Abstract
Free full text

CTDSP1 inhibitor rabeprazole regulates DNA-PKcs dependent topoisomerase I degradation and irinotecan drug resistance in colorectal cancer
Abstract
Irinotecan specifically targets topoisomerase I (topoI), and is used to treat various solid tumors, but only 13–32% of patients respond to the therapy. Now, it is understood that the rapid rate of topoI degradation in response to irinotecan causes irinotecan resistance. We have published that the deregulated DNA-PKcs kinase cascade ensures rapid degradation of topoI and is at the core of the drug resistance mechanism of topoI inhibitors, including irinotecan. We also identified CTD small phosphatase 1 (CTDSP1) (a nuclear phosphatase) as a primary upstream regulator of DNA-PKcs in response to topoI inhibitors. Previous reports showed that rabeprazole, a proton pump inhibitor (PPI) inhibits CTDSP1 activity. The purpose of this study was to confirm the effects of rabeprazole on CTDSP1 activity and its impact on irinotecan-based therapy in colon cancer. Using differentially expressing CTDSP1 cells, we demonstrated that CTDSP1 contributes to the irinotecan sensitivity by preventing topoI degradation. Retrospective analysis of patients receiving irinotecan with or without rabeprazole has shown the effects of CTDSP1 on irinotecan response. These results indicate that CTDSP1 promotes sensitivity to irinotecan and rabeprazole prevents this effect, resulting in drug resistance. To ensure the best chance at effective treatment, rabeprazole may not be a suitable PPI for cancer patients treated with irinotecan.
Introduction
Topoisomerase I (topoI) was identified as a specific target for camptothecin (CPT) and its analogues like irinotecan and topotecan [1]. Irinotecan is frequently used to treat colon, gastric, ovarian, pancreatic, breast, and small cell lung cancer. TopoI reduces DNA supercoiling by cutting and re-ligating DNA, and a controlled rotation between the cutting and re-ligation cycles reduces the supercoiling. However, in the presence of irinotecan, the re-ligation cycle is inhibited, and collision of progressing replication fork with nicked DNA leads to a DNA double strand break (DNA-DSB) and cell death [2]. CPTs are used extensively to treat various solid tumors, however, only 13–32% of patients respond, and the mechanisms of resistance are not well understood [3]. Classical mechanisms of irinotecan resistance potentially involve an ATP-binding cassette (ABC) transporter, ABCG2, which reduces intracellular drug accumulation [4–6]. Inhibition of the ABCG2 drug efflux pump using sorafenib sensitizes both non-resistant cells and resistant cells to irinotecan [7, 8]. Another proposed mechanism for irinotecan resistance is topoI gene mutation [9]. However, DNA sequencing studies have failed to find topoI gene mutations in cancer patients [10, 11]. Thus, none of the proposed drug resistance mechanisms have been validated. In response to irinotecan, topoI is ubiquitinated and degraded by ubiquitin proteasomal pathway (UPP) and rapid topoI degradation is shown to be associated with irinotecan resistance [12]. However, the precise mechanism was not understood and molecular determinants were not defined. Recently we have published the mechanism of topoI degradation by UPP, and have shown that the DNA-PKcs kinase cascade determines the rate of topoI degradation. Importantly, the deregulated kinase cascade keeps the DNA-PKcs constantly active, leading to higher basal levels of phosphorylated topoI-S10 and rapid degradation of topoI that leads to irinotecan resistance. To identify the upstream regulator of DNA-PKcs, we performed a siRNA library screen of 56 reported nuclear phosphatases, and subsequently phosphatase-silenced cells were treated with irinotecan to determine the CPT-induced rate of topoI degradation. Carboxy-terminal domain RNA polymerase II polypeptide small phosphatase 1 (CTDSP1) was identified as one of the strongest upstream regulators of DNA-PKcs-dependent proteasomal degradation of topoI.
The DNA-dependent protein kinase (DNA-PK) is a serine/threonine protein kinase composed of a large catalytic subunit (DNA-PKcs) and Ku 70/80 heterodimer [13]. It is now established that the Ku 70/80 heterodimer binds broken DNA double strands and recruits DNA-PKcs in vitro [14]. Once recruited, DNA-PK stabilizes the broken strand, targets Artemis for end processing, and finally the XLF/XRCC4/Ligase IV complex carries out the ligation. This classic NHEJ is the major eukaryotic pathway for DNA double strand break repair [15]. Although DNA-PKcs is considered a component of the DNA-double strand repair (DDR) pathway, recent findings indicate a variety of other important roles of this kinase in genome maintenance and tumor pathogenesis [16]. However, our understanding of the role of DNA-PKcs in tumor pathogenesis is not complete, and regulation of DNA-PKcs kinase activity is only partly understood. Autophosphorylation at multiple S/T sites is the first mechanism of DNA-PKcs kinase activation and inactivation, particularly in response to DNA-DSB. The second regulatory component is dephosphorylation of DNA-PKcs by phosphatases [13].
DNA-dependent RNA polymerase II consists of 12 polypeptides. The largest polypeptide Rpb1 contains heptad repeats, (Tyr-Ser-Pro-Thr-Ser-Pro-Ser) in the c-terminus domain (CTD), this unique feature of RNAPII distinguishes it from other polymerases [17]. The 52-heptad tandem repeats of CTD determine the regulation of transcription. Post-translational modifications of the 7 amino acid tandem repeats, particularly Ser2 and Ser5, are considered critical in RNAP II—dependent mRNA transcription. The pattern of CTD phosphorylation during the transcription cycle is highly dynamic and requires the activity of dedicated phosphatases and kinases [17]. Several kinases have been identified to phosphorylate CTD, most notably CDK7, CDK8, and CDK9. FCP1, Ssu72, and small CTD phosphatases (SCP) dephosphorylate the CTD during the different phases of MRNA transcription. CTDSP1 is part of the SCP (Small CTD Phosphatases) family, whose loss of function results in tumor development and proliferation [18]. This nuclear phosphatase associates with RNAP II and de-phosphorylates the serine 5 of the heptad repeat [19, 20]. The phosphorylation state of RNAP II affects its transcription activity; therefore CTDSP1 plays a role in gene expression regulation [20]. Recent studies have shown that the activity of CTDSP1 is suppressed by the proton pump inhibitor, rabeprazole [21].
In this study, we demonstrated that CTDSP1 promotes sensitivity to irinotecan and rabeprazole prevents this effect, resulting in drug resistance.
Materials and methods
Cell culture and drug treatment
The human colon cancer cell lines HCT116, HT29, DLD1, and LoVo were obtained from ATCC. HCT116 and DLD1 cells were grown and maintained in RPMI containing 10% FBS and pen-strep. HT29 cells were grown and maintained in McCoy’s medium containing 10% FBS and pen-strep. LoVo cells were grown and maintained in Ham’s F12-K containing 10% FBS and pen-strep. All cells were grown at 37 °C and 5% CO2 in a humidified cell culture incubator. Topo I inhibitor treatment was performed using various concentrations of either irinotecan (Sigma-Aldrich) or SN-38 (Tocris). SN-38 is the irinotecan active metabolite. Cells were also treated with various concentrations of Rabeprazole (Santa Cruz Biotechnology).
CTDSP1 siRNA transfection
CTDSP1-specific siRNA (Silencer Pre-designed siRNA: 5’-CCUCGUGGUUUGACAACAU-3’) and negative control were purchased from Dharmacon. Transfection of HCT116 cells (0.6×106 cells/well in 6-well plate) with siRNA oligonucleotides was performed using Lipofectamine RNAiMAX, following the manufacturer’s instructions (Invitrogen).
CTDSP1 overexpression
For reverse transcription quantitative RT-PCR, the following mRNA sequences were used: CTDSP1, forward primer, 5’-CACCATGGACAGCTCGGCCGTCATTACTC-3’, and reverse primer, 5’-CTAGCTCCCTGGCCGTGGCTGCCTG-3’. The cDNA was synthesized from a total RNA of HT29 cells using Super Script 3 First-Strand Synthesis (Invitrogen). Quantitative PCR amplification was performed using a C1000 Touch Thermal Cycler. The PCR product and the pENTR/D-TOPO vector were mixed and incubated for 5 min at room temperature. The reaction was used to transform competent E. coli cells (Mach1, Invitrogen). The recombinant bacteria were screened using an LB agar medium containing kanamycin. The recombinant plasmid pENTR-CTDSP1 was extracted from positive colonies using the QIAprep Spin Miniprep Kit (Qiagen) and proper orientation of the cloned fragment was verified by PCR and DNA sequencing. The recombinant plasmid was transfected into HT29 cells using 4D-Nucleofecor (Lonza).
