP9 Mediates Expression of Enzymatically Active MMP9 Compared to the KGW Control oHSV
(A) The oHSV vectors were created from the KOS-37 BAC full-length genomic clone of the HSV-1 KOS strain.47 They are also deleted for the internal repeat (joint) region6 and carry two gain-of-function mutations in the external domain of glycoprotein B (gB) that improve virus entry (D285N/A549T, gB:N/T).48 The GFP (EGFP) gene was linked downstream of the glycoprotein C (gC) gene-coding sequence using a T2A skipping sequence in order to track virus replication and spread using fluorescence.53 The vectors were retargeted to tumor cells by modifying glycoprotein D (gD) using a single-chain variable fragment (scFv) antibody that recognizes both EGFR and its constitutively active mutant form EGFRvIII.5 Recognition of the natural gD receptors was ablated by deletion of the N-terminal amino acids 2–24 of gD and deletion of the residue at position 38 (Δ38). A double Red recombination technique was used to insert the Gateway cassette (GW) and the bovine growth hormone polyadenylation sequence (bGHpA) between UL3 and UL4 in order to produce KGWBAC as an MMP9-deficient control vector.54 The oncolytic KMMP9BAC was created by Gateway recombination, replacing the Gateway cassette with the MMP9 gene driven by the highly active CAG promoter. (B) Cell supernatants from U2OS mock-infected cells, U2OS cells transfected with a plasmid expressing MMP9 (+ Control), and U2OS cells infected with KGW or KMMP9 (MOI of 100 gc/cell) were collected 48 hpi and analyzed via a gelatin zymography assay followed by Coomassie blue staining.53 White bands are indicative of gelatinase activity (top panel), signifying enzymatically active MMP9. MMP9 expression in the media was verified by western blot analysis using a monoclonal anti-MMP9 antibody (middle panel). Monoclonal anti-tubulin antibody (bottom panel) was used to detect cellular tubulin gene expression as a loading control.
To confirm MMP9 expression, U2OS cells were infected with KMMP9 or KGW (both with an MOI of 0.01); separate wells were transfected with an MMP9 expression plasmid as a positive control, and untreated U2OS cells were used as negative control. Western blot analysis (Figure 1B, lower panels) showed that MMP9 was detectable only in the supernatants from KMMP9 virus-infected cells (KMMP9 lane) and MMP9 plasmid-transfected cells (+ Control lane); β-tubulin was visualized as a loading control (bottom panel). The biological activity of oHSV-expressed MMP9 was confirmed by a gelatin zymography assay demonstrating the release of active enzyme from KMMP9-infected U2OS cells as well as from the MMP9 plasmid-transfected positive control cells (Figure 1B, top panel, lanes 4 and 2, respectively).
EGFR-Retargeted oHSV Expressing MMP9 Displays Entry Selectivity for EGFR-Bearing Cells and Enhanced Tumor Cell Killing In Vitro
Since glioblastomas often express the HSV gD cognate receptors nectin-1/CD111,9, 10, 11 and HVEM/TNFRSF14,12,13 in addition to EGFR, the specificity of the KMMP9 and KGW vectors for selective infection of EGFR-bearing cells was first confirmed using gD receptor-deficient J1.1-2 cells stably transduced with nectin-1 (J/C), HVEM (J/A), or EGFR (J/EGFR).5 Entry of the EGFR-retargeted KMMP9 and KGW oHSV recombinants into J/EGFR cells was as efficient as entry of a non-retargeted, miR-124-sensitive vector, KG4:T124,6 into J/C cells and more efficient than KG4:T124 entry into J/A cells (Figure 2A). In contrast, neither retargeted vector showed entry into J/C or J/A cells, and KG4:T124 failed to enter into J/EGFR cells. These data confirmed that the retargeted vectors could not use the canonical gD receptors for entry and instead recognized EGFR for efficient infection. We then compared the kinetics of virolysis of both viruses on established (U87, SNB19) and primary (GBM30) EGFR-bearing human glioma cell lines. Neither retargeted oHSV provided effective oncolysis of the target cell lines at 3 days post-infection (dpi) (Figure 2B) at multiplicities of 100 genome copies (gc) per cell. At 7 dpi, however, KMMP9 at 100 gc/cell showed significantly higher oncolytic activity than did KGW on two of the cell lines, consistent with the hypothesis that MMP9 can augment vector oncolysis (Figure 2B).
