Abstract
Free full text

The Ethanolamine-Sensing Transcription Factor EutR Promotes Virulence and Transmission during Citrobacter rodentium Intestinal Infection
ABSTRACT
Enteric pathogens exploit chemical and nutrient signaling to gauge their location within a host and control expression of traits important for infection. Ethanolamine-containing molecules are essential in host physiology and play important roles in intestinal processes. The transcription factor EutR is conserved in the Enterobacteriaceae and is required for ethanolamine sensing and metabolism. In enterohemorrhagic Escherichia coli (EHEC) O157:H7, EutR responds to ethanolamine to activate expression of traits required for host colonization and disease; however, the importance of EutR to EHEC intestinal infection has not been examined. Because EHEC does not naturally colonize or cause disease in mice, we employed the natural murine pathogen Citrobacter rodentium as a model of EHEC virulence to investigate the importance of EutR in vivo. EHEC and C. rodentium possess the locus of enterocyte effacement (LEE), which is the canonical virulence trait of attaching and effacing pathogens. Our findings demonstrate that ethanolamine sensing and EutR-dependent regulation of the LEE are conserved in C. rodentium. Moreover, during infection, EutR is required for maximal LEE expression, colonization, and transmission efficiency. These findings reveal that EutR not only is important for persistence during the primary host infection cycle but also is required for maintenance in a host population.
INTRODUCTION
Nutrient and chemical signaling are fundamental for all cellular processes, including interactions between the mammalian host and the microbiota, which have a significant impact on health and disease. Bacteria respond to metabolites within a host to enhance growth and regulate expression of traits important for colonization (1). Ethanolamine (EA) is a component of phosphatidylethanolamine, an abundant and essential phospholipid in the cell membrane. The turnover of enterocytes and bacteria contributes to a continuously replenished source of EA in the gastrointestinal (GI) tract (2,–4), suggesting that EA sensing is a reliable means of gauging the host intestinal environment. Indeed, in vitro and in vivo studies have revealed that EA influences growth and/or gene expression of diverse commensal and pathogenic bacteria (5,–14).
The role of EA in activating virulence gene expression was first described for enterohemorrhagic Escherichia coli O157:H7 (EHEC) (15,–17). EHEC is a foodborne pathogen that colonizes the colon and causes major outbreaks of bloody diarrhea and hemolytic-uremic syndrome (18). After ingestion, EHEC travels through the GI tract; upon reaching the colon, EHEC expresses virulence factors, including Shiga toxin and the locus of enterocyte effacement (LEE) pathogenicity island. Stx is a potent inhibitor of protein synthesis (19) that is responsible for the severe morbidity and mortality associated with EHEC (19). The LEE encodes a type III secretion system (T3SS) and most effectors required for formation of attaching and effacing lesions on enterocytes (20,–27). AE lesions are characterized by effacement of microvilli and intimate attachment of EHEC to epithelial cells (28).
EHEC senses EA through the transcription factor EutR. EutR directly activates expression of the LEE and promotes expression of other regulatory factors and adhesins that contribute to virulence expression and adherence to epithelial cells (15,–17). To date, the importance of EutR to EHEC virulence during infection of the mammalian GI tract has not been tested. This is mainly due to the lack of convenient animal models that recapitulate EHEC infection. In this study, we employed Citrobacter rodentium to examine the role of EutR in LEE expression and colonization during intestinal infection. C. rodentium is a natural murine pathogen that colonizes the cecum and distal colon in mice (29) and is commonly used as a murine model of EHEC infection (29,–31). C. rodentium carries the LEE and forms AE lesions on epithelial cells (32). We demonstrate that EutR-dependent EA sensing and concomitant regulation of the LEE are conserved in EHEC and C. rodentium. Notably, EutR is required for robust C. rodentium colonization of the colon and transmission efficiency. These data underscore the importance of optimal control of virulence expression in pathogen transmission and persistence in a host population.
RESULTS AND DISCUSSION
C. rodentium possesses a functional eut operon.
In the Enterobacteriaceae, the eut locus is comprised of 17 genes (Fig. 1A) that code for transport and catabolism of EA, as well as a microcompartment that contains toxic breakdown products of EA metabolism (33,–38). Despite the availability of EA in the intestine, at least a subset of enteric E. coli strains have acquired a large phage insertion between the genes encoding the EA ammonia lyase EutBC, resulting in the inability to metabolize EA (17, 39). Our data demonstrate that C. rodentium grows using EA as a nitrogen source (Fig. 1B), indicating that C. rodentium possesses a functional eut locus.
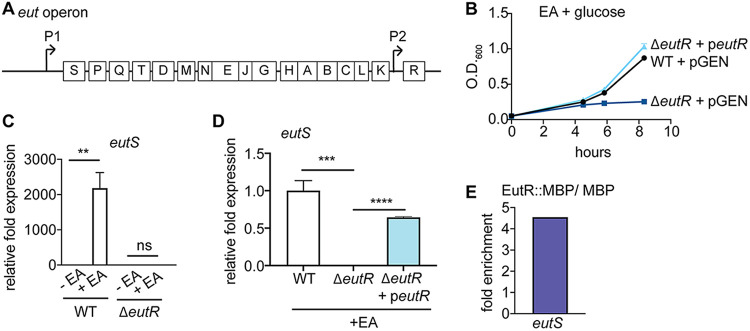
C. rodentium possesses a functional eut operon. (A) Schematic of the eut locus. (B) Growth curve of WT, ΔeutR, and complemented C. rodentium strains grown in minimal medium with EA (n =
= 3). (C) RT-qPCR of eutS expression in WT and ΔeutR
C. rodentium strains grown in the absence or presence of EA (n
3). (C) RT-qPCR of eutS expression in WT and ΔeutR
C. rodentium strains grown in the absence or presence of EA (n =
= 3). (D) RT-qPCR of eutS expression in WT, ΔeutR, and complemented C. rodentium strains grown in the presence of EA (n
3). (D) RT-qPCR of eutS expression in WT, ΔeutR, and complemented C. rodentium strains grown in the presence of EA (n =
= 3). (E) qPCR showing enrichment of eutS from in vivo ChIP of EutR::MBP compared to MBP (n
3). (E) qPCR showing enrichment of eutS from in vivo ChIP of EutR::MBP compared to MBP (n =
= 2). Error bars represent standard deviations (SD). **, P
2). Error bars represent standard deviations (SD). **, P <
< 0.01; ***, P
0.01; ***, P <
< 0.001; ****, P
0.001; ****, P <
< 0.0001. ns, not significant.