Immunoblotting
Cells cultured in 6-well plates were scraped into an ice-cold RIPA buffer. Samples were clarified by centrifugation at 15000 rpm for 15 min at 4°C. We used the iBind Western System (Invitrogen) and performed the imaging using the Amersham Imager 600 instrument (GE Healthcare, Little Chalfont, UK). Primary antibodies included antibodies against β-actin (Cell Signaling Technology), CTDSP1 (Abcam), Topoisomerase I (BD Pharmingen), and DNAPKcs-pS2056 (Abcam). Antigen/antibody complexes were visualized by enhanced chemiluminescence (ECL detection system).
Immunofluorescence
Control and CTDSP1 knockdown cells were grown on sterile coverslips in 100mm plates and after washing with PBS, cells were fixed with 3.7% formaldehyde. After 25 minutes of fixation, coverslips were washed with 0.2% Triton-X-100 and then blocked with 3% BSA for 1 hour. After blocking, each coverslip was incubated with a monoclonal anti-phosphorylated DNAPK antibody, followed by Alexa- fluor 488-conjugated goat anti-rabbit IgG. Nuclei were stained with DAPI and imaging was performed on the Leica SP5 fluorescence microscope.
Integrating EGFP following the hTOPI gene in HCT-116 cells
Summary sequences of plasmids used in this study can be found in S1 Table. Roughly 1000 bases 5’ and 3’ of the genomic sequence flanking the last hTOP1 exon in HCT-116 cells were sequenced to identify and account for any cell-type specific polymorphisms. To do so, multiple PCR products spanning the genomic sequence were amplified using Phusion DNA Polymerase (New England Biolabs). The resulting PCR amplicons were cloned using the Zero Blunt TOPO PCR Cloning Kit (Invitrogen), transformed into E.coli XL-1 blue competent cells, and ~20–25 colonies were grown overnight at 37°C in TB media prior to miniprep and sanger sequencing. A single guide-RNA (sgRNA) targeting the hTOP1 stop codon was designed so that the SpCas9 binding site would be destroyed following gene conversion. To generate the sgRNA plasmid, oligonucleotides corresponding to the spacer sequence of the target site were annealed and ligated into BsmBI cut BPK1520. The homologous recombination donor plasmid designed to create the topoI-EGFP fusion protein was generated by Gibson assembly into the NheI and HindIII sites of pUC19. Regions of the genomic sequence 5’ and 3’ of the hTOP1 stop codon were assembled with an EGFP-P2A-Pac (for puromycin resistance) fusion cassette to generate the final donor plasmid (with 5’ and 3’ homology regions of 934 and 824 bases, respectively). Transfections into HCT-116 cells were performed by lipofectamin (lifetechnology) method with: 1) 2μg of a wild-type Streptococcus pyogenes Cas9 expression plasmid (MSP469); 2) 1μg of the sgRNA expression plasmid (MMW134) and 3) 2μg of the homologous recombination donor plasmid (MMW274). After 7 days of the transfection, cells were selected with 4μg/ml puromycin and after 14 days of selection, the cells were sorted by MoFlo Legacy (Beckman Coulter). The sorted cells were maintained in 2μg/ml puromycin contained media. Precise incorporation of the genome editing cassette in single cell cloned HCT-116 hTOP1-EGFP cells was confirmed by PCR amplifying the genomic locus and sequencing (S1 Data).
Cell viability assay
Cells were seeded in 96-well plates at 700–2000 per well in 180 μl of medium. Next, a 100 μM SN-38 stock solution was used to prepare dilutions in 20 μl of medium before treatment. After incubating for 72 h, cells were incubated CellTiter-Glo 2.0 reagent into each well. Cell viability was then measured by luminescence detection using a Synergy H1 microplate reader (BioTek).
Clonogenic assay
All cells were treated with various concentrations of SN-38 (0, 2.5, 5.0, or 7.5 nM) for 24 h, and then 50 cells per well were seeded in 6-well plates. After 14 days, when colonies were apparent, cells were fixed in 6% glutaraldehyde and 0.5% crystal violet for 15 min, then washed with tap water. The colonies per well were counted.
Patients
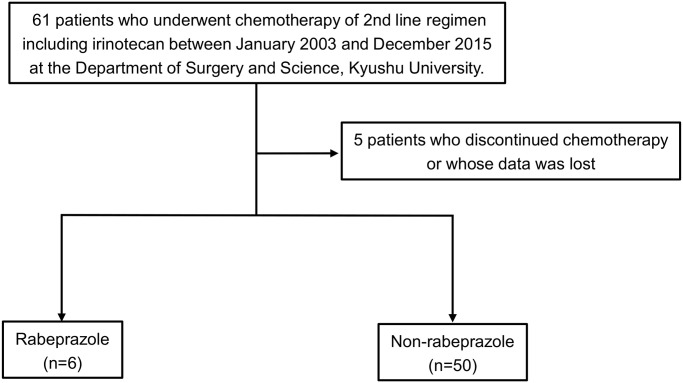
Retrospective data from 61 patients who underwent 2nd-line chemotherapy including irinotecan between January 2003 and December 2015 at the Department of Surgery and Science, Kyushu University, were retrieved. Of the 61 patients, 5 patients who discontinued chemotherapy, or whose data was not complete, were excluded from the study. Finally, 56 patients were eligible for analysis.
Statistical analysis
Each blots were quantified using Image J software (NIH, Bethesda, MD, USA). The statistical analyses were performed by using the JMP 14 statistical software package (SAS institute, Cary, NC, USA). The Student t-test, the chi-square test, Fisher’s exact test and ANOVA one-way test were used where appropriate.
Study approval
All experiments were conducted with approval by the Ethics Committee of the Kyushu University (Approval Number: No. 30–215)
Results
CTDSP1 expression correlated with topoI degradation in colorectal cancer cell line
We have previously reported that topoI is phosphorylated by DNA-PKcs at serine-10, and the resulting phosphoprotein (topoI-pS10) is efficiently ubiquitinated and targeted for rapid proteasomal degradation [3]. Protein phosphatases have been shown to interact with DNA-PKcs and could serve as upstream regulators of DNA-PKcs. To identify potential upstream regulators of DNA-PKcs, we screened a siRNA library comprised of all known human nuclear phosphatases. Our data demonstrated that silencing of CTDSP1 significantly altered the rate of topoI degradation. In the present study, the possible role of CTDSP1 as a regulator of topoI stability and irinotecan sensitivity was tested first by assessing the relationship between CTDSP1 expression and rate of topoI degradation. Cell lysates from four CRC cell lines, HCT116, DLD1, LoVo, and HT29 were subjected to immunoblot analysis with anti-CTDSP1. The results indicate a higher level of CTDSP1 in HCT116 cells and the lowest level in HT29 (Fig 1A). HCT116 and HT29 cells were treated with SN-38 for 90 and 180 min and cell lysates were subjected to determine the rate of topoI degradation. The lower level of topoI protein at 180 min clearly indicates a higher rate of topoI degradation in HT29 cells compared to HCT116 cells (Fig 1B). We then asked the obvious question: does this SN-38-induced rate of topoI degradation have an impact on drug sensitivity? The clonogenic survival assay clearly demonstrated that HT29, with low CTDSP1 and rapid topoI degradation, is more resistant to HCT116 cells (Fig 1C).