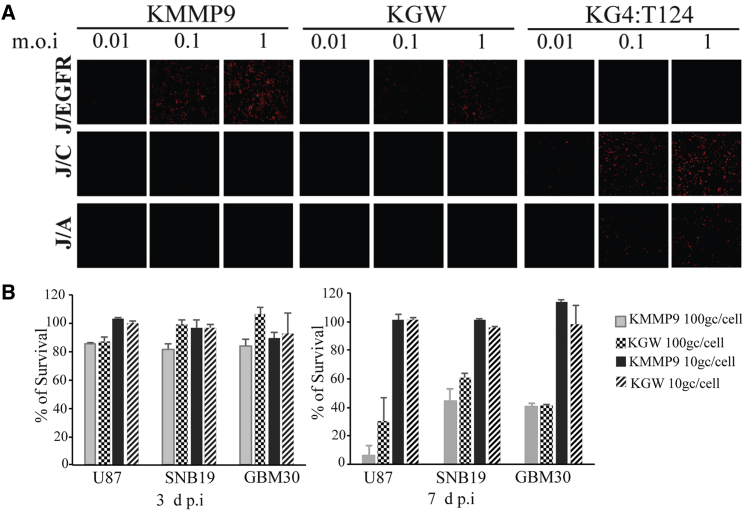
EGFR-Retargeted oHSV Expressing MMP9 Displays Entry Selectivity for EGFR-Bearing Cells and Enhanced Tumor Cell Killing In Vitro
(A) KMMP9, KGW MMP9 negative control oHSV, and the KG4:T124 backbone viruses were employed to confirm EGFR-dependent entry of the EGFR-retargeted KMMP9 and KGW viruses into J cells expressing the EGFR (J/EGFR) compared to J cells expressing either of the two HSV gD cognate receptors nectin-1/HveC (J/C) or TNFRSF14/HVEM/HveA (J/A). Cells were infected at a range of multiplicities (MOI of 0.01, 0.1, and 1.0), and at 6 hpi, infected cell monolayers were immunostained for HSV ICP4 to detect virus entry/infection of the J cells bearing the different cell surface receptors. (B) U87, SNB19, and GBM30 glioma cell lines were infected with KMMP9 and the KGW control virus at an MOI of 10 genome copies (gc)/cell or 100 gc/cell for 3 (left panel) or 7 days (right panel), and the percent cell viability determined by a 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide (MTT) assay was compared for each sample to uninfected cells. Bars represent the mean ± SD of six replicates. Significant differences between the KMMP9 and KGW control viruses (p < 0.05, unpaired Student’s t test) are indicated by an asterisk (*).
oHSV Armed with MMP9 Enhances Virus Spread in GBM30 Spheroids in Culture
To determine whether MMP9 expression would enhance oHSV spread in tumor-cell spheroids in culture, GBM30 spheroids were infected with KMMP9 or KGW at MOIs of 0.01 or 0.05, and virus distribution in specific spheroids was imaged daily for 4 days using EGFP reporter gene expression as a marker of virus spread (Figure 3A). At each time point, KMMP9-infected spheroids displayed more widespread fluorescent signal than did KGW-infected spheroids, consistent with the ability of MMP9 to promote viral spread throughout the tumor cell mass. Virus spread in infected spheroids (MOI of 0.01) was further quantified using confocal microscopy at 72 h post-infection (hpi) (Figure 3B). 3D reconstruction from 5-μm z stack sections revealed enhanced spread of KMMP9 within the spheroid compared to KGW (Figure 3Ba and b). Examination of five consecutive z axis segments (Figure 3Bc) showed significantly higher penetration of KMMP9 into deeper segments within the 3D structure of the spheroid compared to the KGW control (25–50 and 50–85 μm, p < 0.05), demonstrating that MMP9 enhanced vector spread throughout the spheroids. A significant difference was also found when all z stacks were compared between spheroids (unpaired Student’s t test, p = 0.013). Taken together, these in vitro experiments suggested that the KMMP9 vector might be more efficacious as an anti-tumor agent in vivo by enabling better oHSV spread within the tumor mass.