0.0001. ns, not significant.
The last gene in the eut locus encodes the transcriptional regulator EutR, which is 96% similar at the amino acid level between EHEC and C. rodentium (see Fig. S1 in the supplemental material). To examine how EutR functions in C. rodentium, we generated an eutR deletion strain. The ΔeutR C. rodentium strain was unable to grow using EA as a nitrogen source, and this defect could be complemented when eutR was expressed in trans (Fig. 1B). The ΔeutR growth defect was specific to EA utilization, as the wild-type (WT) and ΔeutR strains grew similarly in LB (Fig. S2). Genetic and biochemical studies with Salmonella and EHEC demonstrated that in response to EA and adenosylcobalamin, EutR promotes EA metabolism by binding to the eutS promoter to activate eut expression (17, 34, 36). Consistent with these data, in C. rodentium, EutR was required to sense EA and promote eut expression (Fig. 1C and andD).D). Moreover, the eutS promoter of C. rodentium contains the EutR recognition sequence (17, 40). To further examine EutR regulation of the eut locus, we generated a plasmid expressing C. rodentium EutR fused to maltose-binding protein (MBP). After growth of the ΔeutR strain carrying the EutR::MBP or the empty MBP vector, EutR-DNA interactions were assessed using in vivo chromatin immunoprecipitation (ChIP). Using quantitative PCR (qPCR), we measured an enrichment of eutS in EutR::MBP samples compared to MBP alone (Fig. 1E). These data indicate that C. rodentium has maintained the ability to metabolize EA and that EutR is required for EA sensing and eut expression.
EutR promotes EA-dependent LEE expression in C. rodentium.
EutR is important for EHEC to sense EA and activate expression of the LEE (16, 17). The LEE includes five major operons (41,–43) (Fig. 2A). The T3SS filament protein is encoded by espA, which is carried in LEE4. Significantly, EA promoted C. rodentium virulence by enhancing expression of espA (Fig. 2B). Moreover, EutR was required for C. rodentium to sense EA and activate LEE expression, as the ΔeutR strain was unresponsive to the addition of EA to the culture medium (Fig. 2B). ler is the first gene within the LEE1 operon (Fig. 2A) and encodes the master regulator of the LEE (43,–46). Consistent with the case with EHEC, EutR was required for EA-dependent ler expression (Fig. 2C), and complementation of the ΔeutR strain restored LEE expression similar to WT levels (Fig. 2C).
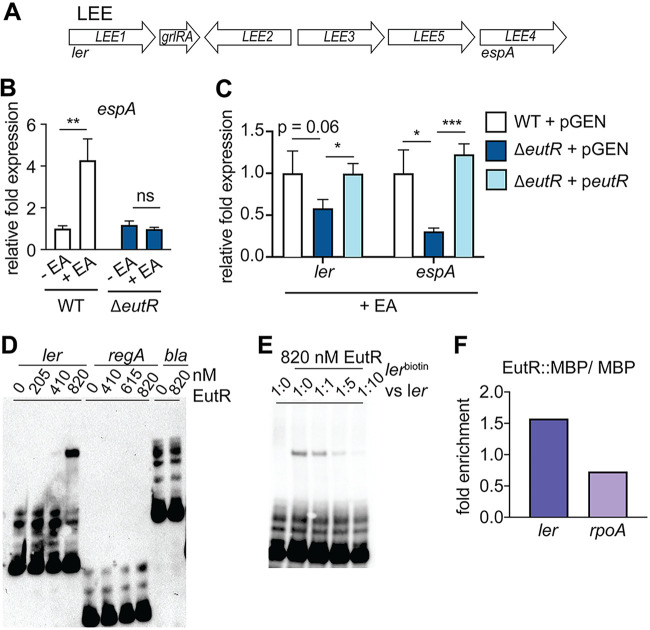
EA sensing and EutR-dependent LEE expression in C. rodentium. (A) Schematic of the LEE pathogenicity island. (B) RT-qPCR of espA expression in WT and ΔeutR
C. rodentium in the absence or presence of EA (n =
= 3). (C) RT-qPCR of ler and espA expression in WT, ΔeutR, and complemented C. rodentium strains grown in the presence of EA (n
3). (C) RT-qPCR of ler and espA expression in WT, ΔeutR, and complemented C. rodentium strains grown in the presence of EA (n =
= 3). (D) EMSA of ler, regA, and bla with EutR::MBP. (E) Competition EMSA using biotin-labeled and/or unlabeled ler probes. Ratios compare amount of labeled probe to amount of unlabeled probe. (F) qPCR showing enrichment of ler and rpoA from in vivo ChIP of EutR::MBP compared to MBP (n
3). (D) EMSA of ler, regA, and bla with EutR::MBP. (E) Competition EMSA using biotin-labeled and/or unlabeled ler probes. Ratios compare amount of labeled probe to amount of unlabeled probe. (F) qPCR showing enrichment of ler and rpoA from in vivo ChIP of EutR::MBP compared to MBP (n =
= 2). Error bars represent SD. *, P
2). Error bars represent SD. *, P <
< 0.05; **, P
0.05; **, P <
< 0.01; ***, P
0.01; ***, P <
< 0.001.
