Abstract
Free full text

Somatic variants in epilepsy - advancing gene discovery and disease mechanisms
Abstract
In the last ten years, there has been increasing recognition that cells can acquire genetic variants during cortical development that can give rise to brain malformations as well as nonlesional focal epilepsy. These often brain tissue-specific, de novo variants can result in highly variable phenotypes based on the burden of a variant in specific tissues and cells. By discovering these variants, shared pathophysiological mechanisms are being revealed between clinically distinct disorders. Beyond informing disease mechanisms, mosaic variants also offer a powerful research tool to trace cellular lineages, to study the roles of specific cell types in disease presentation, and to establish the cell-type specific genomic consequences of a variant.
Introduction
Genome-wide genotyping and sequencing technologies facilitated the discovery of more than 100 specific epilepsy genes and revealed many novel aspects of the underlying genetic architecture. This work has revealed a significant role of de novo variants in the genetic risk of epilepsy. In fact, the vast majority of epilepsy genes identified to date were discovered either by sequencing trios to directly isolate de novo variants or by sequencing individual probands to ascertain the ultra-rare variants with dramatic effects on proteins critical to brain development that presumably arose de novo [1–5]. Most de novo variants associated with epilepsy were discovered in DNA derived from blood, and therefore likely to have arisen largely in parental gametes and transmitted to the germline of their offspring.
De novo variants can also arise post-zygotically and result in a mosaic organism if the cells continue to divide after the mutation occurs. The distribution and extent of mosaicism depends on the timing of the mutation. Mutations that happen post-zygotically within the first few cell divisions will result in the variant being present in the majority of the cells comprising the organism, whereas variants arising much later in embryogenesis after gastrulation may be found in only a small fraction of cells and localized to specific tissue or cell types. Studies have estimated that neural progenitors accumulate mutations at a rate of at least 1.6 per cell division [6], suggesting genetic risk for epilepsy may be in part conferred by brain tissue specific genetic variants that would be overlooked in traditional studies of leukocyte-derived DNA. This possibility is particularly plausible for focal epilepsy types where localization of the variant may in part explain the focal disease presentation. Refractory focal epilepsy, where surgical resections are relatively common, offers a unique opportunity to much more comprehensively study the role of de novo variants in epilepsy, including the analysis of post-zygotically acquired de novo variants localized to epileptogenic neural tissue. These collective observations and opportunities motivated extensive research in recent years towards the identification of the role of somatic variants in epilepsy disorders [7–11]. This review focuses on the recent discoveries of somatic variants in epilepsy, highlights novel insights gleaned from work in somatic genetics, and discusses future avenues to continue to advance the field.
Somatic mosaicism in epilepsy
Very early evidence for somatic mosaicism in epilepsy was identified in monozygotic twins who were discordant for Dravet syndrome, a severe sporadic disorder that is typically caused by de novo variants in SCN1A [12]. A pathogenic SCN1A variant was detected in all tissue types analyzed in the affected twin that was undetectable in the healthy twin, providing clear evidence for a very early post-zygotically acquired variant. Since this finding a number of early postzyotically-acquired variants have been identified in leukocyte-derived DNA in a range of epilepsy disorders. Somatic variants arising after the formation of the germ layers and enriched in the neural tissue have also been shown in a range of epilepsy disorders, and in particular brain malformations associated with seizures (Table 1) [2,3,13–16]. In some cases, the somatic variant is a second hit, in which a post-zygotically acquired variant arises in combination with an inherited variant, whereas in other instances only a single somatic variant is sufficient to give rise to the phenotype (Table 1).
Table 1.