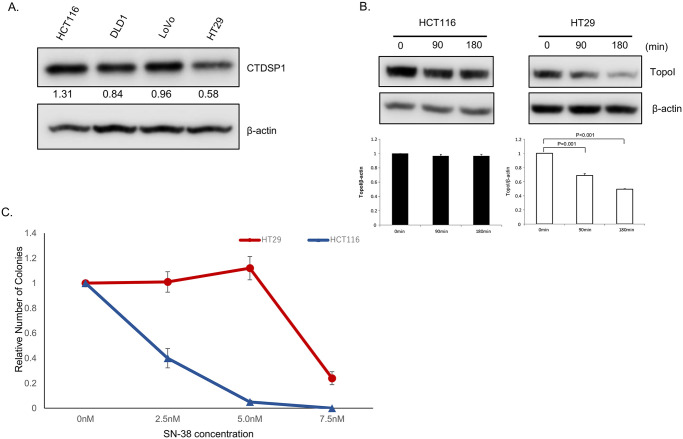
A, Cell lysates from four colon cancer lines, HCT15, DLD1, LoVo and HT29, were immunoblotted with anti-CTDSP1 and anti-β-actin. B, HCT116 and HT29 cells were treated with 2.5 μM SN-38 and harvested after 90 and 180 min. Cell lysates were immunoblotted with anti-topoI and anti-β-actin. C, HCT116 and HT29 cells were treated with various concentrations of SN-38 and clonogenic assays were performed to determine the relative number of colonies.
Silencing of CTDSP1 enhances topoI degradation and irinotecan resistance
We asked if reducing CTDSP1 expression would change the rate of topoI degradation and impart cellular resistance to SN-38 in HCT116 cells. CTDSP1 expression was reduced significantly by siRNA (Fig 2A). Control and siCTDSP1- HCT116 cells were treated with SN38 and harvested at 90 and 180 min. The cell lysates were analyzed by immunoblotting with anti-topoI and anti-β-actin (for protein loading). The quantitative analysis of the immunoblot demonstrated a statistically significant reduction in topoI level in siCTDSP1-HCT116 cells. No appreciable reduction in topoI protein level was observed in control-HCT116 cells (Fig 2B). To further understand the CTDSP1-mediated rate of topoI proteasomal degradation in response to SN38, we silenced CTDSP1 in genomically edited HCT116 cells expressing topoI-GFP protein. Cells were treated with SN38 and relative florescence of topoI-GFP was visualized by confocal microscope. Untreated control and siCTDSP1 cells demonstrated similar topoI-GFP florescence, however, when cells were treated with SN-38, siCTDSP1 cells had lower topoI-GFP florescence, indicating a higher rate of degradation (Fig 2C). Comparative clonogenic assays and cell survival assays demonstrated that siCTDSP1 was significantly more resistant to SN38 (Fig 2D–2F).
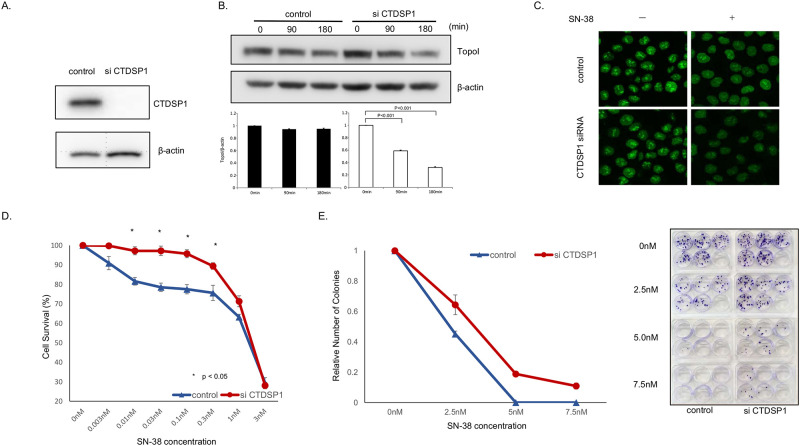
A, Cells transfected with CTDSP1 or control siRNA were lyzed and the cells’ lysates were immunoblotted with anti-CTDSP1 and anti-β-actin antibodies. B, HCT116-siRNA CTDSP1 or control siRNA, treated with 2.5 μM SN-38 were harvested after 90 and 180 min. Cell lysates were immunoblotted with anti-topoI and anti-β-actin antibodies. Cells’ lysates were immunoblotted with anti-topoI and anti-β-actin antibodies. C, EGFP was integrated with topoI in HCT116 cells using CRISPR/Cas9 system and CTDSP1 was knocked down in this cell line by siRNA. Cells were treated with 2.5uM SN-38 for 60 min and the topoI-EGFP signal was imaged by Leica SP5 confocal microscope. D, HCT116-siRNA-CTDSP1 or control siRNA were plated in a 96-well plate and treated with various concentrations of SN-38 for 72 h. Cell viability was determined by detecting the luminescence. E, HCT116-siRNA-CTDSP1 or control siRNA cells were plated in a 6-well plate and treated with various concentrations of SN-38 for 24 h. Then, 50 cells per well were plated in a 6-well plate. After 14 days, when colonies were apparent (right panel), colonies were counted and the relative number of colonies was determined (left panel).
The higher expression of CTDSP1 in HT29 cells inhibits topoI degradation and restores SN-38 sensitivity
The basal level of CTDSP1 protein expression in HT29 is low compared to other CRC cells and SN38-induced proteasomal degradation in these cells is high. To better understand the role of CTDSP1 in SN-38-induced proteasomal degradation of topoI, we overexpressed CTDSP1 in HT29 cells. The stably transfected HT29 cells showed significantly higher CTDSP1 compared to vector-only cells (Fig 3A). Vector-only (control) and CTDSP1 overexpressing HT29 cells were treated with SN-38 and the cells were harvested at 90 and 180 min. The cell lysates were immunoblotted with anti-topoI and anti-β-Actin. The topoI proteins in vector alone cells were significantly reduced both at 90 and 180 min after SN-38 treatment. In contrast, very little or no topoI degradation was observed in cells overexpressing CTDSP1 (Fig 3B). A comparative estimation of cell survival assays demonstrated a significantly increased SN-38 sensitivity in cells overexpressing CTDSP1. The drug sensitivity difference was most pronounced at 10 nm SN-38 concentration (Fig 3C).
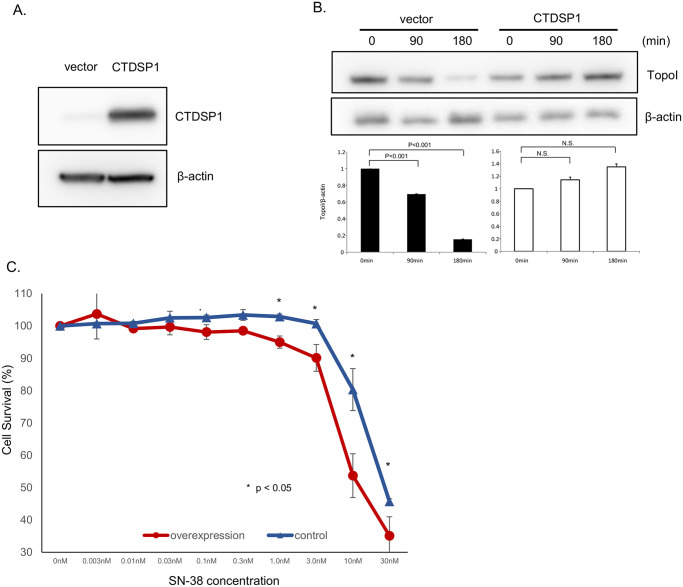
A, CTDSP1-overexpressing and control cells were lyzed and the cell lysates were immunoblotted with anti-CTDSP1 and anti-β-actin antibodies. B, CTDSP1-overexpressing and control cells were treated with 2.5 μM SN-38 and harvested after 90 or 180 min. Cells lysates were immunoblotted with anti-topo I and anti-β-actin antibodies. C, CTDSP1-overexpressing and control cells were plated in a 96-well plate and treated with various concentrations of SN-38 for 72 h. Cell viability was determined by luminescence detection.