0.001.
To examine EutR interaction with the ler promoter, we purified EutR::MBP and performed electrophoretic mobility shift assays (EMSAs) using biotinylated ler, regA (previously shown to influence LEE expression in C. rodentium [47]), and bla (negative control) probes. These assays indicated that EutR directly binds the ler promoter to activate expression of the LEE in a specific manner, as EutR did not bind either of the control probes regA and bla (Fig. 2D). Furthermore, MBP alone did not shift the ler promoter (Fig. S3). To confirm specificity of EutR interaction with the ler promoter, we performed a competition EMSA. Addition of an equal concentration of unlabeled ler probe competed with the lerbiotin probe for EutR binding, as visualized by decreased band intensity in shifted DNA compared to that for labeled probe alone (Fig. 2E). Furthermore, when added at increasing concentrations, the unlabeled probe completely outcompeted the labeled ler probe for EutR binding (Fig. 2E). To substantiate the in vitro data, we performed in vivo ChIP. In these experiments, we measured an enrichment of ler in EutR::MBP samples compared to MBP alone as well as compared to the negative control rpoA (Fig. 2F). These data demonstrate that EutR-dependent regulation of LEE expression is conserved in EHEC and C. rodentium.
EutR promotes C. rodentium ler expression during infection.
The in vitro studies identified the LEE genes as EutR regulatory targets. We next tested the importance of EutR in controlling ler expression during intestinal infection. To do this, we constructed a transcriptional reporter in which the ler promoter was cloned upstream of the bacterial luciferase genes (Fig. S4A) and verified that this plasmid was retained during infection (Fig. S4B). At 10 days postinfection (dpi), ler expression was significantly decreased in the ΔeutR strain compared to that in the WT (Fig. 3A and andB).B). To substantiate these data and control for potential differences in bacterial colonization, we infected mice with the WT or ΔeutR strain and harvested tissue at 10
days postinfection (dpi), ler expression was significantly decreased in the ΔeutR strain compared to that in the WT (Fig. 3A and andB).B). To substantiate these data and control for potential differences in bacterial colonization, we infected mice with the WT or ΔeutR strain and harvested tissue at 10 dpi for transcriptional analyses of ler expression normalized to 16S rRNA expression. These data were consistent with data shown in Fig. 2, in which the eutR deletion resulted in a statistically significant (~2-fold) decrease in ler expression (Fig. 3C). Altogether, these data indicate that EutR is important for sensing the host environment and promoting LEE expression in the GI tract. Moreover, the magnitude differences in ler gene expression between WT and ΔeutR measured in whole colons versus normalized to C. rodentium CFU suggest that EutR affects pathogen burden in the intestine.
dpi for transcriptional analyses of ler expression normalized to 16S rRNA expression. These data were consistent with data shown in Fig. 2, in which the eutR deletion resulted in a statistically significant (~2-fold) decrease in ler expression (Fig. 3C). Altogether, these data indicate that EutR is important for sensing the host environment and promoting LEE expression in the GI tract. Moreover, the magnitude differences in ler gene expression between WT and ΔeutR measured in whole colons versus normalized to C. rodentium CFU suggest that EutR affects pathogen burden in the intestine.
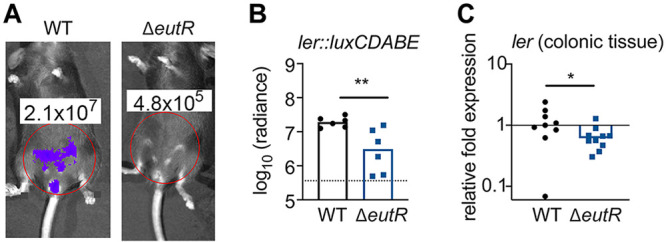
EutR enhances C. rodentium virulence gene expression in vivo. (A) Representative bioluminescent image of ler expression in mice infected with WT or ΔeutR
C. rodentium at 10 dpi. (B) Quantification of bioluminescent data. (C) RT-qPCR of ler expression in WT and ΔeutR
C. rodentium associated with colonic tissue. *, P
dpi. (B) Quantification of bioluminescent data. (C) RT-qPCR of ler expression in WT and ΔeutR
C. rodentium associated with colonic tissue. *, P <
< 0.05; **, P
0.05; **, P <
< 0.01.
0.01.
EutR is required for maximal C. rodentium fitness in the intestinal tract.
The C. rodentium infection cycle in C57BL/6 mice is characterized by initial low levels of C. rodentium, followed by proliferation, and steady state, before clearance (Fig. 4A) (29). To examine how EutR affects C. rodentium colonization, we determined numbers of WT and ΔeutR strain CFU shed in stool throughout the infection cycle. Similar levels of ΔeutR and WT C. rodentium CFU were measured during the initial stages of infection. However, starting at 8 dpi and continuing until the end of the experiment, the ΔeutR strain was recovered at lower numbers than the WT (Fig. 4B). These findings reveal an important role for EutR in promoting colonization and persistence in the mammalian GI tract.
dpi and continuing until the end of the experiment, the ΔeutR strain was recovered at lower numbers than the WT (Fig. 4B). These findings reveal an important role for EutR in promoting colonization and persistence in the mammalian GI tract.