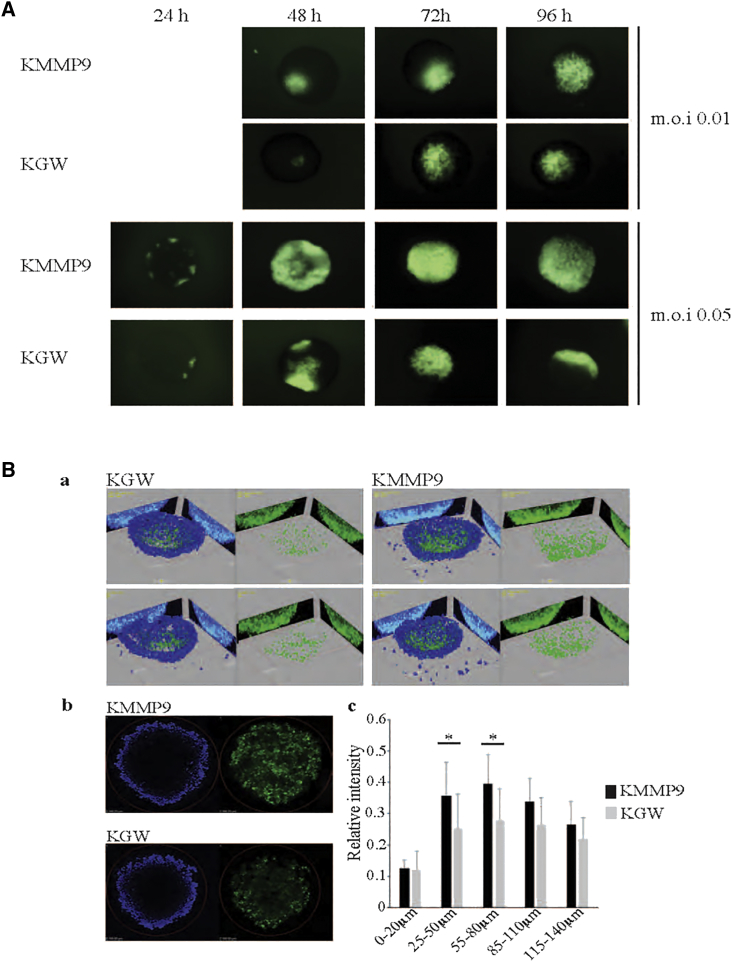
MMP9 Improves oHSV Spread in KMMP9-Infected GBM30 Spheroids
(A) GBM30 cells were grown in suspension and infected with either KMMP9 or KGW (MOI of 0.01 or 0.05), both expressing the EGFP fluorescent reporter that is only expressed when the virus is actively replicating within the tumor cells. Viral replication and spread were monitored daily by visualization of EGFP expression in whole-mount spheroids. (B) GBM30 spheroids were infected with either KMMP9 or KGW (MOI of 0.01) and imaged by confocal microscopy at 72 hpi: (Ba) representative images of two KMMP9- and two KGW-infected spheroids using 3D reconstruction from 0 to 150 μm; (Bb) z sections of two KMMP9- and two KGW-infected spheroids at z = 100 μm. (Bc) Each spheroid was divided into five segments along the z axis. Relative signal intensity of each segment of the spheroid was calculated by averaging EGFP signal (green) divided by DAPI signal (blue) (n = 7; *p < 0.05).
oHSV Armed with MMP9 Enhances Animal Survival in GBM30 Tumors in Nude Mice
We examined the in vivo anti-tumor efficacy of KMMP9 oHSV in our previously described GBM30 orthotopic animal tumor model that generates aggressive intra-axial GBM in nude mice.5,6 Mice injected with PBS died between 2 and 4 weeks of tumor-cell implantation (median survival, 21 days; Figure 4). Animals injected with a low dose of infectious KGW virus (104 plaque-forming units [PFU]; n = 4) showed a median overall survival of 47 days after tumor implantation. In contrast, approximately 70% of animals treated with the same dose of KMMP9 oHSV (n = 8) remained alive and symptom-free throughout the length of the experiment (log-rank test for trend, p < 0.0001). This represents a dramatic improvement in vector performance compared to our previous observation that a dose of 106 PFU of the KGW-comparable vector KGE-4:T124 achieved long-term survival of approximately 50% of tumor-bearing animals in a very similar experiment.6 These results indicated that MMP9 expression enhances the oHSV efficacy in this GBM30 xenograft model, presumably by improving intratumoral virus spread.
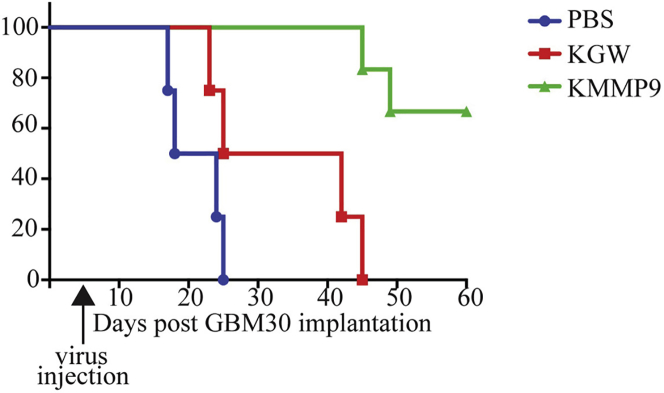
KMMP9 Treatment of Nude Mice Bearing GBM30 Intracranial Tumors Increases Animal Survival
Kaplan-Meier survival plot for nude mice receiving intracranial implant of GBM30 cells and 5 days later injected at the same coordinates with PBS or 104 PFU of KMMP9 or KGW. Animals treated with KMMP9 or KGW survived significantly longer than did those treated with PBS (KMMP9, p < 0.001; KGW, p < 0.01). KMMP9 also showed significantly improved efficacy compared to KGW (p < 0.01).