CTDSP1 activates DNA-PKcs and enhances SN-38 induced proteasomal degradation of topoI and drug resistance
Recently we have published that DNA-PKcs dependent phosphorylation of topoI at Serine 10 is critical for SN-38- induced proteasomal degradation of topoI. We have also demonstrated that CTDSP1 is one of the upstream effector nuclear phosphatases that regulates DNA-PKcs activation and topoI proteasomal degradation. To better understand the role of CTDSP1 in this pathway, we first asked if deferential expression of CTDSP1 in HCT116 and HT29 affects DNA-PKcs kinase activity. Phosphorylation of DNA-PKcs-pS2056 is indicative of activated DNA-PKcs kinase. Cell lysates from HCT116 and HT29 cells were immunoblotted with anti-DNA-PKcs-pS2056 and the data clearly demonstrated a higher phosphorylation status of S2056 only in HT29 cells. Importantly HCT116 cells demonstrated minimal DNA-PKcs kinase activity. The GAPDH immunoblotting indicated that a similar amount of protein was analyzed (Fig 4A). To better understand the CTDSP1 dependent activation of DNA-PKcs, DNA-PKcs-S2056 phosphorylation status was determined in CTDSP1-silence HCT116 cells. The cells were also treated with SN38 to determine the effect of DNA-PKcs activation due to SN-38-induced DNA-DSB. CTDSP1 silencing resulted in remarkable activation of DNA-PKcs and there was not much difference in control and treated cells (Fig 4B). Control and siCTDSP1-HCT116 cells were also immunoflorescentely stained with anti-phosphospecific DNA-PKcs, and results indicated a higher basal level of phospho-DNA-PKcs compared to control cells (Fig 4C).
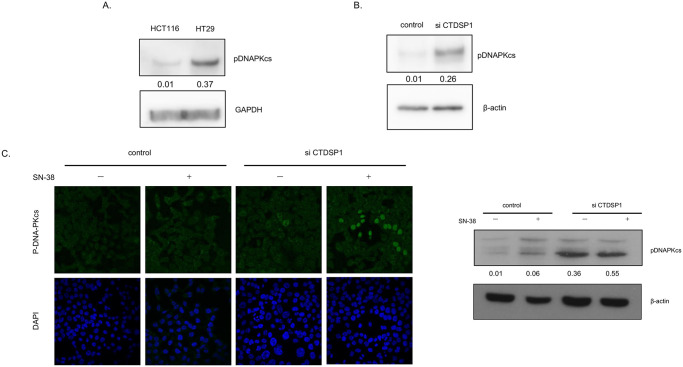
A, HCT116 and HT29 cell lysates were immunoblotted with anti-DNAPKcs-pS2056 and anti-GAPDH antibodies. B, CTDSP1 was silenced by CTDSP1 siRNA in HCT116 cells, and control as well as CTDSP1-silenced cell lysates were subjected to immunoblot with anti-CTDSP1 and anti-β-actin. C, Control and CTDSP1 knockdown cells were treated with SN38 and analyzed by immunofluorescence staining with anti-DNA-PKcs–pS2056). The cells were analyzed by Leica SP5 confocal microscope.
Rabeprazole inhibits CTDSP1 and enhances topoI degradation and irinotecan resistance
Rabeprazole binds to the hydrophobic pocket of CTDSP1 and inhibits its phosphatase activity. This hydrophobic pocket is adjacent to the active site in CTDSP1 and rabeprazole most likely acts as a direct competitor of the natural substrate, i.e., the CTD phosphorylated peptide [21]. We treated HCT116 cells with rabeprazole and then with SN-38. Cells were harvested at 90, 180, and 270 min post-SN-38 treatment, and cell lysates were analyzed to determine the rate of topoI degradation. The results indicate that the topoI degradation in response to SN-38 increased when cells were pretreated with rabeprazole (Fig 5A). The quantitative analysis of the immunoblot demonstrated significantly lower topoI protein at 90 min that continued til the 270 min time point. The rate of topoI degradation was enhanced when cells were either pretreated with 10 or 20μm rabeprazole. However, the degradation rate was more pronounced in cells pretreated with 20μm rabeprazole. In contrast, no appreciable topoI protein degradation was observed in HCT116 cells that were not pretreated with rabeprazole (Fig 5A). To further confirm these findings, genomically edited HCT116 cells with topoI-GFP protein were first treated with 5 and 10 μm of rabeprazole, and after 48 h the cells were treated with SN-38. The relative fluorescence intensity of GFP was observed by confocal microscope to determine SN-38-induced rate of topoI degradation. The data indicates that the rate of topoI degradation increased when cells were pretreated with rabeprazole. GFP fluorescence intensity was lower when cells were pretreated with 10μm compared to 5μm rabeprazole. Importantly cells not pretreated with rabeprazole topoI-GFP protein level remained high in the response to SN-38 (Fig 5B).
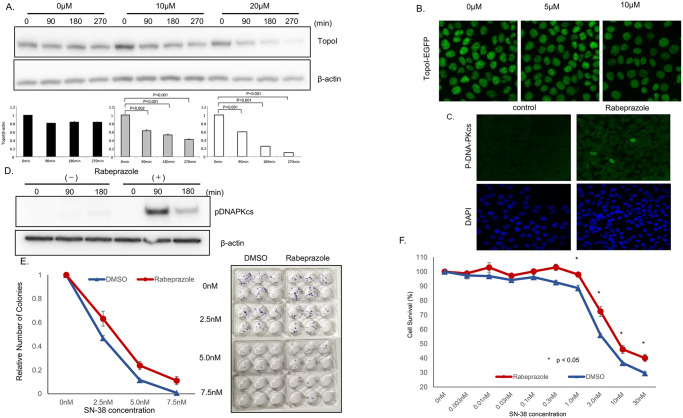
A, HCT116 cells were plated in a 6-well plate and treated with various concentrations of rabeprazole (0, 10, 20 μM) for 72 h, and then with 2.5 μM SN-38 and harvested after 90 or 180 min. Cell lysates were immunoblotted with anti-topoI and anti-β-actin. B, Genomically edited HCT116 cells with TopoI-EGFP fusion proteins were treated with 5 and 10 μM of Rabeprazole for 48 hours and topoI-GFP protein level was analyzed by confocal microscope. C. HCT116 cells were plated in a 6-well plate, treated with 40 μM rabeprazole or DMSO for 72 h, and then with 2.5 μM SN-38, and harvested after 90 or 180 min. Cell lysates were immunoblotted with anti-pDNA-PKcs and anti-β-actin. D, HCT 116 cells were treated with rabeprazole, control and treated cells were analyzed by immunofluorescence analysis with anti-phospho-DNA-PKcs-pS2056 and confocal microscopy. E. HCT116 cells were plated in a 6-well plate and treated with rabeprazole or DMSO for 72 h. Then, 50 cells were plated in each well of a 6-well plate and treated with various concentrations of SN-38 for 24 hours. Cell colonies were counted after 14 days. F. HCT116 cells were plated in a 6-well plate and treated with 40 μM rabeprazole or DMSO for 72 h. Then, cells were plated in a 96-well plate and treated with various concentrations of SN-38 for 72 h. Cell viability was determined by luminescence detection.
To further understand this pathway and the role of rabeprazole in SN-38-induced topoI degradation, immunofluorescence analysis of control and rabeprazole-treated HCT116 cells were immunostained with anti-DNA-PKcs-pS2056. Cells treated with rabeprazole demonstrated higher immunofluorescence levels, indicating kinase activation in response to rabeprazole (Fig 5C). Rabeprazole-treated cells were also analyzed by immunoblotting analysis with anti-DNA-PKcs-pS2056. The data clearly demonstrated the activation of DNA-PKcs at 90 min post SN38 treatment. The activation was transient and phosphorylated S2016 of DNA-PKcs was reduced at 180 min. Immunoblot analysis with anti-β-actin showed equal protein loading. To determine the effect of rabeprazole-induced proteasomal degradation of topoI and irinotecan resistance, HCT116 cells were treated with rabeprazole, and after 48 hours, cells were treated with DMSO (control) or various concentrations of SN-38 and clonogenic and cell survival assays were performed. The results demonstrated that the cells treated with rabeprazole had a higher number of colonies, and a higher percentage of cells was viable in rabeprazole-treated cells. The difference is statistically significant (Fig 5E and 5F).
Treatment with rabeprazole may reduce the effect of irinotecan in colorectal cancer patients
We next examined clinical data to investigate rabeprazole-induced irinotecan resistance in patients with colon cancer. Of 56 eligible patients who underwent chemotherapy with irinotecan, 50 (89.3%) had not received rabeprazole during or before chemotherapy, while 6 (10.7%) had been treated with rabeprazole (Fig 6). We compared the overall response in the two groups. Partial response (PR) was observed in only one patient (16.7%) of the rabeprazole group, while 14 (28%) in the non-rabeprazole group had PR (Table 1). Similarly, the stable disease (SD) percent was higher and progressive disease was lower in patients not receiving rabeprazole. The results suggest that rabeprazole had an effect on the response to irinotecan in clinical settings.