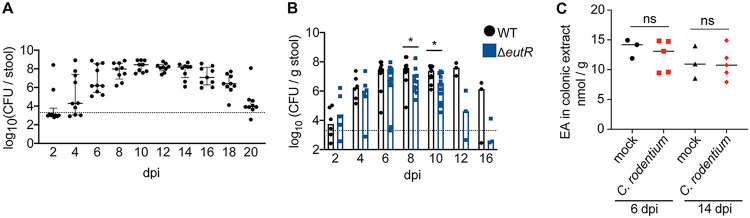
EutR is required for robust C. rodentium colonization independent of changes in EA availability. (A) CFU of WT C. rodentium shed in stool during infection. Data are presented as medians ± interquartile ranges. (B) CFU of WT and ΔeutR
C. rodentium shed in stool. The dotted line represents the limit of detection. (C) EA levels in colons of mock-infected or WT C. rodentium-infected mice. *, P <
< 0.05.
0.05.
C. rodentium infection might affect EA levels in the intestine, which could be a contributing factor in the differences in colonization between the WT and ΔeutR strains measured during later stages of infection. To examine this possibility, we harvested whole colons from mock- or C. rodentium-infected mice at 6 and 14 dpi and measured EA levels in the tissue homogenates using liquid chromatography-mass spectrometry (LC-MS). At each time point, similar EA concentrations were measured from the colons of mice infected with 109 CFU of C. rodentium and mock-infected mice (Fig. 4C). These data suggest that C. rodentium infection does not alter EA concentrations in the intestine and that differences in WT and ΔeutR strain colonization are not due to changes in EA availability in the colon.
dpi and measured EA levels in the tissue homogenates using liquid chromatography-mass spectrometry (LC-MS). At each time point, similar EA concentrations were measured from the colons of mice infected with 109 CFU of C. rodentium and mock-infected mice (Fig. 4C). These data suggest that C. rodentium infection does not alter EA concentrations in the intestine and that differences in WT and ΔeutR strain colonization are not due to changes in EA availability in the colon.
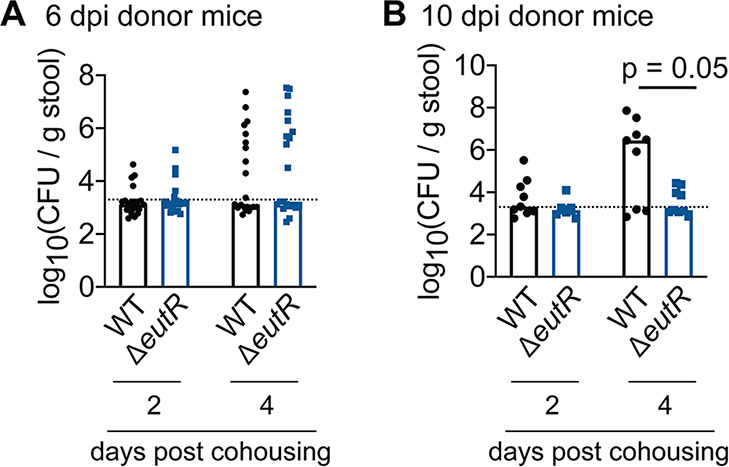
EutR is important for transmission efficiency.
Passage through the GI tract results in a hypertransmissible state in which significantly fewer C. rodentium CFU are sufficient to cause infection than for bacteria cultivated in vitro (48). In C57BL/6 mice, ingestion of 107 to 108 CFU of freshly shed bacteria results in infection (48), compared to 109 CFU of C. rodentium grown in broth. At 10 dpi, the median CFU of the ΔeutR strain shed in stool was ~10-fold lower than the reported threshold that results in efficient transmission between animals. Therefore, we next investigated how EutR affects transmission. Following 24 h of cohousing with 6-dpi donor mice, equal numbers of WT and ΔeutR
C. rodentium organisms were recovered in stool from recipient mice (Fig. 5A). In contrast, following cohousing with 10-dpi donor mice, the ΔeutR strain was recovered at significantly lower levels in stool of (formerly) naive mice (Fig. 5B and Fig. S5), indicating that EutR is important for pathogen persistence not only during primary infection but also within a host population.
dpi, the median CFU of the ΔeutR strain shed in stool was ~10-fold lower than the reported threshold that results in efficient transmission between animals. Therefore, we next investigated how EutR affects transmission. Following 24 h of cohousing with 6-dpi donor mice, equal numbers of WT and ΔeutR
C. rodentium organisms were recovered in stool from recipient mice (Fig. 5A). In contrast, following cohousing with 10-dpi donor mice, the ΔeutR strain was recovered at significantly lower levels in stool of (formerly) naive mice (Fig. 5B and Fig. S5), indicating that EutR is important for pathogen persistence not only during primary infection but also within a host population.
Conclusions.
Proper virulence gene expression in response to particular microenvironments is essential for pathogens to colonize a host (49). Our findings demonstrate that EutR is important for EA sensing and activation of LEE expression in C. rodentium, similar to its role in EHEC. Moreover, our findings reveal that EutR enhances virulence gene expression and colonization during infection of the GI tract.
EA is abundant in the intestinal tract due to the turnover of bacterial cells and exfoliation of enterocytes as well as through the host diet (2, 3). Although inflammation and infection have been suggested and/or shown to influence EA concentrations in the GI tract (13, 50), our data indicate that C. rodentium infection does not affect EA levels (Fig. 4C). Despite the consistent EA levels during infection, EutR affects C. rodentium virulence gene expression and colonization at later time points during infection (e.g., day 8 postinfection [Fig. 4B]), suggesting that spatiotemporal regulation of EutR expression and/or activity occurs in the GI tract. It is possible that EutR is part of a feed-forward regulatory circuit that integrates other environmental cues. For example, although eutR is constitutively expressed from the internal P2 promoter (36) (Fig. 1A), transcriptional regulation of this promoter may be more complex and include other transcription factors. Posttranscriptional regulation of eutR may also contribute to EutR expression. Alternatively, although EutR is required for EA sensing (6), EutR may not specifically respond to EA. In the complex in vivo environment, EutR may sense additional signals that affect spatiotemporal virulence gene regulation.