Brain tissue localized/enriched somatic variants in epilepsy
| Epilepsy-associated disorder | Gene(s) | References |
|---|---|---|
| brain arteriovenous malformation | BRAF, KRAS, MAP2K1, NRAS | [17–19] |
| cerebral cavernous malformation | CCM2*, KRIT*, PDCD10* | [20–22] |
| focal cortical dysplasia type Ia/mild malformation of cortical development | SLC35A2 | [23–25] |
| focal cortical dysplasia type IIb/hemimegalencephaly | ATK1, AKT3, DEPDC5*, MTOR, PIK3CA, RHEB, TSC1*, TSC2* | [24,26–32] |
| hypothalamic hamartoma | DYNC2H1*, GLI3, OFD1, PRKACA, PTPN11 | [33–36] |
| non-lesional epilepsy (radiographically non-lesional) | SLC35A2 | [23–25] |
| Sturge-Weber syndrome/leptomeningeal angiomatosis | GNAQ | [37–39] |
Focal cortical dysplasia Type II (FCD II)
Brain malformations with FCD II pathology, including hemimegalencephaly, represent the best documented examples of lesional epilepsy caused by somatic variants. The pathogenic somatic variants identified in brain malformations with FCD II pathology affect components of the PI3K-AKT-mTOR signaling pathway (Table 1) [24,26–32]. Prior to identification of the somatic variants in these disorders, a shared histopathological phenotype had been observed across these brain malformations, including the presence of cytomegalic neurons and cortical dyslamination [40]. The identification of somatic variants across PI3K-AKT-mTOR signaling pathway genes, many of which were proven to activate mTOR signaling in independent model system, validated the suggested shared pathophysiology. The majority of the somatic variants identified in the PI3K-AKT-mTOR signaling pathway are exclusively present or are greatly enriched in the malformed brain regions compared to blood [10].
Hypothalamic hamartomas
Brain tissue localized variants have also been identified in hypothalamic hamartomas (HH), noncancerous tumors of the hypothalamus associated with intractable gelastic seizures. The first somatic variants to be discovered in HH were found in GLI3, a gene encoding a key protein in the Sonic Hedgehog signaling pathway [34,36]. Somatic variants in GLI3 in HH tissue were confirmed in subsequent studies along with additional somatic variants in genes implicated the Hedgehog signaling pathway, including PRKACA and OFD1 [33,35]. In one study, thirty-eight cases were found to have a somatic variant that disrupted a Hedgehog signaling gene, although it should be noted that many of the variants were large multi-genic deletions that also included many genes not known to be involved in the Hedgehog signaling pathway that could contribute to the phenotype [35]. More recently, HH cases with two-hit (germline and somatic) variants were identified in DYNC2H1, encoding a key protein in the dynein-2 complex critical for the formation of cilia [41]. This study also identified somatic variants in PTPN11, encoding a protein involved in the RAS/mitogen-activated protein kinase (MAPK) pathway that also contributes to signaling in the primary cilium [41]. This suggests that disruption of Hedgehog signaling in HH may be specifically related to its role in proper cilia development functioning, and that the mechanisms underlying HH may be attributed more broadly to ciliary defects through multiple different pathways.
Interestingly, one group showed that cortical dyslamination caused by somatic MTOR variants in FCD II patients may be caused in part by defective neuronal ciliogenesis resulting from impaired autophagy [42]. Furthermore, a recurrent early post-zygotically-acquired somatic mutation in SMO that leads to constitutive activation of Hedgehog signaling has been identified in Curry Jones Syndrome, associated with multiple congenital malformations that sometimes includes hemimegalencephaly [43]. Collectively, this suggests that abnormal cilia formation and activity may be critical across a range of brain malformations associated with epilepsy, and not just limited to HH.
Brain arteriovenous malformations
Cerebral vascular malformations are another group of epilepsy associated lesions attributed to mosaic pathogenic variants. Somatic variants have been identified in the MAPK signaling pathway specifically as the cause of arteriovenous malformations [17–19]. Cerebral cavernous malformations are thought to arise from a combined somatic and inherited variant, two-hit model, involving CCM2, KRIT, and PDCD10 [20–22]. Sturge Weber, a sporadic neurocutaneous syndrome associated with leptomeningeal angiomatosis, port-wine stain, and vascular glaucoma, has been associated with a brain tissue enriched recurrent variant in GNAQ [37–38]. This recurrent variant was also later identified in isolated leptomeningeal angiomatosis [39].