Table 1
| Best overall response | ||
|---|---|---|
| Rabeprazole | Non-rabeprazole | |
| CR | 0(0%) | 1(2.0%) |
| PR | 1(16.7%) | 14(28.0%) |
| SD | 2(33.3%) | 22(44.0%) |
| PD | 3(50%) | 13(26.0%) |
RECIST: Response Evaluation Criteria Inn Solid Tumors; CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease.
Discussion
One of the most remarkable phenomena observed in the cellular response to irinotecan is degradation of topoI. It was first observed in leucocytes of patients treated with 9-nitro-camptothecin in phase 1 clinical trials. The leucocytes isolated after 24, 48, and 72 hours demonstrated decreasing topoI protein levels [22]. Later, these findings were reproduced in cancer cells [23]. More importantly, Desai et al, in a series of publications, demonstrated that the irinotecan-induced degradation of topoI is mediated by a ubiquitin proteasomal pathway, cells degrade topoI differentially, and the cells that degrade topoI rapidly are resistant to irinotecan [24]. More recently we have demonstrated that: i) topoI associates with DNA-PK, and DNA-PKcs phosphorylates topoI at Serine 10; ii) Phosphorylated topoI is ubiquitinated by BRCA1; iii) cells with higher basal levels of topoI-pS10 degrade topoI rapidly and are resistant to irinotecan; iv) the higher basal level of topoI-pS10 is maintained by phosphatase dependent activation of DNA-PKcs v) nuclear phosphatase siRNA library screen identified PTEN and CTDSP1 enhances irinotecan-induced topoI degradation and vi) silencing of PTEN enhanced DNA-PKcs activity and irinotecan resistance [3]. Phosphorylation-dependent activation and inactivation of DNA-PKcs is well documented. Autophosphorylation of several S/T moieties localized in ABCD and PQR clusters is at the core of regulation of DNA-PKcs kinase activity in response to DNA-DSB and phosphorylation of serine 2506 indicates the activation of DNA-PKcs [13]. The second proposed mechanism of DNA-PKcs regulation depends on the dephosphorylation by phosphatases. In one of the early reports, protein phosphatase 1 or protein phosphatase 2A were shown to reactivate autophosphorylated inactive DNA-PKcs. Furthermore, the addition of a phosphatase inhibitor reverses this process [25]. PP2A was also shown to play a role in NHEJ by directly dephosphorylating DNA-PKcs [26]. Another phosphatase, PP6, was shown to regulate the kinase activity of DNA-PKcs in response to DNA-DSB. Both catalytic units (PP6C) and regulatory subunits (PPR1) of PP6 interact with DNA-PKcs, and silencing of PP6C induced IR sensitivity and delayed release from the G2/M checkpoint [27, 28].
Our siRNA library screen of nuclear phosphatase identified PTEN and CTDSP1 as two phosphatases that significantly enhance CPT-induced topoI degradation. Importantly, we have demonstrated that PTEN regulates DNA-PKcs kinase activity in this pathway and PTEN deletion ensures DNA-PKcs dependent higher topoI-pS10, rapid topoI degradation and irinotecan resistance [3]. We asked if CTDSP1 also dephosphorylates DNA-PKcs, and is the upstream regulator of CPT-induced topoI degradation. CTDSP1 is a SCP1 family of phosphatases that dephosphorylates RNAP II and plays a regulatory role in mRNA transcription. Our novel finding indicates that CTDSP1 dephosphorylates DNA-PKcs, changes its kinase activity, and regulates irinotecan-induced topoI degradation. Our library screen demonstrated that silencing of CTDSP1 enhances irinotecan-induced topoI degradation in HCT15 cells. HCT15 cells degrade topoI rapidly and are resistant to irinotecan. To determine the activation of this pathway in cells that do not degrade topoI, we used WT and genomically edited HCT116 cells. We impaired CTDSP1 phosphatase activity either by silencing by siRNA or using a specific inhibitor. The results clearly demonstrated that either CTDSP1 inhibition or silencing caused the activation of DNA-PKcs, indicated by phosphorylation of DNA-PKcs-S-2056. Importantly, the knocking down of CTDSP1 resulted in higher basal phosphorylation of DNA-PKcs-S-2056. Similar results were observed when cells were treated with rabeprazole. The activated state changed in response to irinotecan when the DNA-PKcs-S2056 protein level was analyzed by immunoblotting and immunofluorescence. One of the important physiological functions attributed to DNA-PKcs in irinotecan-induced pathways is the degradation of topoI. HCT116 cells do not degrade topoI, however, a rapid topoI degradation was observed when CTDSP1 function was inhibited. Immunoblot analysis of topoI and reduction in the florescence intensity of topoI-EGFP clearly demonstrated enhanced topoI degradation in CTDSP1-deficient cells. Others and we have (change this) shown that enhanced rates of topoI degradation cause irinotecan resistance. To further validate this pathway, we asked if CTDSP1-mediated activation of DNA-PKcs and rapid degradation of topoI causes irinotecan resistance. Cells that were treated with rabeprazole did show irinotecan resistance in both cell survival and clonogenic assay.
CTDSP1 is a member of phosphatases that rely on the DxDx motif and Mg2+ to catalyze the phosphoryl-transfer [20, 29]. Rabeprazole binds to a unique hydrophobic pocket of CTDSP1, located in the insertion domain and adjacent to the DxDx motif. Digestive symptoms are the most feared complications of chemotherapy [30, 31] and a proton pump inhibitor (PPI) like rabeprazole, is frequently prescribed to patients. PPI. including rabeprazole, was recently reported to enhance the antitumor effects of both docetaxel-cisplatin combinations [32] and 5-FU [33] in cells. PPI can restore drug sensitivity in drug-resistant cells by preventing the increase of extracellular and lysosomal pH and increasing the cytoplasmic retention of doxorubicin [34]. However, another study highlighted a potential adverse effect of PPI on overall survival in colorectal cancer [35]. Treatment with a PPI may result in hypochlorhydria in the stomach, which causes hypersecretion of the hormone gastrin from the gastric antrum [36]. Moreover, in vitro studies have shown that hypergastrinemia promotes cell proliferation and migration, and inhibits apoptosis [37–39]. The relationship between PPI therapy and chemo-sensitivity is not clear. Notably, none of these studies examined the impact of rabeprazole on irinotecan sensitivity. This study is the first to demonstrate that rabeprazole inhibited CTDSP1 activity and caused resistance to irinotecan. Based on our findings, the concurrent use of rabeprazole should be avoided in cancer patients under treatment with irinotecan. If PPI therapy cannot be discontinued due to the severity of the gastrointestinal symptoms, alternative PPIs should be employed.
This study has some limitations. All analyzed patients underwent irinotecan chemotherapy as a second-line regimen and this analysis was retrospective. Prospective studies focusing on 1st-line irinotecan regimens would be necessary to definitively establish the relationship between rabeprazole and irinotecan.
In conclusion, we showed that CTDSP1 and its inhibitor, rabeprazole, play important roles in topoI regulation, particularly in response to irinotecan. This study clearly indicated that rabeprazole induces irinotecan resistance by enhancing proteasomal degradation of topoI and may not be a suitable PPI for cancer patients receiving irinotecan-based therapy.
Supporting information
S1 Fig
The pictures that performed clonogenic assays of HCT116 and HT29 cells.HCT116 and HT29 cells were treated with various concentrations of SN-38 and clonogenic assays were performed to determine the relative number of colonies.
(TIF)
S1 Table
Sequences of plasmids used for genome editing.(DOCX)
Acknowledgments
The authors thank S. Tsurumaru and A. Nakamura for technical assistance with the experiments.
References
Decision Letter 0
17 Feb 2020
PONE-D-20-00259
CTDSP1 inhibitor rabeprazole regulates DNA-PKcs dependent topoisomerase I degradation and irinotecan drug resistance in colorectal cancer
PLOS ONE
Dear Dr. Koji Ando,
Thank you for submitting your manuscript to PLOS ONE. After careful consideration, we feel that it has merit but does not fully meet PLOS ONE’s publication criteria as it currently stands. Therefore, we invite you to submit a revised version of the manuscript that addresses the points raised during the review process.
