Abstract
Free full text

Species-specific oligonucleotide probes for rRNA of Clostridium difficile and related species.
Abstract
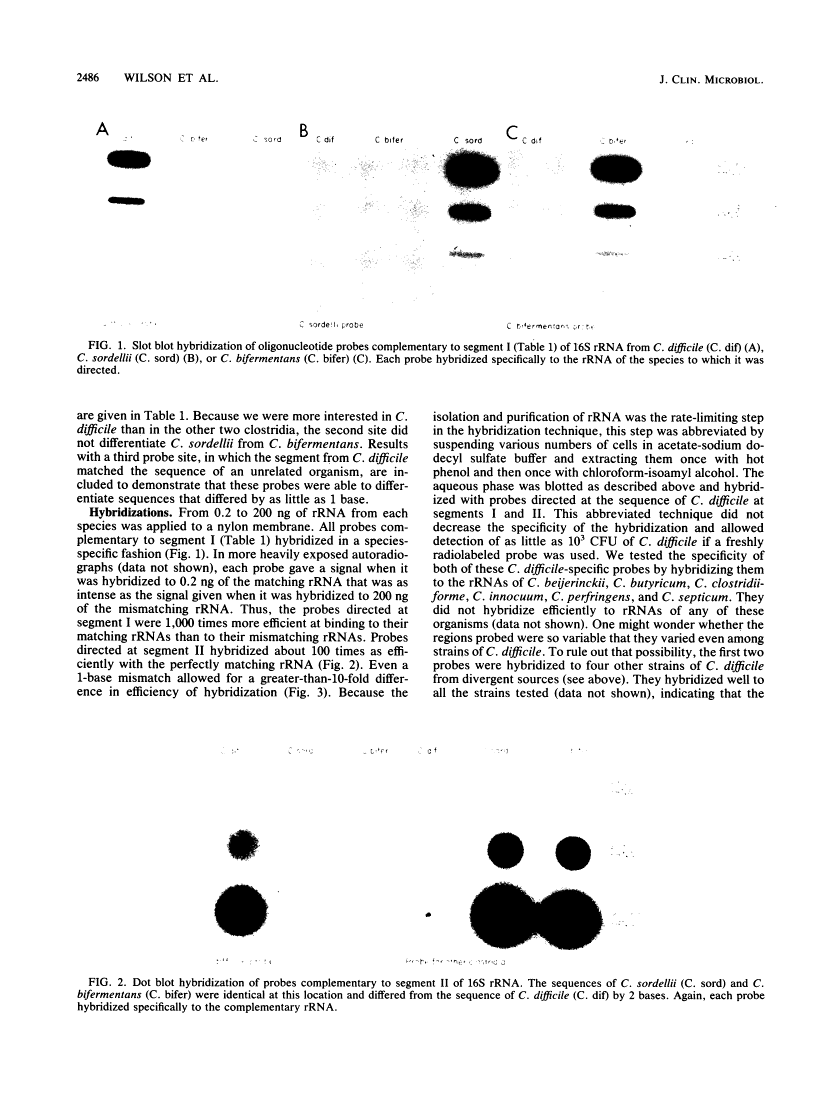
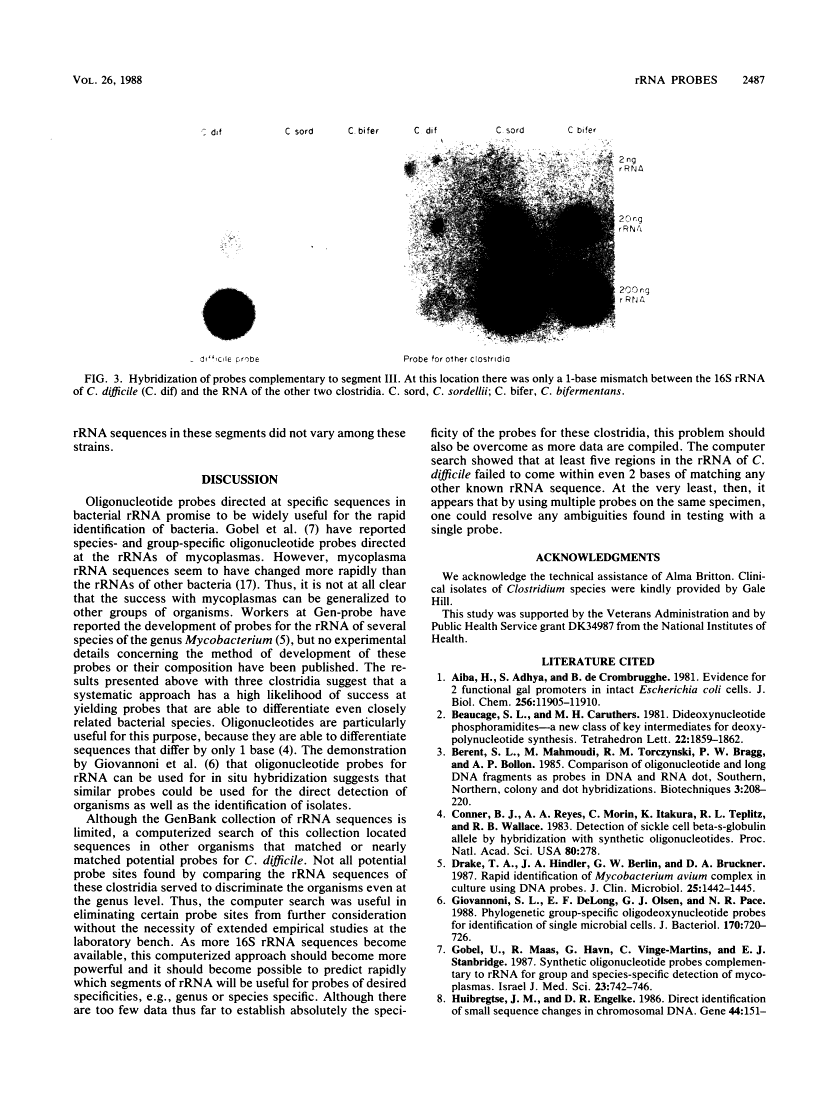
The large copy number of rRNA makes it an appealing target for oligonucleotide probes designed to identify microorganisms. Given that nucleotide sequences in rRNA are known to reflect phylogeny, species-specific rRNA probes should be feasible if the sequences found in closely related species are different. We sequenced portions of the 16S rRNA of three closely related clostridia found in the human colonic microflora: Clostridium bifermentans, C. sordellii, and C. difficile. The rRNAs of these three species showed 97 to 98% sequence similarity. Five oligonucleotide probes complementary to unique segments of the sequences were end labeled with 32P and hybridized on a nylon filter to the immobilized rRNA of each clostridium. Each probe efficiently hybridized only to the rRNA of the species to which it was directed. Complementary probes emitted a signal that exceeded by a factor of 100 to 1,000 the signal of probes that mismatched the target rRNA by 2 to 5 bases. Even a 1-base difference in rRNA sequence allowed a clear distinction between species. A systematic approach can efficiently yield taxon-specific oligonucleotide probes directed at rRNA.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.2M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981 Nov 25;256(22):11905–11910. [Abstract] [Google Scholar]
- Conner BJ, Reyes AA, Morin C, Itakura K, Teplitz RL, Wallace RB. Detection of sickle cell beta S-globin allele by hybridization with synthetic oligonucleotides. Proc Natl Acad Sci U S A. 1983 Jan;80(1):278–282. [Europe PMC free article] [Abstract] [Google Scholar]
- Drake TA, Hindler JA, Berlin OG, Bruckner DA. Rapid identification of Mycobacterium avium complex in culture using DNA probes. J Clin Microbiol. 1987 Aug;25(8):1442–1445. [Europe PMC free article] [Abstract] [Google Scholar]