One of the most recent somatic variant discoveries in epilepsy was found in SLC35A2 in individuals with radiographically non-lesional focal epilepsy sometimes with FCD Ia pathology, also referred to as a “mild malformation of cortical development” [23–25]. Across the three independent studies reporting mosaic SLC35A2 variants in intractable neocortical epilepsy, more than 15% of individuals undergoing surgical resection for either non-lesional focal epilepsy or a mild malformation of cortical development had a pathogenic somatic variant in this gene. SLC35A2, located on the X chromosome, encodes a UDP-Gal transporter that moves cytosolic UDP-Gal into the Golgi apparatus to serve as a substrate in glycan formation for the post-translational modification of cellular proteins and lipids. De novo germline or very early post-zygotically acquired SLC35A2 variants had been implicated in congenital disorders of glycosylation and in a rare form of X-linked developmental and epileptic encephalopathy [44,45]. This discovery suggests that glycosylation defects may explain a much larger fraction of intractable neocortical epilepsy that previously thought. As there was no evidence of activated mTOR-signaling in any of the individuals with a mosaic SLC35A2 variant, the mechanism is likely novel and independent of that implicated in FCD II associated phenotypes [23–25].
Tissue and cellular burden contribute to phenotypic variability in mosaic epilepsy
There is extensive discussion in the genetics community about the reasons for the highly variable phenotypic spectrum associated with nearly all genes implicated in neurodevelopmental disorders. These reasons include the potential contributions of modifier loci, differential effects of the variants on the encoded protein, and influential environmental factors. While all possible factors contributing to variable expressivity for germline variants also apply in somatic genetics, there is additional complexity in the case of post-zygotically acquired variants in that tissue and cell-type specific burden also likely contribute to the variability in phenotype (Figure 1).
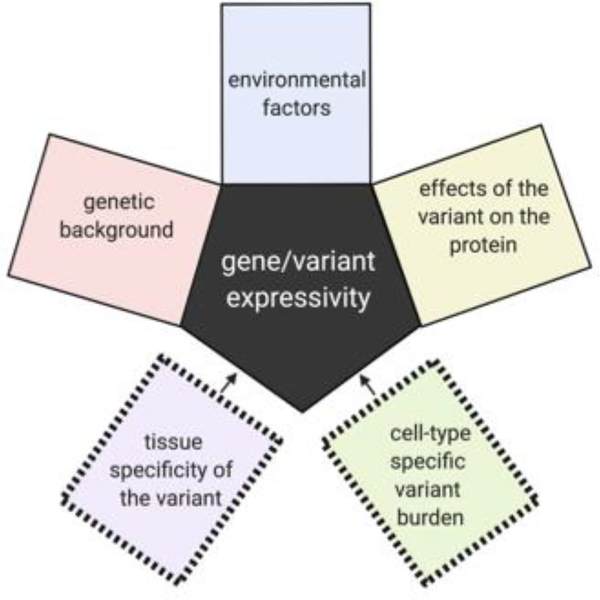
Additional complexities affecting the variable expressivity of genes harboring pathogenic post-zygotically acquired genetic variants.