This manuscript identified CTDSP1 as a primary upstream regulator of DNA-PKcs in response to topo I inhibitors. The article is interesting. However, there are some major concerns for this study. The description for M & M is unclear. It have to be wrote the detail design. In addition, The quality for the results (Western blot) is poor. It have to be improved and followed the instruction for PLOS ONE to uploaded the original image during the revising submission. Moreover, the statistic is lacked in the manuscript. Overall, the article have to improve to fit the scientific requirement.
We would appreciate receiving your revised manuscript by Apr 02 2020 11:59PM. When you are ready to submit your revision, log on to https://www.editorialmanager.com/pone/ and select the 'Submissions Needing Revision' folder to locate your manuscript file.
If you would like to make changes to your financial disclosure, please include your updated statement in your cover letter.
To enhance the reproducibility of your results, we recommend that if applicable you deposit your laboratory protocols in protocols.io, where a protocol can be assigned its own identifier (DOI) such that it can be cited independently in the future. For instructions see: http://journals.plos.org/plosone/s/submission-guidelines#loc-laboratory-protocols
Please include the following items when submitting your revised manuscript:
A rebuttal letter that responds to each point raised by the academic editor and reviewer(s). This letter should be uploaded as separate file and labeled 'Response to Reviewers'.
A marked-up copy of your manuscript that highlights changes made to the original version. This file should be uploaded as separate file and labeled 'Revised Manuscript with Track Changes'.
An unmarked version of your revised paper without tracked changes. This file should be uploaded as separate file and labeled 'Manuscript'.
Please note while forming your response, if your article is accepted, you may have the opportunity to make the peer review history publicly available. The record will include editor decision letters (with reviews) and your responses to reviewer comments. If eligible, we will contact you to opt in or out.
We look forward to receiving your revised manuscript.
Kind regards,
Yu-Jia Chang
Academic Editor
PLOS ONE
Journal Requirements:
When submitting your revision, we need you to address these additional requirements.
1. Please ensure that your manuscript meets PLOS ONE's style requirements, including those for file naming. The PLOS ONE style templates can be found at
http://www.journals.plos.org/plosone/s/file?id=wjVg/PLOSOne_formatting_sample_main_body.pdf and http://www.journals.plos.org/plosone/s/file?id=ba62/PLOSOne_formatting_sample_title_authors_affiliations.pdf
2. PLOS requires an ORCID iD for the corresponding author in Editorial Manager on papers submitted after December 6th, 2016. Please ensure that you have an ORCID iD and that it is validated in Editorial Manager. To do this, go to ‘Update my Information’ (in the upper left-hand corner of the main menu), and click on the Fetch/Validate link next to the ORCID field. This will take you to the ORCID site and allow you to create a new iD or authenticate a pre-existing iD in Editorial Manager. Please see the following video for instructions on linking an ORCID iD to your Editorial Manager account: https://www.youtube.com/watch?v=_xcclfuvtxQ
3. PLOS ONE now requires that authors provide the original uncropped and unadjusted images underlying all blot or gel results reported in a submission’s figures or Supporting Information files. This policy and the journal’s other requirements for blot/gel reporting and figure preparation are described in detail at https://journals.plos.org/plosone/s/figures#loc-blot-and-gel-reporting-requirements and https://journals.plos.org/plosone/s/figures#loc-preparing-figures-from-image-files. When you submit your revised manuscript, please ensure that your figures adhere fully to these guidelines and provide the original underlying images for all blot or gel data reported in your submission. See the following link for instructions on providing the original image data: https://journals.plos.org/plosone/s/figures#loc-original-images-for-blots-and-gels.
In your cover letter, please note whether your blot/gel image data are in Supporting Information or posted at a public data repository, provide the repository URL if relevant, and provide specific details as to which raw blot/gel images, if any, are not available. Email us at gro.solp@enosolp if you have any questions.
[Note: HTML markup is below. Please do not edit.]
Reviewers' comments:
Reviewer's Responses to Questions
Comments to the Author
1. Is the manuscript technically sound, and do the data support the conclusions?
The manuscript must describe a technically sound piece of scientific research with data that supports the conclusions. Experiments must have been conducted rigorously, with appropriate controls, replication, and sample sizes. The conclusions must be drawn appropriately based on the data presented.
Reviewer #1: Partly
Reviewer #2: Partly
Reviewer #3: Partly
**********
2. Has the statistical analysis been performed appropriately and rigorously?
Reviewer #1: No
Reviewer #2: N/A
Reviewer #3: No
**********
3. Have the authors made all data underlying the findings in their manuscript fully available?
The PLOS Data policy requires authors to make all data underlying the findings described in their manuscript fully available without restriction, with rare exception (please refer to the Data Availability Statement in the manuscript PDF file). The data should be provided as part of the manuscript or its supporting information, or deposited to a public repository. For example, in addition to summary statistics, the data points behind means, medians and variance measures should be available. If there are restrictions on publicly sharing data—e.g. participant privacy or use of data from a third party—those must be specified.
Reviewer #1: Yes
Reviewer #2: Yes
Reviewer #3: No
**********
4. Is the manuscript presented in an intelligible fashion and written in standard English?
PLOS ONE does not copyedit accepted manuscripts, so the language in submitted articles must be clear, correct, and unambiguous. Any typographical or grammatical errors should be corrected at revision, so please note any specific errors here.
Reviewer #1: No
Reviewer #2: Yes
Reviewer #3: No
**********
5. Review Comments to the Author
Please use the space provided to explain your answers to the questions above. You may also include additional comments for the author, including concerns about dual publication, research ethics, or publication ethics. (Please upload your review as an attachment if it exceeds 20,000 characters)
Reviewer #1: This study claimed that CTDSP1 inhibitor rabeprazole regulates DNA-PKcs dependent topoisomerase I degradation and irinotecan drug resistance in colorectal cancer. The findings are likely useful for overcoming irinotecan resistance in colorectal cancer, as well as other solid tumors. However, experimental designs and descriptions are need to be improved. I have some comments as follows.
Major comments:
1. In Figure 1, the authors should quantify the intensity of each blots compared to beta-actin protein levels. In Figure 1B, the authors should determine the phosphorylated Topo I levels.
2. The authors treated cells with 2.5 �M SN-38 in Figure 1B but treated cells with SN-38 at concentrations lower than 7.5 nM in Figure 1C. Similar views also found in Figures 2, 3 and 5. Is it a typo? Otherwise, it is hard to believe that Topo I protein levels determine the therapeutic effectiveness of SN-38. Unless the authors show that Topo I protein degradation is visible at the treatment with SN-38 at low nM range.
3. In Figure 2B, the remarkable protein degradation should be detected in CTDSP1 knockdown cells at time 0 in comparison with control cells. However, it seems to be not. Please describe it.
4. The experimental procedure to generate Topo I-EGFP fusion by CRISPR/Cas9 system is not clearly presented. Please provide the sequence of sgRNA used in this study and sequencing result for the gene fusion.
5. What is the protein band for pDNA-PKcs in Figure 4B?
6. In Figure 5A, the protein levels of Topo 1 at time 0 seem to be not equal. Why?
7. In Table 1, if the clinical data from a small sample size is capable of supporting the conclusion that rabeprazole had an effect on the response to irinotecan in clinical settings?
Minor comments:
1. The authors should describe statistical tools they used in MM section
2. I found many typos and grammar errors in the manuscript. So, English editing of this manuscript should be needed.
Reviewer #2: This manuscript identified CTDSP1 as a primary upstream regulator of DNA-PKcs in response to topo I inhibitors. The authors used HCR116 and HT29 to identify the effect of CTDSP1 in irinotecan response. The results imply that CTDSP1 is related to drug sensitivity of irinotecan and rabeprazole treatment, resulting in drug resistant occurrence. In general, the idea of this manuscript is interesting. In order to improve the quality of this study, several suggestions are given in the following list.
1: The expressions of CTDSP1 and Topo I should be evaluated and quantified in western blot and Q-PCR analysis. This includes figure 1, 2, 3 and 5.
2: The appropriate internal control of pDNAPKcs is total form of DNAPKcs. Please add this important data with individual pDNAPKcs western blot.
3: p-ATM, ATM, p-ATR, ATR, gimma H2AX and XRCC4 protein expressions are always changed, followed by the activation of pDNAPKcs. The authors should also evaluate these proteins.