Notably, EutR not only is important for regulating expression of virulence traits in the attaching and effacing pathogens EHEC and C. rodentium but also plays a key role in regulating expression of Salmonella virulence traits required for survival and replication in macrophages (5, 6). These findings indicate a shared mechanism of host sensing and adaptation among distinct bacterial pathogens. Additionally, our findings suggest that EutR-dependent defects in pathogen colonization can be amplified during transmission and significantly impact the persistence of a pathogen within a host population. Together, these findings highlight the conserved role for EutR in promoting bacterial virulence and reveal new insights into the importance of EutR in host-to-host transmission, which is a poorly understood aspect of bacterial pathogenesis.
MATERIALS AND METHODS
Strains, plasmids, and recombinant DNA.
C. rodentium DBS100 (51, 52) was used in this study. Primers are listed in Table S1. C. rodentium deletion strains were generated using λ-red mutagenesis (53). Briefly, PCR products were generated using pKD3 (ΔlacZ) or pKD4 (ΔeutR). The PCR products were transformed into C. rodentium expressing the λ-red genes from plasmid pKD46. The deletions were confirmed by sequencing. C. rodentium DBS100 lacks antibiotic resistance; therefore, we generated unresolved mutant strains for mouse infections. The ΔlacZ strain was used as the WT strain. Disruption of lacZ did not affect C. rodentium colonization of the GI tract (Fig. S6).
The eutR mutant was complemented using pGEN::eutR, which expressed eutR from the native promoter. This plasmid was constructed by amplifying C. rodentium genomic DNA (gDNA) using primers specific to the eutR gene, including 206 nucleotides upstream of the ATG start site. Amplified DNA was digested with NheI and SacI and inserted into pGEN-MCS (54) (Addgene MTA). As a control, the WT and ΔeutR strains were transformed with the empty pGEN-MCS vector.
Culture media.
Bacteria were grown in Luria-Bertani medium (LB), Dulbecco’s modified Eagle medium (DMEM; Invitrogen), or M9 minimal medium (55) that was supplemented with 10 mM EA hydrochloride (Sigma) as the sole nitrogen source. Cyanocobalamin (150
mM EA hydrochloride (Sigma) as the sole nitrogen source. Cyanocobalamin (150 nM; Sigma) was added to the medium whenever EA was added. In general, bacteria were grown overnight (OVN) with shaking in LB at 37°C and then diluted 1:100 into fresh medium for experimental analysis.
nM; Sigma) was added to the medium whenever EA was added. In general, bacteria were grown overnight (OVN) with shaking in LB at 37°C and then diluted 1:100 into fresh medium for experimental analysis.
EutR amino acid alignment.
EutR sequences were obtained using the annotated EHEC EDL933 (56) and C. rodentium (57) genome sequences. The EMBOSS Needle program (58) was used to generate the alignment.
Transcriptional fusions.
C. rodentium gDNA including the ler promoter, including 400 upstream and 43 nucleotides downstream of the ATG start site, was amplified by PCR. Resulting PCR products were cloned into the pGEN-luxCDABE vector (54) (Addgene MTA).
Purification of EutR under native conditions.
The plasmid expressing EutR::MBP was constructed by amplifying the eutR gene from C. rodentium strain DBS100 using primers EutRMBP_F1 and EutRMBP_R1 and cloning the resulting PCR product into the NcoI/BamHI cloning sites of vector pMAL-c5X (New England BioLabs [NEB]). In order to purify MBP and EutR::MBP, E. coli strain BL-21(DE3) containing MBP or EutR::MBP was grown at 37°C in LB with glucose (0.2% [final concentration]) and ampicillin (100 μg/ml) to an optical density at 600
μg/ml) to an optical density at 600 nm (OD600) of 0.5. Then, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.3
nm (OD600) of 0.5. Then, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.3 mM, and protein expression was induced OVN at 18°C. Cells were harvested by centrifugation at 4,000
mM, and protein expression was induced OVN at 18°C. Cells were harvested by centrifugation at 4,000 ×
× g for 20
g for 20 min, then resuspended in 25
min, then resuspended in 25 ml of column buffer (20
ml of column buffer (20 mM Tris-HCl, 200
mM Tris-HCl, 200 mM NaCl, 1
mM NaCl, 1 mM EDTA), and lysed by homogenization. The lysed cells were centrifuged, and the lysate was loaded onto a gravity column (Qiagen) with amylose resin. The column was washed with column buffer and then eluted with column buffer containing 10
mM EDTA), and lysed by homogenization. The lysed cells were centrifuged, and the lysate was loaded onto a gravity column (Qiagen) with amylose resin. The column was washed with column buffer and then eluted with column buffer containing 10 mM maltose. Fractions containing purified proteins were confirmed by SDS-PAGE and Western blot analysis. Protein concentration was determined using a NanoDrop spectrophotometer.
mM maltose. Fractions containing purified proteins were confirmed by SDS-PAGE and Western blot analysis. Protein concentration was determined using a NanoDrop spectrophotometer.
ChIP-qPCR.