Somatic genetics has revealed multiple instances of how tissue distribution of post-zygotically acquired variants can lead to different disease presentations. GLI3 variants in the germline give rise to Pallister Hall, a syndrome that includes multiple congenital anomalies including HH [46], whereas variants restricted to (or enriched in) the hypothalamus can result in isolated HH. PIK3CA variants can lead to megalencephaly-polymicrogyria-polydactyly-hydrocephalus (MPPH) or congenital lipomatous overgrowth, vascular malformations, and epidermal nevi (CLOVE) syndromes when the mutations arise in the germline or early post-zygotic development, whereas variants in these genes that are acquired later result in hemimegalencephaly [32,47]. Beyond the more obvious tissue localization effects of the variant on phenotype, overall burden within the region with the variant has also been shown to contribute to severity or extent of the clinical presentation for some genes. For hemimegalencephaly, the mutation burden of PI3K-AKT-mTOR pathway mutations tended to be higher compared to that found in more isolated FCD II presentations [27]. A similar pattern is observed for somatic SLC35A2 associated epilepsy where radiographically non-lesional epilepsy patients had on average 6% variant allele burden compared to 22% on average for mild malformations of cortical development [23–25]. In one study that evaluated five individuals with brain-tissue specific SLC35A2 variants, samples with the lowest variant allele burden also showed no evidence of cognitive impairment and later ages of onset compared to that observed in either cases with higher variant burden or cases with germline de novo variants in SLC35A2 (Figure 2). It should be noted, however, that analyses of the landscape of mutation burden across the epileptogenic or dysplastic region are extremely challenging in human tissue given than only small sections are available for research. Much more comprehensive studies in in vitro or ex-vivo analyses of resected tissue would be required to make definitive conclusions about the relationships of variant burden and localization to the clinical phenotype.
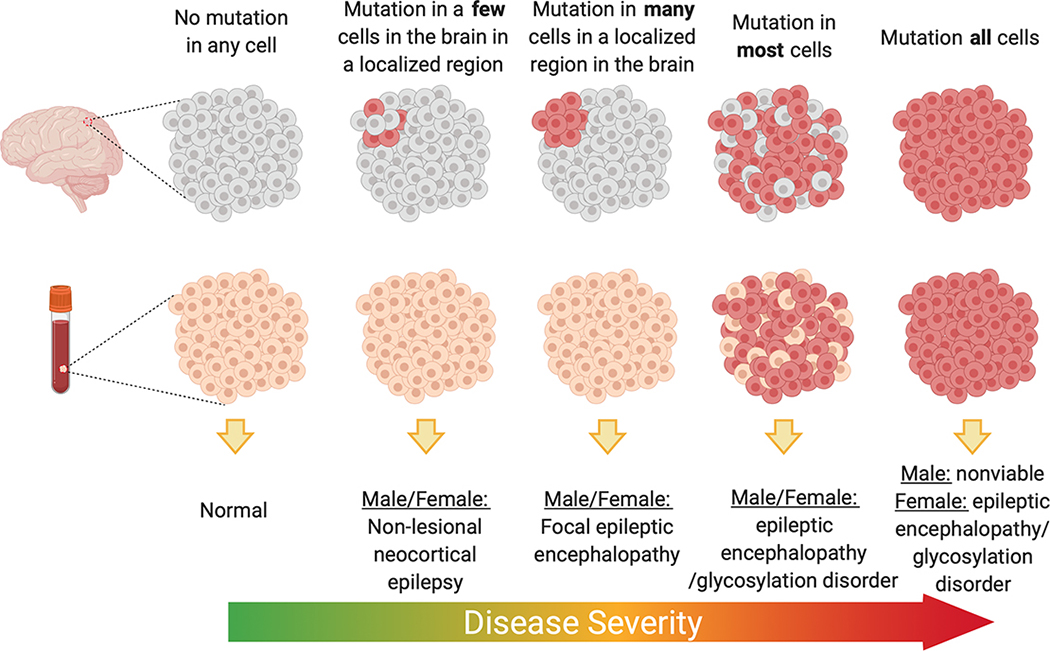
Suggested model for SLC35A2 epilepsy whereby variant burden and localization dictate phenotypic severity. Figure made with Biorender.com
In addition to overall burden of mutation, there are several examples where the effects of cell-type-specific variant burden has been explored. For a recurrent pathogenic KRAS variant in arteriovenous malformations, the variants are highly enriched in brain endothelial cells and virtually absent from neurons [19]. For hemimegalencephaly and FCD, the somatic variants in the PI3K-AKT-mTOR pathway are always found in neurons and in some cases also present in glia, indicating that neuronal involvement is critical to pathogenesis [30]. Notably, both of these studies demonstrate how somatic variants can also be used more generally a research tool to study the role of various cell types in diseases.