4: Several expressions of control are not equally presented. I would suggest to repeat these data. Example: figure 2b, time point 0 of control and siCTDSP1. Figure 2b, time point 0 of control and CTDSP1. figure 5, time point 0 of 0, 10 and 20uM drug treatment.
5: Immunofluorescence (IF) is not the appropriate method to quantify protein expression changes. All the protein expression of IF should be measured by flow cytometry, in order to understand both protein expression and cell population.
Reviewer #3: The presented study investigated the role of CTDSP1 in topoisomerase I associated irinotecan resistance in human colorectal cancer cells and effects of rabeprazeole, a CTDSP1 inhibitor, for reducing irinotecan resistance. Although experiments performed in this study were well organized and conducted, this manuscript was not prepared in an appropriate academic format with a plenty of missing information. This manuscript therefore is required a major revision before the acceptance is considered.
Comments:
1. Instruments, chemicals and reagents used in experiments shall be clearly described with information manufacturers including name of manufacturers and location in the section of materials and methods. Some of them only stated in results and figure legends such as Leica SP5 confocal microscope and EGFP.
2. Line 182, cell density for each cell type shall be described clear.
3. Replication numbers of each experiment shall be described by using proper statistical analysis to verify the differences among treatment conditions. Particularly the results of immunoblottings shall contain quantification data (E.g. Measurement of intensity) to compare the differences between cell types, treatments and experimental conditions with statistical analysis as well.
4. Methodology of statistics for this manuscript is completely missed. Statistical analysis for both in vitro and in vivo studies shall be clearly described in an independent paragraph of materials and methods. Authors claimed many significant differences and effects among experiments but gave no statistical results for verification.
5. This manuscript was not written in a proper academic English and organization. For example, the aims of study was not clearly described in introduction, and too many active voices and questing among writing. Professional English editing service therefore is required to make this manuscript more readable.
**********
6. PLOS authors have the option to publish the peer review history of their article (what does this mean?). If published, this will include your full peer review and any attached files.
If you choose “no”, your identity will remain anonymous but your review may still be made public.
Do you want your identity to be public for this peer review? For information about this choice, including consent withdrawal, please see our Privacy Policy.
Reviewer #1: No
Reviewer #2: Yes: Chia-Hwa Lee
Reviewer #3: No
[NOTE: If reviewer comments were submitted as an attachment file, they will be attached to this email and accessible via the submission site. Please log into your account, locate the manuscript record, and check for the action link "View Attachments". If this link does not appear, there are no attachment files to be viewed.]
While revising your submission, please upload your figure files to the Preflight Analysis and Conversion Engine (PACE) digital diagnostic tool, https://pacev2.apexcovantage.com/. PACE helps ensure that figures meet PLOS requirements. To use PACE, you must first register as a user. Registration is free. Then, login and navigate to the UPLOAD tab, where you will find detailed instructions on how to use the tool. If you encounter any issues or have any questions when using PACE, please email us at gro.solp@serugif. Please note that Supporting Information files do not need this step.
Author response to Decision Letter 0
5 Apr 2020
We also appreciate the time and effort you and each of the reviewers have dedicated to providing insightful feedback on ways to strengthen our paper. Thus, it is with great pleasure that we resubmit our article for further consideration. We have incorporated changes that reflect the detailed suggestions you have graciously provided.
Decision Letter 1
11 May 2020
PONE-D-20-00259R1
CTDSP1 inhibitor rabeprazole regulates DNA-PKcs dependent topoisomerase I degradation and irinotecan drug resistance in colorectal cancer
PLOS ONE
Dear Dr. Ando_Koji,
Thank you for submitting your manuscript to PLOS ONE. After careful consideration, we feel that it has merit but does not fully meet PLOS ONE’s publication criteria as it currently stands. Therefore, we invite you to submit a revised version of the manuscript that addresses the points raised during the review process.
==============================
ACADEMIC EDITOR: There are some major concerns which need to be improved. Please check the reviewers' question.
==============================
We would appreciate receiving your revised manuscript by Jun 25 2020 11:59PM. When you are ready to submit your revision, log on to https://www.editorialmanager.com/pone/ and select the 'Submissions Needing Revision' folder to locate your manuscript file.
If you would like to make changes to your financial disclosure, please include your updated statement in your cover letter.
To enhance the reproducibility of your results, we recommend that if applicable you deposit your laboratory protocols in protocols.io, where a protocol can be assigned its own identifier (DOI) such that it can be cited independently in the future. For instructions see: http://journals.plos.org/plosone/s/submission-guidelines#loc-laboratory-protocols
Please include the following items when submitting your revised manuscript:
A rebuttal letter that responds to each point raised by the academic editor and reviewer(s). This letter should be uploaded as separate file and labeled 'Response to Reviewers'.
A marked-up copy of your manuscript that highlights changes made to the original version. This file should be uploaded as separate file and labeled 'Revised Manuscript with Track Changes'.
An unmarked version of your revised paper without tracked changes. This file should be uploaded as separate file and labeled 'Manuscript'.
Please note while forming your response, if your article is accepted, you may have the opportunity to make the peer review history publicly available. The record will include editor decision letters (with reviews) and your responses to reviewer comments. If eligible, we will contact you to opt in or out.
We look forward to receiving your revised manuscript.
Kind regards,
Yu-Jia Chang
Academic Editor
PLOS ONE
[Note: HTML markup is below. Please do not edit.]
Reviewers' comments:
Reviewer's Responses to Questions
Comments to the Author
1. If the authors have adequately addressed your comments raised in a previous round of review and you feel that this manuscript is now acceptable for publication, you may indicate that here to bypass the “Comments to the Author” section, enter your conflict of interest statement in the “Confidential to Editor” section, and submit your "Accept" recommendation.
Reviewer #1: (No Response)
Reviewer #2: (No Response)
Reviewer #3: All comments have been addressed
**********
2. Is the manuscript technically sound, and do the data support the conclusions?
The manuscript must describe a technically sound piece of scientific research with data that supports the conclusions. Experiments must have been conducted rigorously, with appropriate controls, replication, and sample sizes. The conclusions must be drawn appropriately based on the data presented.
Reviewer #1: Yes
Reviewer #2: Partly
Reviewer #3: Yes
**********
3. Has the statistical analysis been performed appropriately and rigorously?
Reviewer #1: Yes
Reviewer #2: Yes
Reviewer #3: Yes
**********
4. Have the authors made all data underlying the findings in their manuscript fully available?
The PLOS Data policy requires authors to make all data underlying the findings described in their manuscript fully available without restriction, with rare exception (please refer to the Data Availability Statement in the manuscript PDF file). The data should be provided as part of the manuscript or its supporting information, or deposited to a public repository. For example, in addition to summary statistics, the data points behind means, medians and variance measures should be available. If there are restrictions on publicly sharing data—e.g. participant privacy or use of data from a third party—those must be specified.
Reviewer #1: Yes
Reviewer #2: No
Reviewer #3: Yes
**********
5. Is the manuscript presented in an intelligible fashion and written in standard English?
PLOS ONE does not copyedit accepted manuscripts, so the language in submitted articles must be clear, correct, and unambiguous. Any typographical or grammatical errors should be corrected at revision, so please note any specific errors here.
Reviewer #1: Yes
Reviewer #2: No
Reviewer #3: Yes
**********
6. Review Comments to the Author
Please use the space provided to explain your answers to the questions above. You may also include additional comments for the author, including concerns about dual publication, research ethics, or publication ethics. (Please upload your review as an attachment if it exceeds 20,000 characters)
Reviewer #1: The authors have fully answered my questions. I suggest the manuscript is acceptable for publication.
Reviewer #2: This is the second-round review of this manuscript. Unfortunately the authors failed to appropriately response the comment from the first-round review.
4. The experimental procedure to generate TopoI-EGFP fusion by CRISPR/Cas9 system is not clearly
presented. Please provide the sequence of sgRNA used in this study and sequencing result for the gene
fusion.
A well hTOP1 gene edit efficiency analysis and protein expression analysis should be performed in this manuscript. A previous study may be taken as an example. HDAC1,2 Knock-Out and HDACi Induced Cell Apoptosis in Imatinib-Resistant K562 Cells. Int J Mol Sci. 2019 May 8;20(9). pii: E2271. 10.3390/ijms20092271.