Chromatin immunoprecipitation (ChIP) was performed using the C. rodentium ΔeutR strain transformed with pEutR::MBP or pMBP. Strains were grown in DMEM supplemented with EA, cyanocobalamin, and 2.5 μM IPTG until cells reached an OD600 of ~0.8. Cross-linking and ChIP were performed based on established methods (5, 59). After growth, formaldehyde was added (1% [final concentration]), and cells were incubated for 15
μM IPTG until cells reached an OD600 of ~0.8. Cross-linking and ChIP were performed based on established methods (5, 59). After growth, formaldehyde was added (1% [final concentration]), and cells were incubated for 15 min at room temperature. Reactions were quenched with 0.5 M glycine, and then samples were pelleted, resuspended in phosphate-buffered saline (PBS), and washed. Cells were lysed with 2
min at room temperature. Reactions were quenched with 0.5 M glycine, and then samples were pelleted, resuspended in phosphate-buffered saline (PBS), and washed. Cells were lysed with 2 mg/ml of lysozyme at 37°C for 30
mg/ml of lysozyme at 37°C for 30 min. Subsequently, samples were placed on ice and sonicated. Insoluble cell debris was removed by centrifugation, and supernatants were collected. RNase A was added to the samples, which were then incubated at 37°C for 1 h. One hundred units of Benzonase nuclease (EMD Millipore) was added to digest DNA into smaller fragments (60). Immunoprecipitation was performed using amylose beads (NEB) for 2 h at 4°C with gentle mixing. Beads were pelleted and washed. Then samples were incubated for 10
min. Subsequently, samples were placed on ice and sonicated. Insoluble cell debris was removed by centrifugation, and supernatants were collected. RNase A was added to the samples, which were then incubated at 37°C for 1 h. One hundred units of Benzonase nuclease (EMD Millipore) was added to digest DNA into smaller fragments (60). Immunoprecipitation was performed using amylose beads (NEB) for 2 h at 4°C with gentle mixing. Beads were pelleted and washed. Then samples were incubated for 10 min at 65°C in elution buffer with occasional gentle mixing. Samples were centrifuged and supernatants were collected. To reverse the cross-link, samples were boiled for 10
min at 65°C in elution buffer with occasional gentle mixing. Samples were centrifuged and supernatants were collected. To reverse the cross-link, samples were boiled for 10 min and DNA was purified using the Qiagen PCR purification kit. For ChIP-qPCR, an aliquot of each reaction was taken prior to immunoprecipitation, de-cross-linked by boiling for 10
min and DNA was purified using the Qiagen PCR purification kit. For ChIP-qPCR, an aliquot of each reaction was taken prior to immunoprecipitation, de-cross-linked by boiling for 10 min, and purified for use as the input control. The fold enrichment of each promoter represents the value of the immunoprecipitated DNA divided by the input unprecipitated DNA. These values were normalized to the values obtained for each promoter precipitated using MBP empty vector in order to account for nonspecific enrichment.
min, and purified for use as the input control. The fold enrichment of each promoter represents the value of the immunoprecipitated DNA divided by the input unprecipitated DNA. These values were normalized to the values obtained for each promoter precipitated using MBP empty vector in order to account for nonspecific enrichment.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed basically as described previously (5). We modified the protocol to use 5′-biotinylated primers to generate the labeled DNA probes. EMSAs were performed by adding increasing amounts of purified EutR protein to end-labeled probe (30 ng) in binding buffer (10
ng) in binding buffer (10 mM Tris-HCl [pH 8.0], 1
mM Tris-HCl [pH 8.0], 1 mM Na-EDTA, 80
mM Na-EDTA, 80 mM NaCl, 10
mM NaCl, 10 mM β-mercaptoethanol, and 4% glycerol) (61), and reactions were incubated for 20
mM β-mercaptoethanol, and 4% glycerol) (61), and reactions were incubated for 20 min at 37°C. For competition EMSAs, 25
min at 37°C. For competition EMSAs, 25 ng of labeled probe was used with 0
ng of labeled probe was used with 0 ng, 25
ng, 25 ng, 125
ng, 125 ng, and 250
ng, and 250 ng of unlabeled probe added. Immediately before the samples were loaded, Ficoll solution was added to the reaction mixtures. The samples were electrophoresed for approximately 4 to 5 h at 175 V on a 6% polyacrylamide gel, transferred to Zeta-Probe membranes, and visualized using the chemiluminescent nucleic acid detection module kit (Thermo Scientific).
ng of unlabeled probe added. Immediately before the samples were loaded, Ficoll solution was added to the reaction mixtures. The samples were electrophoresed for approximately 4 to 5 h at 175 V on a 6% polyacrylamide gel, transferred to Zeta-Probe membranes, and visualized using the chemiluminescent nucleic acid detection module kit (Thermo Scientific).
Animal infections.
All experiments were approved by the Institutional Animal Care and Use Committee at the University of Virginia School of Medicine. Female C57BL/6J (6- to 10-week-old) mice were inoculated by oral gavage of 1 ×
× 109 CFU of C. rodentium. Mice were monitored daily for weight loss and other signs of distress. At the end of each experiment, mice were euthanized by CO2 asphyxiation followed by cervical dislocation.
109 CFU of C. rodentium. Mice were monitored daily for weight loss and other signs of distress. At the end of each experiment, mice were euthanized by CO2 asphyxiation followed by cervical dislocation.
Enumeration of CFU from stool.
For enumeration of CFU from stool, fresh fecal pellets were collected at the desired time points, weighed, and homogenized in 1 ml of PBS. Serial dilutions were plated on LB plates containing appropriate antibiotic selectivity to evaluate CFU. Enumerated CFU per gram of stool were log transformed prior to statistical analysis, as previously described (62).
ml of PBS. Serial dilutions were plated on LB plates containing appropriate antibiotic selectivity to evaluate CFU. Enumerated CFU per gram of stool were log transformed prior to statistical analysis, as previously described (62).