While the effects of variant burden and cellular distribution have only been explored to a very limited extent, the expectation is that variant enrichment in specific brain cell types in conjunction with cell-specific burden will be major contributors to the individual’s phenotype and the phenotypic spectrum associated with a gene harboring somatic variants (Figure 1).
Challenges and explaining the unexplained
Despite remarkable progress in somatic genetics in epilepsy, many cases of presumed somatic epilepsies remain genetically unexplained. For hemimegalencephaly where there is a high likelihood of a pathogenic somatic variant, the percent explained across all published studies approaches only 20% [10]. For FCD II, it is less than 15% [10]. The high fraction of genetically unexplained are likely due to three major contributing factors. The first is the lack of data on the genetic landscape of somatic variants in the normal brain limiting the ability to distinguish benign and pathogenic somatic variation in abnormal brains. It is likely that a significant number of pathogenic variants have been identified that are uninterpretable without more information about how exceptional this type of variant is in normal human brain tissue.
Similar to germline genetic variant detection, the field of somatic genetics is also limited by the technological constraints of short read exome sequencing, specifically the inability to comprehensively detect structural variants, non-coding variants, and short tandem repeat variation. Short tandem repeats are highly mutable DNA sequences that have been associated with many neurological disorders. Given their high potential for mutability, short tandem repeats will likely be important de novo contributors to the genetics of epilepsy both in terms of somatic and germline risk. While in the short term there are budgetary constraints to large scale deployment, long-read sequencing will likely ultimately advance somatic gene discovery in epilepsy by allowing more comprehensive detection of genetic variants (e.g., short tandem repeats, structural variants) and long-range haplotypes that have been largely overlooked by short-read next-generation sequencing technology, the mainstay of studies performed to date.
The third aspect that is likely contributing to the large unexplained fraction in genetic epilepsies due to somatic variation is the inability to detect very low burden variants in bulk tissue and the challenges of determining which cells to pursue for single cell analyses. Single cell DNA and RNA sequencing has experienced tremendous technical advancement in recent years making genome-wide analyses much more affordable. These advancements likely will allow this limitation to be overcome in the near future.
Conclusions
Revealing pathogenic somatic variants concealed in human brain tissue has proven to be a impactful approach to identifying new pathways involved in epilepsy pathogenesis and to linking known genes to novel phenotypes. Beyond this obvious advancement, somatic variation serves as endogenous barcodes and offers an extremely valuable tool for retrospective tracing of cellular origins and fate during brain and tumor development [48–51]. Somatic mutations also enable a powerful means to study the downstream effects of a variant in cells from the same individual, correcting for potential confounding factors that exists when comparing across individuals. Coupling single cell genetic and genomic analyses will likely provide high resolution assessments of the impact of genetic variants in individual cell types in the future. While there remains much work to be done to continue to further somatic gene discovery in the epilepsies and learn about the somatic mutation landscape in the brain, the field is technologically poised to make significant advancements in the near future.
Acknowledgements
The author would like to thank Dulcie Lai, Tristan Sands, and Jinfeng Lu for the helpful comments and discussions. ELH is supported by the NINDS (R01-NS094596), and previously by NINDS (R01-NS089552) and by the Irving Institute for Clinical and Translational Research at Columbia University.
Footnotes
Competing Interests
The author declares no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
* The Epi25 Collaborative reports the results of the largest exome sequencing study performed to date oncomparing 9,170 individuals with epilepsy, including individuals with developmental and epileptic encephalopathies, genetic generalized epilepsy, and nonacquired focal epilepsy, to ~8,500 controls. In all types of epilepsy there was an enrichment of ultra-rare deleterious variants in genes intolerant to functional variation suggesting that ultra-rare variants contribute to epilepsy risk. This study confirms the findings from an earlier study showing overlapping genetic contributors in both rare and common epilepsies.