In order to model a preclinical therapeutic environment that closely resembles human GBM, we performed a parallel experiment in which mice were intracranially injected with GBM30 cells, and oncolytic vector treatment was initiated only when MRI depicted large tumor masses. Mice were paired into groups of comparable tumor volume based on MRI data, and they were subsequently imaged at different time points following treatment with KMMP9, KGW, or PBS. Figure 5 shows T2-weighted MRI images obtained 1 day before treatment and 3, 6, 9, and 13 days after injection of PBS (Figure 5A), KGW (Figure 5B), or KMMP9 (Figure 5C) (n = 3 mice/group). Based on the averages of tumor volumes in each treatment group by time point (Figure 5D), KMMP9 and the KGW control oHSV both exhibited potent anti-tumor activity compared to PBS alone. In particular, despite the relatively large tumor volumes, both KMMP9 and KGW were able to dramatically reduce the rate of tumor growth compared to PBS treatment (p < 0.0001, unpaired Student’s t test). However, no significant difference between the two virus-treated groups was apparent in this analysis. The apparent discordance between these results and those of Figure 4 are discussed below.
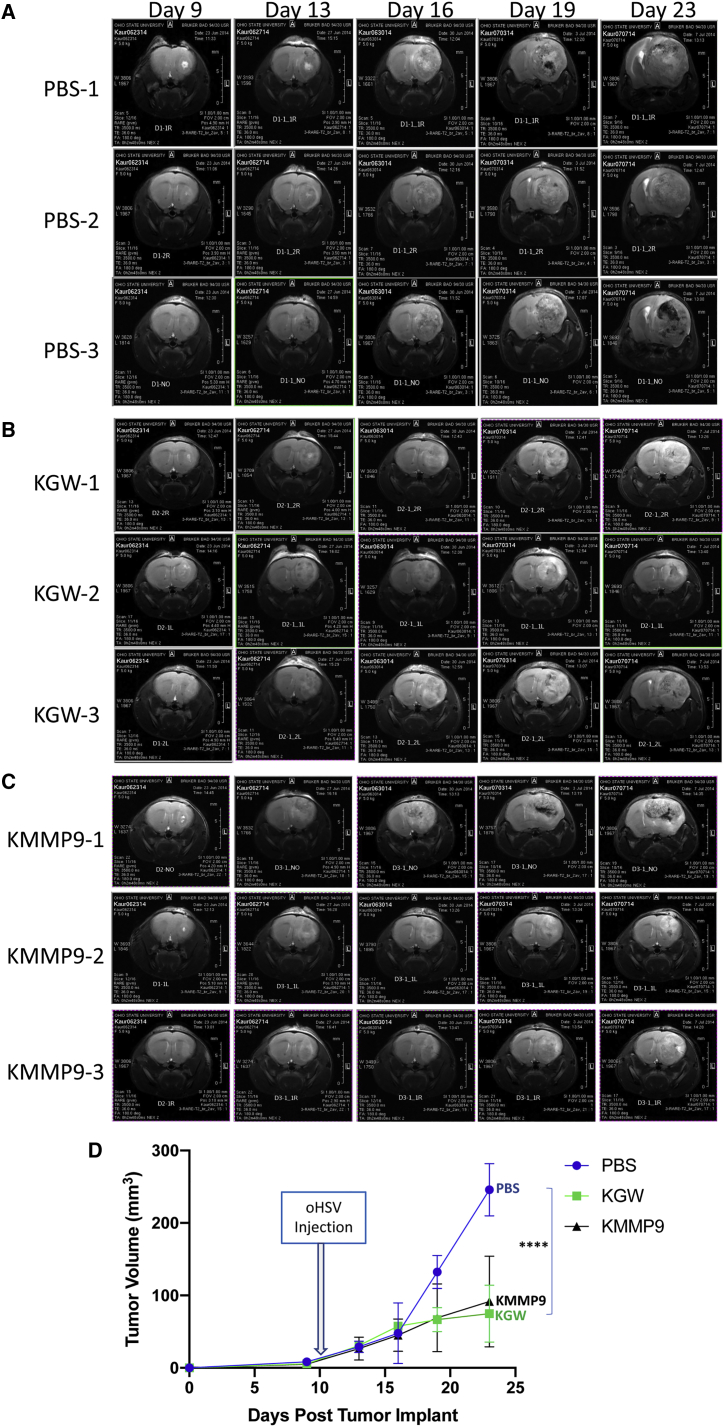
MMP9 Improves Oncolysis Even in Well-Established Brain Tumors
(A–C) T2-weighted brain MRI images of the three animals from each treatment group (A, PBS; B, KGW; and C, KMMP) on day 9 after tumor cell implantation and 1 day before virus injection (day 10) and on days 13, 16, 19, and 23 after tumor cell implantation (4, 7, 9, and 13 days after virus injection). (D) The overall tumor volume calculated based according to MRI imaging is plotted at different time points (n = 3; ****p < 0.0001).