RNA extraction and RT-qPCR.
Bacterial cells grown in vitro were resuspended in TRIzol (Life Technologies). Colonic tissue was harvested, washed with PBS to remove luminal contents, and then homogenized in TRIzol. For all samples, RNA was extracted using a PureLink RNA minikit (Invitrogen). Reverse transcription-qPCR (RT-qPCR; for in vitro and colonic samples) was performed as previously described (63) in a one-step reaction using an ABI 7500-FAST sequence detection system and software (Applied Biosystems). For each 10-μl reaction mixture, 5 μl of 2× SYBR master mix (Ambion), 0.05
μl of 2× SYBR master mix (Ambion), 0.05 μl of Multi-Scribe reverse transcriptase (Invitrogen), and 0.05
μl of Multi-Scribe reverse transcriptase (Invitrogen), and 0.05 μl of RNase inhibitor (Invitrogen) were added. For stool samples, cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen) with random primers prior to qPCR to overcome low RNA yields. Reactions are identical to those described for RT-qPCR omitting reverse transcriptase and adjustment for a final 10-μl volume. Primers were designed using Primer BLAST (NCBI) to ensure no cross-reactivity to other genes in the C. rodentium chromosome. Amplicon length was approximately 100
μl of RNase inhibitor (Invitrogen) were added. For stool samples, cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen) with random primers prior to qPCR to overcome low RNA yields. Reactions are identical to those described for RT-qPCR omitting reverse transcriptase and adjustment for a final 10-μl volume. Primers were designed using Primer BLAST (NCBI) to ensure no cross-reactivity to other genes in the C. rodentium chromosome. Amplicon length was approximately 100 bp. Amplification efficiency of each primer pair was verified using standard curves of known DNA concentrations. Melting-curve analysis was used to ensure template specificity by heating products to 95°C for 15 s, followed by cooling to 60°C and heating to 95°C while monitoring fluorescence. After the amplification efficiency and template specificity were determined for each primer pair, relative quantification analysis was used to analyze the samples using the following conditions for cDNA generation and amplification: 1
bp. Amplification efficiency of each primer pair was verified using standard curves of known DNA concentrations. Melting-curve analysis was used to ensure template specificity by heating products to 95°C for 15 s, followed by cooling to 60°C and heating to 95°C while monitoring fluorescence. After the amplification efficiency and template specificity were determined for each primer pair, relative quantification analysis was used to analyze the samples using the following conditions for cDNA generation and amplification: 1 cycle at 48°C for 30
cycle at 48°C for 30 min, 1
min, 1 cycle at 95°C for 10
cycle at 95°C for 10 min, and 40 cycles at 95°C for 15 s and 60°C for 1
min, and 40 cycles at 95°C for 15 s and 60°C for 1 min. Two technical replicates of each biological replicate were included for each gene target. Data were normalized to the reference controls rpoA (in vitro samples) or 16S rRNA specific to C. rodentium (tissue samples) and analyzed using the comparative critical threshold (CT) method (64). The expression levels of the target genes were compared using the relative quantification method (64).
min. Two technical replicates of each biological replicate were included for each gene target. Data were normalized to the reference controls rpoA (in vitro samples) or 16S rRNA specific to C. rodentium (tissue samples) and analyzed using the comparative critical threshold (CT) method (64). The expression levels of the target genes were compared using the relative quantification method (64).
EA measurements.
EA levels were measured as previously described (13), with minor modifications. Colons were harvested, weighed, and homogenized in 1.5 ml of ultrapure water. Homogenized tissue was incubated with 150
ml of ultrapure water. Homogenized tissue was incubated with 150 μl of 40% sulfosalicylic acid (Sigma) for 15
μl of 40% sulfosalicylic acid (Sigma) for 15 min and then centrifuged at 11,000
min and then centrifuged at 11,000 ×
× g for 15
g for 15 min at room temperature. Then 1
min at room temperature. Then 1 ml of the supernatant was mixed with 300
ml of the supernatant was mixed with 300 μl of 0.5 M NaHCO3 (Sigma), 2
μl of 0.5 M NaHCO3 (Sigma), 2 ml of 20-mg/ml dansyl chloride solution (Sigma) in acetone, and 200
ml of 20-mg/ml dansyl chloride solution (Sigma) in acetone, and 200 μl of 1 M NaOH (Fisher). Following incubation in the dark for 20
μl of 1 M NaOH (Fisher). Following incubation in the dark for 20 min at room temperature, 200
min at room temperature, 200 μl of 25% NH4OH (Sigma) was added. The volume was adjusted to 5
μl of 25% NH4OH (Sigma) was added. The volume was adjusted to 5 ml with acetonitrite (Sigma), and 1
ml with acetonitrite (Sigma), and 1 ml was centrifuged at 11,000
ml was centrifuged at 11,000 ×
× g for 1
g for 1 min. The supernatant was taken for analysis. The LC-MS system consists of a ThermoElectron Orbitrap ID-X mass spectrometer with a Heated Electrospray Ionization source interfaced to a Thermo Accucore Vanquish C18 1.5-μm, 2.1-
min. The supernatant was taken for analysis. The LC-MS system consists of a ThermoElectron Orbitrap ID-X mass spectrometer with a Heated Electrospray Ionization source interfaced to a Thermo Accucore Vanquish C18 1.5-μm, 2.1- by
by 100-mm column. Two microliters of the extract was injected and the compounds were eluted from the column by a methanol-0.1% formic acid gradient at a flow rate of 250
100-mm column. Two microliters of the extract was injected and the compounds were eluted from the column by a methanol-0.1% formic acid gradient at a flow rate of 250 μl/min (total time, 15 min). The nanospray ion source was operated at 3.2
μl/min (total time, 15 min). The nanospray ion source was operated at 3.2 kV. The sample was analyzed by MS and tandem MS (MS/MS). The dansyl-EA was detected as a peak at ~4.36
kV. The sample was analyzed by MS and tandem MS (MS/MS). The dansyl-EA was detected as a peak at ~4.36 min and a mass of 295.111+. A 1 M in vitro sample of EA hydrochloride (Sigma) taken through the dansylation process was used to generate a standard curve.
min and a mass of 295.111+. A 1 M in vitro sample of EA hydrochloride (Sigma) taken through the dansylation process was used to generate a standard curve.