* Nikolaev et al. identified somatic activating KRAS variants in >60% of individuals with arteriovenousmalformations studied. These variants were found to be selective enriched in endothelial cells populations suggesting that MAPK-ERK pathway activation in endothelial cells leads to arteriovenous malformations.
** This study reported the discovery of somatic SLC35A2 variants in both nonlesional focal epilepsy and individuals with a mild malformation of cortical development, implicating glycosylation pathway as a cause for intractable focal epilepsy. Seventeen percent of cases studied with nonlesional focal epilepsy were found to have a somatic variant in SLC35A2.
** This study showed using next-generation sequencing that the same mutations are implicated in both FCD II and hemimegalencephaly depending on the timing of the mutational event. All individuals studied were found to have the variants in neurons suggesting that neuronal involvement is critical to the pathology. In a mouse model they were able to show that activation of mTOR in excitatory neurons specifically was associated with the development cortical dysplasia.
* Park et al. showed that human resected brain tissue from individuals with an FCD II associated brainmalformations harboring a somatic MTOR variant showed defective neuronal ciliogenesis. This finding was confirmed in a mouse model and in vitro using a human cell line. The MTOR variant associated altered ciliogenesis was found to be due disruptions in autophagy leading to accumulation of OFD1 at centriolar satellites; and that defective ciliogenesis accounted for the observed cortical dyslamination.
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.gde.2020.04.004
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7666655
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.gde.2020.04.004
Article citations
Somatic mosaicism in schizophrenia brains reveals prenatal mutational processes.
Science, 386(6718):217-224, 10 Oct 2024
Cited by: 0 articles | PMID: 39388546 | PMCID: PMC11490355
Emerging roles of long non-coding RNAs in human epilepsy.
Epilepsia, 65(6):1491-1511, 30 Apr 2024
Cited by: 1 article | PMID: 38687769
Review
Open Sesame: Door to Enriched Somatic Variants Underlying Sporadic mTLE.
Epilepsy Curr, 23(6):383-385, 28 Sep 2023
Cited by: 0 articles | PMID: 38269352
Toward the use of novel alternative methods in epilepsy modeling and drug discovery.
Front Neurol, 14:1213969, 31 Aug 2023
Cited by: 3 articles | PMID: 37719765 | PMCID: PMC10501616
Review Free full text in Europe PMC
GWAS meta-analysis of over 29,000 people with epilepsy identifies 26 risk loci and subtype-specific genetic architecture.
Nat Genet, 55(9):1471-1482, 31 Aug 2023
Cited by: 32 articles | PMID: 37653029 | PMCID: PMC10484785
Go to all (9) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The role of somatic mutational events in the pathogenesis of epilepsy.
Curr Opin Neurol, 32(2):191-197, 01 Apr 2019
Cited by: 4 articles | PMID: 30762606 | PMCID: PMC6424360
Review Free full text in Europe PMC
Dissecting the genetic basis of focal cortical dysplasia: a large cohort study.
Acta Neuropathol, 138(6):885-900, 23 Aug 2019
Cited by: 129 articles | PMID: 31444548 | PMCID: PMC6851393
Pathway-driven discovery of epilepsy genes.
Nat Neurosci, 18(3):344-350, 24 Feb 2015
Cited by: 98 articles | PMID: 25710836 | PMCID: PMC4852130
Review Free full text in Europe PMC
Somatic variants in new candidate genes identified in focal cortical dysplasia type II.
Epilepsia, 61(4):667-678, 26 Mar 2020
Cited by: 11 articles | PMID: 32216069
Funding
Funders who supported this work.
Irving Institute for Clinical and Translational Research
NINDS (2)
Grant ID: R01-NS089552
Grant ID: R01-NS094596
NINDS NIH HHS (2)
Grant ID: R01 NS094596
Grant ID: R01 NS089552