Discussion
The tumor extracellular matrix has been shown to affect oncolytic virus infection and spread throughout the tumor mass.14, 15, 16 Specifically, the physical interference of the ECM with virus diffusion or its masking of viral receptors reduces the effectiveness of viral oncolysis.17, 18, 19, 20 Enzymatic breakdown of the ECM may therefore be a useful strategy to facilitate oncolytic vector distribution throughout the tumor mass, as supported by previous studies using a variety of different ECM-degrading enzymes and OVs.17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 Previous studies have shown that oHSV armed with E-cadherin can enhance viral spread and survival of animals bearing GBM tumors that overexpress E-cadherin.27 Degrading the hyaluronan-rich ECM with hyaluronidase resulted in improved infection and spread of oncolytic adenovirus within the tumor mass.17,22,23,25 Chondroitinase-ABC (Chase-ABC), which degrades chondroitin sulfate proteoglycans, was shown to enhance oHSV spread in glioma spheroids in culture, inhibit GBM tumor growth, and mediate a significant increase in overall survival of mice bearing intracranial tumors.21 oHSV-expressed Chase-ABC enhanced glioma tumor killing in combination with the chemotherapeutic agent temozolomide,18 suggesting that breakdown of the ECM by Chase-ABC led to increased distribution of not only oncolytic virus but also of the chemotherapeutic drug.18 Degradation of collagen within the tumor ECM by injection of collagenase in conjunction with oHSV enhanced oHSV spread within the tumor mass and produced a significant decrease in the growth of human melanoma xenografts,24 suggesting that fibrillar collagen is an important ECM component deterring oHSV spread. Similar observations were reported using an oncolytic adenovirus in GBM xenografts.19,20 Collagenase co-injection with oncolytic Newcastle disease virus (NDV) was reported to enable virus spread to the center of the tumor mass while the center remained virus-free in the absence of collagenase.28
Previous studies have also indicated that unmasking of HSV receptors enhances infection and lateral spread. Depletion of calcium in the media of polarized MDCK cells resulted in the disruption of adherens junctions, enabling efficient HSV-1 attachment and entry.29 Likewise, infection of epithelial cells with HIV-1, or expression of the HIV-1 TAT or gp120 proteins, led to disruption of tight and adherens junctions, thereby exposing the HSV receptor nectin-1 and dramatically increasing cell-to-cell spread.30 This effect results from MMP9 activation by the HIV proteins.31 MMP1 and MMP8 have previously been shown to improve oHSV spread throughout HSTS26T fibrosarcomas,32 while MMP9 expressed from a vaccinia virus (VACV) vector increased spread of the virus and led to enhanced oncolysis in PC-3 prostate tumor-bearing mice.26 We have previously shown that ectopic expression of MMP9 in human SK-N-AS neuroblastoma cells yielded increased oHSV infection and spread,7 supporting the arming of oHSV with MMP9 to degrade the ECM in order to facilitate oHSV spread and potentially enhance animal survival.