In vivo imaging system (IVIS) analysis.
For in vivo bioluminescence on living mice, an IVIS Spectrum (Caliper Lifesciences, Alameda, CA) was used. Luminescence (radiance) was quantified using the software program Living Image (Xenogen). Radiance is reported as photons per second.
Transmission studies.
Donor mice were infected with WT or ΔeutR
C. rodentium for 6 or 10 days. At these time points, each donor mouse was placed in a fresh cage with 3 naive mice for 24 h. For data shown in Fig. 5, at 24 h after cohousing, all mice were separated and placed in fresh, individual cages. For data shown in Fig. S5, respective recipient mice were cohoused after exposure to donor mice. At 2 or 4
days. At these time points, each donor mouse was placed in a fresh cage with 3 naive mice for 24 h. For data shown in Fig. 5, at 24 h after cohousing, all mice were separated and placed in fresh, individual cages. For data shown in Fig. S5, respective recipient mice were cohoused after exposure to donor mice. At 2 or 4 days after cohousing with donor mice, stool was collected from each recipient mouse, homogenized in PBS, and plated on appropriate selective media to determine CFU of transmitted C. rodentium.
days after cohousing with donor mice, stool was collected from each recipient mouse, homogenized in PBS, and plated on appropriate selective media to determine CFU of transmitted C. rodentium.
Statistical analyses.
Statistical analyses were performed using GraphPad Prism software version 8.0 (GraphPad Software Inc.). Group comparisons were performed using Student’s t test for in vitro analyses and the Wilcoxon signed-rank test for all in vivo analyses.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Kendall lab for thoughtful discussions on this work and Hervé Agaisse for feedback on the manuscript. We thank Sarah Ewald for advice on the IVIS experiments and James Nataro for the Citrobacter rodentium DBS100 strain. We thank the Biomolecular Analysis Facility at the University of Virginia for LC-MS and analysis services.
This work was supported by National Institutes of Health NIAID grants AI118732, AI130439, and AI146888 to M.M.K. C.A.R. received support from NIH training grant 5T32AI007046 and a UVA Wagner Fellowship. A.B.S. received support from NIH training grant 5T32AI055432.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
Articles from Infection and Immunity are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/iai.00137-20
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7440760
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/iai.00137-20
Article citations
The yad and yeh fimbrial loci influence gene expression and virulence in enterohemorrhagic Escherichia coli O157:H7.
mSphere, 9(7):e0012424, 21 Jun 2024
Cited by: 0 articles | PMID: 38904402 | PMCID: PMC11287998
Ethanolamine metabolism through two genetically distinct loci enables Klebsiella pneumoniae to bypass nutritional competition in the gut.
PLoS Pathog, 20(5):e1012189, 07 May 2024
Cited by: 0 articles | PMID: 38713723 | PMCID: PMC11101070
Enterobacteriaceae in the Human Gut: Dynamics and Ecological Roles in Health and Disease.
Biology (Basel), 13(3):142, 23 Feb 2024
Cited by: 5 articles | PMID: 38534413 | PMCID: PMC10967970
Review Free full text in Europe PMC
Cooperation between physiological defenses and immune resistance produces asymptomatic carriage of a lethal bacterial pathogen.
Sci Adv, 9(25):eadg8719, 23 Jun 2023
Cited by: 1 article | PMID: 37352357 | PMCID: PMC10289649
Food for thought-The link between Clostridioides difficile metabolism and pathogenesis.
PLoS Pathog, 19(1):e1011034, 05 Jan 2023
Cited by: 10 articles | PMID: 36602960 | PMCID: PMC9815643
Review Free full text in Europe PMC
Go to all (7) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Bacterial Adrenergic Sensors Regulate Virulence of Enteric Pathogens in the Gut.
mBio, 7(3):e00826-16, 07 Jun 2016
Cited by: 67 articles | PMID: 27273829 | PMCID: PMC4959670
A pathogen-specific sRNA influences enterohemorrhagic Escherichia coli fitness and virulence in part by direct interaction with the transcript encoding the ethanolamine utilization regulatory factor EutR.
Nucleic Acids Res, 49(19):10988-11004, 01 Nov 2021
Cited by: 8 articles | PMID: 34591974 | PMCID: PMC8565329
A master regulator of central carbon metabolism directly activates virulence gene expression in attaching and effacing pathogens.
PLoS Pathog, 20(10):e1012451, 15 Oct 2024
Cited by: 0 articles | PMID: 39405360 | PMCID: PMC11508082
Citrobacter rodentium: infection, inflammation and the microbiota.
Nat Rev Microbiol, 12(9):612-623, 04 Aug 2014
Cited by: 297 articles | PMID: 25088150
Review
Funding
Funders who supported this work.
HHS | National Institutes of Health (4)
Grant ID: AI118732
Grant ID: 5T32AI007046
Grant ID: AI130439
Grant ID: AI146888
NIAID NIH HHS (5)
Grant ID: R03 AI146888
Grant ID: R01 AI118732
Grant ID: R21 AI130439
Grant ID: T32 AI055432
Grant ID: T32 AI007046
 a
a