With the second review, some critical issues raised in the revised manuscript and the comments need to be appropriately responded.
1: The photos of clonogenic assay in figure 1C, 3C should be attached in supplementary data.
2: It is very confusing of the following description “A comparison of two CRC cell lines, HCT116 and HT29, indicated higher expression of CTDSP1 in HCT116. A comparison of two CRC cell lines, HCT116 and HT29, indicated higher expression of 3 CTDSP1 in HCT116. We also observed minimal proteasomal degradation and HCT116 and HCT116 cells are significantly more sensitive to SN-38.” Page 9, line 2-4.
3: The significant TopoI protein expression drop is occurred after 2.5 μM SN-38 treatment in HCT116 cell (figure 1B). However, the bar figure doesn’t reflect this protein change. The results from blot and bar figure are not consistent.
4: The protein expression blot of topoI-GFP protein in figure 2C and 5B should be revealed in figures.
5: The CTDSP1 expression in figure 1B, 2B, 3B, 4B, and 5A should also be revealed.
6: In figure 3B, a very significant topoI increase was observed in cells overexpressing CTDSP1, the description in result section is misleading.
7: A western blot result of pDNAPKcs expression in figure 4C should be revealed in the same figure.
8: The orders of the cell input in blot should consistent in this manuscript.
For example: figure 2A is different from other figures (control, si or overexpression).
9: IRB approval number is nor provided in the revised manuscript.
Reviewer #3: This reviewer is satisfied with this revised manuscript and suggest an acceptance decision to be considered by PLOS One.
**********
7. PLOS authors have the option to publish the peer review history of their article (what does this mean?). If published, this will include your full peer review and any attached files.
If you choose “no”, your identity will remain anonymous but your review may still be made public.
Do you want your identity to be public for this peer review? For information about this choice, including consent withdrawal, please see our Privacy Policy.
Reviewer #1: No
Reviewer #2: Yes: Chia-Hwa Lee
Reviewer #3: No
[NOTE: If reviewer comments were submitted as an attachment file, they will be attached to this email and accessible via the submission site. Please log into your account, locate the manuscript record, and check for the action link "View Attachments". If this link does not appear, there are no attachment files to be viewed.]
While revising your submission, please upload your figure files to the Preflight Analysis and Conversion Engine (PACE) digital diagnostic tool, https://pacev2.apexcovantage.com/. PACE helps ensure that figures meet PLOS requirements. To use PACE, you must first register as a user. Registration is free. Then, login and navigate to the UPLOAD tab, where you will find detailed instructions on how to use the tool. If you encounter any issues or have any questions when using PACE, please email us at gro.solp@serugif. Please note that Supporting Information files do not need this step.
Author response to Decision Letter 1
31 May 2020
Thank you for inviting us to submit a revised draft of our manuscript entitled, “CTDSP1 inhibitor rabeprazole regulates DNA-PKcs dependent topoisomerase I degradation and irinotecan drug resistance in colorectal cancer.” to PLOS ONE. We also appreciate the time and effort you and each of the reviewers have dedicated to providing insightful feedback on ways to strengthen our paper. Thus, it is with great pleasure that we resubmit our article for further consideration. We have incorporated changes that reflect the detailed suggestions you have graciously provided.
Attachment
Submitted filename: Response to Reviewers.docx
Decision Letter 2
1 Jul 2020
CTDSP1 inhibitor rabeprazole regulates DNA-PKcs dependent topoisomerase I degradation and irinotecan drug resistance in colorectal cancer
PONE-D-20-00259R2
Dear Dr. Koji Ando
We’re pleased to inform you that your manuscript has been judged scientifically suitable for publication and will be formally accepted for publication once it meets all outstanding technical requirements.
Within one week, you’ll receive an e-mail detailing the required amendments. When these have been addressed, you’ll receive a formal acceptance letter and your manuscript will be scheduled for publication.
An invoice for payment will follow shortly after the formal acceptance. To ensure an efficient process, please log into Editorial Manager at http://www.editorialmanager.com/pone/, click the 'Update My Information' link at the top of the page, and double check that your user information is up-to-date. If you have any billing related questions, please contact our Author Billing department directly at gro.solp@gnillibrohtua.
If your institution or institutions have a press office, please notify them about your upcoming paper to help maximize its impact. If they’ll be preparing press materials, please inform our press team as soon as possible -- no later than 48 hours after receiving the formal acceptance. Your manuscript will remain under strict press embargo until 2 pm Eastern Time on the date of publication. For more information, please contact gro.solp@sserpeno.
Kind regards,
Yu-Jia Chang
Academic Editor
PLOS ONE
Additional Editor Comments (optional):
Reviewers' comments:
Acceptance letter
9 Jul 2020
PONE-D-20-00259R2
CTDSP1 inhibitor rabeprazole regulates DNA-PKcs dependent topoisomerase I degradation and irinotecan drug resistance in colorectal cancer
Dear Dr. Ando:
I'm pleased to inform you that your manuscript has been deemed suitable for publication in PLOS ONE. Congratulations! Your manuscript is now with our production department.
If your institution or institutions have a press office, please let them know about your upcoming paper now to help maximize its impact. If they'll be preparing press materials, please inform our press team within the next 48 hours. Your manuscript will remain under strict press embargo until 2 pm Eastern Time on the date of publication. For more information please contact gro.solp@sserpeno.
If we can help with anything else, please email us at gro.solp@enosolp.
Thank you for submitting your work to PLOS ONE and supporting open access.
Kind regards,
PLOS ONE Editorial Office Staff
on behalf of
Dr Yu-Jia Chang
Academic Editor
PLOS ONE
Articles from PLOS ONE are provided here courtesy of PLOS
Full text links
Read article at publisher's site: https://doi.org/10.1371/journal.pone.0228002
Read article for free, from open access legal sources, via Unpaywall:
https://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0228002&type=printable
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1371/journal.pone.0228002
Article citations
Network-based elucidation of colon cancer drug resistance mechanisms by phosphoproteomic time-series analysis.
Nat Commun, 15(1):3909, 09 May 2024
Cited by: 1 article | PMID: 38724493 | PMCID: PMC11082183
Targeting SMAD-Dependent Signaling: Considerations in Epithelial and Mesenchymal Solid Tumors.
Pharmaceuticals (Basel), 17(3):326, 01 Mar 2024
Cited by: 1 article | PMID: 38543112 | PMCID: PMC10975212
Review Free full text in Europe PMC
Tumor Suppressor Properties of Small C-Terminal Domain Phosphatases in Clear Cell Renal Cell Carcinoma.
Int J Mol Sci, 24(16):12986, 19 Aug 2023
Cited by: 2 articles | PMID: 37629167 | PMCID: PMC10455398
Progress in the studies on the molecular mechanisms associated with multidrug resistance in cancers.
Acta Pharm Sin B, 13(3):982-997, 07 Oct 2022
Cited by: 15 articles | PMID: 36970215 | PMCID: PMC10031261
Review Free full text in Europe PMC
Proton Pump Inhibitors and Cancer: Current State of Play.
Front Pharmacol, 13:798272, 14 Mar 2022
Cited by: 11 articles | PMID: 35359844 | PMCID: PMC8963837
Review Free full text in Europe PMC
Go to all (8) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Camptothecin resistance is determined by the regulation of topoisomerase I degradation mediated by ubiquitin proteasome pathway.
Oncotarget, 8(27):43733-43751, 01 Jul 2017
Cited by: 14 articles | PMID: 28415827 | PMCID: PMC5546437
Developing a Phosphospecific IHC Assay as a Predictive Biomarker for Topoisomerase I Inhibitors.
J Histochem Cytochem, 66(8):549-561, 27 Mar 2018
Cited by: 1 article | PMID: 29587004 | PMCID: PMC6071180
Epigenetic changes in histone acetylation underpin resistance to the topoisomerase I inhibitor irinotecan.
Nucleic Acids Res, 45(3):1159-1176, 01 Feb 2017
Cited by: 31 articles | PMID: 28180300 | PMCID: PMC5388393
Resistance to TOP-1 Inhibitors: Good Old Drugs Still Can Surprise Us.
Int J Mol Sci, 24(8):7233, 13 Apr 2023
Cited by: 5 articles | PMID: 37108395 | PMCID: PMC10138578
Review Free full text in Europe PMC