Heparanase rapidly degrades syndecan-1 and heparan sulfate,24,25 and it has been shown to enable active release of HSV-1 particles from infected cells,33, 34, 35, 36 which may be expected to increase oHSV spread in tumors.29 However, increased heparanase activity has been associated with increased angiogenesis, tumor cell migration/invasion, and disease progression that ultimately resulted in decreased survival, thereby cautioning its therapeutic use in human clinical trials.37,38 We have recently reported that MMP9 can release VEGF from the ECM of syngeneic tumors in a mouse model system, suggesting that metalloproteinases can have unwanted side effects.39 However, the growth of blood vessels can be reversed by the use of an anti-VEGF antibody (B20), indicating that countermeasures can be taken to achieve better virus spread without enhancing tumor growth.39
The arming of our EGFR-targeted/miR-124-controlled oHSV with MMP9 showed that virus-mediated expression of MMP9 did not affect the selectivity of viral infection for EGFR+ cancer cells (Figure 2B), but rather improved virus spread in GBM30 neurospheres derived from primary glioma tumors (Figure 3). GBM 3D neurospheres represent an in vitro model system that mimics the GBM tumor to the extent that the spheroids display both oxygen and nutrient gradients in the outer edge and central necrotic regions, making them ideal for testing novel therapeutic approaches.40, 41, 42, 43 GBM neurospheres have previously been employed to assess the destruction of GBM ECM components in efforts to enhance oHSV spread within the tumor mass.7,18,21 Our current results using the GBM30 neurosphere model comparing the KMMP9 oHSV to the parental KGW oHSV displayed high similarity to glioma studies using oHSV armed with Chase-ABC,18,21 and our prior work using neuroblastoma spheroids and tumors expressing MMP9.7 In both cases, the expressed enzyme promoted deeper oHSV penetration into the spheroid mass.
Animal survival studies employing a highly invasive primary human tumor line, GBM30, that causes animal death in approximately 20–30 days under our experimental conditions5 showed that a dose of only 104 PFU of our MMP9-expressing oHSV was sufficient to provide nearly 70% long-term survival in tumor-bearing nude mice (Figure 4). Even though the control oHSV lacking MMP9 at this low vector dose improved the average survival time compared to PBS injection, all animals eventually succumbed to disease. Previous studies using a very similar unarmed vector showed substantial animal survival in the same tumor model, but at a 100-fold higher vector dose.6 Thus, MMP9 expression improved performance, providing effective tumor killing after low-dose treatment.
Since GBM patients present with diffusely infiltrated disease at the time of diagnosis, we assessed the effect of MMP9 expression on therapeutic efficacy in advanced brain tumors. Using nude mice, we injected primary GBM30 tumor cells intracranially and imaged the growth of the tumor every other day by MRI. The animals only received treatment when the tumors appeared well defined, hypervascularized, and could be sorted into groups of three (one animal for each treatment) by similar tumor mass. MRI imaging confirmed that within the first 2 weeks after oHSV injection, MMP9-expressing oHSV controlled tumor progression to the same extent as the control oHSV (KGW) lacking MMP9 (Figure 5). Both oHSVs conferred improved survival compared to the PBS controls, which succumbed to disease within a few weeks. Although KMMP9 yielded a similar reduction in tumor growth as the unarmed vector, the armed vector was shown to be more effective in allowing for animal survival. We note that similar results have been seen using Vstat120, an oHSV armed with vasculostatin that blocks tumor angiogenesis. Here too, MRI showed reduced tumor volumes that were similar between the Vstat120 and backbone oHSV viruses compared to PBS-injected mice while Vstat120 treatment produced 80%–100% animal survival compared to 20% of mice treated with the control vector.44 Further studies using dynamic contrast-enhanced MRI (DCE-MRI) showed that most of the GBM tumor-bearing mice treated with the Vstat120 displayed regions of necrosis within their tumor masses while those injected with the control oHSV did not. We have not investigated whether similar events occurred using our MMP9 vector-infected tumors.
Overall, our results demonstrate that MMP9 greatly improves the therapeutic efficacy of our EGFR-retargeted/miR-124 replication-controlled oHSV in a brain tumor model of human GBM. By extension, this oHSV may also prove to be efficacious in non-brain solid tumors in which collagen type IV is a major component of the ECM.45,46 Vector-mediated expression of genes that degrade the tumor ECM may also improve the infiltration of inflammatory and antigen-presenting cells into the tumor mass and thus may promote NK cell activity and ultimately the development of anti-tumor immunity. The utility of metalloproteinase expression to promote intratumoral virus spread may depend on the tumor type and ECM composition and must be weighed against the potential release of tumor-promoting factors.
Materials and Methods
Cell Culture
U2OS (ATCC HTB-96), Vero (ATCC CCL-81), Vero-Cre,47 J/A,48 J/C,49 J/EGFR,50 and GBM30 cells5 have been previously described.