- Giovannoni SJ, DeLong EF, Olsen GJ, Pace NR. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988 Feb;170(2):720–726. [Europe PMC free article] [Abstract] [Google Scholar]
- Göbel U, Maas R, Haun G, Vinga-Martins C, Stanbridge EJ. Synthetic oligonucleotide probes complementary to rRNA for group- and species-specific detection of mycoplasmas. Isr J Med Sci. 1987 Jun;23(6):742–746. [Abstract] [Google Scholar]
- Huibregtse JM, Engelke DR. Direct identification of small sequence changes in chromosomal DNA. Gene. 1986;44(1):151–158. [Abstract] [Google Scholar]
- Huysmans E, De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1986;14 (Suppl):r73–118. [Europe PMC free article] [Abstract] [Google Scholar]
- Johnson JL, Francis BS. Taxonomy of the Clostridia: ribosomal ribonucleic acid homologies among the species. J Gen Microbiol. 1975 Jun;88(2):229–244. [Abstract] [Google Scholar]
- Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6955–6959. [Europe PMC free article] [Abstract] [Google Scholar]
- Poxton IR, Byrne MD. Immunological analysis of the EDTA-soluble antigens of Clostridium difficile and related species. J Gen Microbiol. 1981 Jan;122(1):41–46. [Abstract] [Google Scholar]
- Tenover FC. Diagnostic deoxyribonucleic acid probes for infectious diseases. Clin Microbiol Rev. 1988 Jan;1(1):82–101. [Europe PMC free article] [Abstract] [Google Scholar]
- Woese CR. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from Journal of Clinical Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jcm.26.12.2484-2488.1988
Read article for free, from open access legal sources, via Unpaywall:
https://jcm.asm.org/content/jcm/26/12/2484.full.pdf
Free to read at intl-jcm.asm.org
http://intl-jcm.asm.org/cgi/content/abstract/26/12/2484
Free after 4 months at intl-jcm.asm.org
http://intl-jcm.asm.org/cgi/reprint/26/12/2484.pdf
Citations & impact
Impact metrics
Article citations
Simple lateral flow assays for microbial detection in stool.
Anal Methods, 10(45):5358-5363, 20 Sep 2018
Cited by: 8 articles | PMID: 31241058 | PMCID: PMC6253687
Inside Out: HIV, the Gut Microbiome, and the Mucosal Immune System.
J Immunol, 198(2):605-614, 01 Jan 2017
Cited by: 43 articles | PMID: 28069756
Review
Reprint of New opportunities for improved ribotyping of C. difficile clinical isolates by exploring their genomes.
J Microbiol Methods, 95(3):425-440, 16 Sep 2013
Cited by: 0 articles | PMID: 24050948
Review
New opportunities for improved ribotyping of C. difficile clinical isolates by exploring their genomes.
J Microbiol Methods, 93(3):257-272, 29 Mar 2013
Cited by: 3 articles | PMID: 23545446
Review
Nondigestible oligosaccharides enhance bacterial colonization resistance against Clostridium difficile in vitro.
Appl Environ Microbiol, 69(4):1920-1927, 01 Apr 2003
Cited by: 88 articles | PMID: 12676665 | PMCID: PMC154806
Go to all (22) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Sequence determination of rRNA genes of pathogenic Vibrio species and whole-cell identification of Vibrio vulnificus with rRNA-targeted oligonucleotide probes.
Int J Syst Bacteriol, 44(2):330-337, 01 Apr 1994
Cited by: 56 articles | PMID: 8186099
Use of subtractive hybridization to design habitat-based oligonucleotide probes for investigation of natural bacterial communities.
Appl Environ Microbiol, 64(1):185-191, 01 Jan 1998
Cited by: 7 articles | PMID: 9435075 | PMCID: PMC124691
The 16s/23s ribosomal spacer region as a target for DNA probes to identify eubacteria.
PCR Methods Appl, 1(1):51-56, 01 Aug 1991
Cited by: 140 articles | PMID: 1726852
Development and application of oligonucleotide probes for molecular identification of Xenorhabdus species.
Appl Environ Microbiol, 56(1):181-186, 01 Jan 1990
Cited by: 16 articles | PMID: 2310180 | PMCID: PMC183270