Plasmids and oHSV Viral Vector Constructs
KGWBAC was derived from KGE-4:T124BAC, as follows. A previously described plasmid containing the GW cassette from Gateway conversion plasmid A (Thermo Fisher) between the CMV promoter and bovine growth hormone (bGH) polyadenylation region in pcDNA3.151 was used as a template to amplify the GW-bGH sequence with primers 5′-TGCCCGTCGCGCGTGTTTGATGTTAATAAATAACACATAAATTTGGCTGGCCACTA GTCCAGTGTGGTGG-3′ and 5′-CTGAAATGCCCCCCCCCCCTTGCGGGCGGTCCATTAA AGACAACAAACAAATCCCCAGCATGCCTGCTATTGT-3′ that add 50-bp homology arms (underlined) targeting the HSV-1 KOS UL3/UL4 intergenic region. The PCR product was introduced into the UL3/4 intergenic region of KGE-4:T124BAC DNA by Red/ET-mediated recombination in E. coli (Gene Bridges, Heidelberg, Germany) and bacterial selection for resistance to 25 μg/mL zeocin.
To create KMMP9, MMP9 cDNA was isolated from plasmid pCM6-XL4-MMP9 (OriGene) and inserted into pENTR1A. A fragment containing the CAG promoter was isolated from pCAgH:KV48 and inserted 5′ to the MMP9 sequence in pENTR1A-MMP9, generating plasmid pEnCM. Clonase-mediated in vitro recombination between KGWBAC DNA and pEnCM was then performed, electroporated E. coli was selected for resistance to 15 μg/mL chloramphenicol, and resistant colonies were screened for zeocin sensitivity. Selected KGWBAC and KMMP9BAC isolates were verified by restriction enzyme digestion, field inversion gel electrophoresis (FIGE), and DNA sequencing,6 and one confirmed isolate of each was used for virus production.
Virus Growth and Purification
BAC-containing viruses were converted to infectious virus as previously described,47 and biological titers of virus stocks (PFU/mL) were established on U2OS cells; physical titers in genome copies (gc)/mL were determined by qPCR for the viral gD gene.6
Western Blot Analysis
At 48 hpi, cell lysates and supernatants were analyzed by SDS-PAGE, transferred to Immobilon-P membranes, and probed separately either with rabbit polyclonal anti-MMP9 or rabbit anti-tubulin antibodies and imaged on X-Omat XAR-5 X-ray film.6
Gelatin Zymography
The supernatants of oHSV-infected cells were tested by gelatin zymography as previously described.52 Coomassie brilliant blue R-250 staining was used to detect gelatinolytic activity.52
Spheroid Culture and Confocal Imaging
3 × 103 GBM30 cells were grown in suspension for 2 days. Each spheroid was infected with either KMMP9 or KGW (MOI of 0.01 or 0.05). Viral replication and spread were monitored via fluorescence microscopy. At 3 dpi, spheroids were fixed in 2% paraformaldehyde (PFA), and mounting medium with DAPI was added to visualize the samples. The z stack section images were obtained with a FV1000 confocal imaging system.
In Vivo Studies
Intracranial implantation of 2 × 105 GBM30 cells into BALB/c nude mice was performed as previously described.5 At 5 (University of Pittsburgh [UPIT]) or 10 days (Ohio State University [OSU]) after tumor cell injection, 104 PFU of either KMMP9 or KGW, or PBS, were inoculated at the same coordinates to which tumor cells were injected. Mice were euthanized when demonstrating signs of morbidity as per the University of Pittsburgh Institutional Animal Care and Use Committee and OSU-approved protocols.
MRI Imaging
Mice were randomly selected from each treatment group from the survival experiment (Figure 4) for MRI imaging. Animals were imaged 1 day prior to treatment (9 days after GBM30 implantation) and on days 3, 6, 9, and 13 post-treatment (days 13, 16, 19, and 23 of the study). Imaging was performed using a Bruker BioSpec 94/30 magnet, a 2.0-cm-diameter receive-only mouse brain coil, and a 70-mm-diameter linear volume coil.5 Anesthetized mice were injected with 0.1 mmol/kg Magnevist intraperitoneally, and T2-weighted images (repetition time, 3,500 ms; echo time, 12 ms; rare factor, 8; navgs, 4) were acquired coronally across the region of interest on a 400-MHz Bruker horizontal bore magnet running Paravision 4.0.5
Statistical Analysis
Unpaired Student’s t test with Welch’s correction was performed using GraphPad Prism version 8.1.0 for Mac OS X. Animal survival data were charted as Kaplan-Meier plots and compared by a log-rank test for trend using the same software.
Author Contributions
Study Conception and Design, P.S., N.A., W.F.G, B.K., T.P.C., J.Y., E.A.C., J.C.G., P.G.; Methodology, P.S., N.A., A.L., M.M., D.L., M.Z., W.F.G., C.B., J.Y., P.G.; Acquisition of Data, P.S., N.A., A.L., M.M., D.L., M.Z., W.F.G., C.B., J.Y., P.G.; Analysis and Interpretation of Data, P.S., N.A., W.F.G., B.K., T.P.C., J.Y., E.A.C., J.C.G., P.G.; Writing, Reviewing, Editing of the Manuscript, P.S., N.A., W.F.G., B.K., T.P.C., E.A.C., J.C.G., P.G.; Resources, N.A, W.F.G., B.K., T.P.C., J.Y., E.A.C., J.C.G., P.G.; Study Supervision, W.F.G, B.K., T.P.C., J.Y., E.A.C., J.C.G., P.G.
Conflicts of Interest
P.G., J.C.G., W.F.G., and N.A. are inventors of intellectual property that is licensed to Oncorus. P.G., J.C.G., and W.F.G. also serve as consultants for Oncorus, J.C.G. is Chair of the Scientific Advisory Board, P.G. is the Chief Scientific Officer at CG Oncology. E.A.C. holds ownership interest (including patents) in DNAtrix and is a consultant/advisory board member for Advantagene, DNAtrix, and NanoTherapeutics. The remaining authors declare no competing interests.
Acknowledgments
This work was supported by grants from the NIH (1R01 1A175052 to P.G.; P01 CA163205 to J.C.G., B.K., T.P.C., W.F.G., and E.A.C.; C.B. was supported by T32 CA009338). We thank Dr. D. Leib for KOS-HSV BAC and Vero-Cre cells.
References
Articles from Molecular Therapy Oncolytics are provided here courtesy of American Society of Gene & Cell Therapy
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.omto.2019.10.005
Read article for free, from open access legal sources, via Unpaywall:
https://www.cell.com/article/S2372770519300956/pdf
Citations & impact
Impact metrics
Article citations
An Update on the Clinical Status, Challenges, and Future Directions of Oncolytic Virotherapy for Malignant Gliomas.
Curr Treat Options Oncol, 25(7):952-991, 19 Jun 2024
Cited by: 3 articles | PMID: 38896326
Review
Tutorial: design, production and testing of oncolytic viruses for cancer immunotherapy.
Nat Protoc, 19(9):2540-2570, 20 May 2024
Cited by: 1 article | PMID: 38769145
Review
Overcoming Resistance to Temozolomide in Glioblastoma: A Scoping Review of Preclinical and Clinical Data.
Life (Basel), 14(6):673, 24 May 2024
Cited by: 1 article | PMID: 38929657
Review
Targeted Glioma Therapy-Clinical Trials and Future Directions.
Pharmaceutics, 16(1):100, 11 Jan 2024
Cited by: 6 articles | PMID: 38258110 | PMCID: PMC10820492
Review Free full text in Europe PMC
Novel Therapies in Glioblastoma Treatment: Review of Glioblastoma; Current Treatment Options; and Novel Oncolytic Viral Therapies.
Med Sci (Basel), 12(1):1, 23 Dec 2023
Cited by: 5 articles | PMID: 38249077 | PMCID: PMC10801585
Review Free full text in Europe PMC
Go to all (23) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Stem cells loaded with multimechanistic oncolytic herpes simplex virus variants for brain tumor therapy.
J Natl Cancer Inst, 106(6):dju090, 16 May 2014
Cited by: 66 articles | PMID: 24838834
Blockade of transforming growth factor-β signaling enhances oncolytic herpes simplex virus efficacy in patient-derived recurrent glioblastoma models.
Int J Cancer, 141(11):2348-2358, 26 Aug 2017
Cited by: 22 articles | PMID: 28801914 | PMCID: PMC5765440
Oncolytic herpes simplex virus-based strategies: toward a breakthrough in glioblastoma therapy.
Front Microbiol, 5:303, 20 Jun 2014
Cited by: 34 articles | PMID: 24999342 | PMCID: PMC4064532
Review Free full text in Europe PMC
Human glioblastoma-derived cancer stem cells: establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors.
Cancer Res, 69(8):3472-3481, 07 Apr 2009
Cited by: 252 articles | PMID: 19351838 | PMCID: PMC2785462
Funding
Funders who supported this work.
NCI NIH HHS (7)
Grant ID: R01 CA247550
Grant ID: T32 CA009338
Grant ID: P01 CA163205
Grant ID: R01 CA175052
Grant ID: R01 CA222804
Grant ID: R21 CA223400
Grant ID: R01 CA185301
NIAID NIH HHS (1)
Grant ID: R01 AI129582
NINDS NIH HHS (1)
Grant ID: R01 NS106170






