Abstract
Free full text

Human germinal centres engage memory and naïve B cells after influenza vaccination
Summary
Influenza viruses remain a major public health threat. Seasonal influenza vaccination in humans primarily stimulates pre-existing memory B cells, leading to a transient wave of circulating antibody-secreting plasmablasts1–3. This recall response contributes to “original antigenic sin,” the selective boosting of antibody specificities from prior exposures to influenza virus antigens4. It remains unclear whether such vaccination can also induce germinal centre (GC) reactions in the draining lymph node (LN) where diversification and maturation of recruited B cells can occur5. Here we used ultrasound-guided fine needle aspiration to serially sample the draining LNs and investigate the dynamics and specificity of GC B cell responses after influenza vaccination in humans. We show that influenza vaccine-binding GC B cells can be detected as early as 1 week after vaccination. In 3 out of 8 participants, we detected vaccine-binding GC B cells up to 9 weeks after vaccination. Between 12% and 88% of the responding GC B cell clones overlapped with those detected among early circulating plasmablasts. These shared B cell clones had high frequencies of somatic hypermutation (SHM) and encoded broadly cross-reactive monoclonal antibodies (mAbs). In contrast, vaccine-induced B cell clones detected only in the GC compartment exhibited significantly lower SHM frequencies and predominantly encoded strain-specific mAbs, suggesting a naïve B cell origin. Electron microscopy-based epitope mapping revealed that some of these strain-specific mAbs recognized epitopes that were not targeted by the early plasmablast response. Our results indicate that influenza virus vaccination of humans can elicit a GC reaction to which B cell clones targeting novel epitopes are more likely to be recruited, thereby broadening the spectrum of vaccine-induced protective antibodies against this rapidly mutating pathogen.
Introduction
Seasonal influenza viruses kill 290,000 to 650,000 people globally every year6. As the virus drifts, novel antigenic targets emerge, creating a pressing need for the annual vaccine to engage new B cell clones that recognize such targets. The germinal centre (GC) reaction is critical for generating high-affinity and durable B cell responses5. It is currently unknown whether seasonal influenza virus immunization of humans can elicit a GC response in the draining lymph nodes (LN) where diversification and maturation of recruited B cells can occur. Studies examining human B cell responses have traditionally focused on sampling the easily accessible blood compartment, but ultrasound-guided fine needle aspiration (FNA) has enabled sampling of LNs with good representation of cell populations recovered by excisional biopsy, including GC B cells7–9.
Results
Vaccine-induced B cell responses in blood and lymph nodes
Eight healthy young adults were enrolled in a seasonal influenza vaccination study. Blood and FNA specimens were collected prior to vaccination and at 1, 2, approximately 4, and 9 weeks after vaccination with the 2018/2019 quadrivalent inactivated influenza virus vaccine (QIV) (Fig. 1a). QIV-binding antibody-secreting PBs were measured in blood by enzyme-linked immune absorbent spot (ELISpot). PBs peaked in blood during the first week after vaccination in all participants, with the frequency varying from 160 to 3,400 IgG-secreting QIV-binding PBs per mL (Fig. 1b, Extended Data Fig. 1a). Haemagglutinin (HA)-binding PB were also measured by flow cytometry and peaked 1-week post-vaccination (CD20lo HA+) and activated B cells (ABC, CD20hi HA+) peaked during the second week before declining (Fig. 1d, Extended Data Fig. 1b,,ff)10. Four weeks after vaccination, anti-QIV IgG plasma antibody titers were elevated compared to those at baseline as measured by enzyme-linked immunosorbent assay (ELISA), along with haemagglutination-inhibiting antibody titers against the four constituent viruses of the vaccine as measured by the haemagglutination inhibition (HAI) assay (Extended Data Fig. 1g, ,hh).
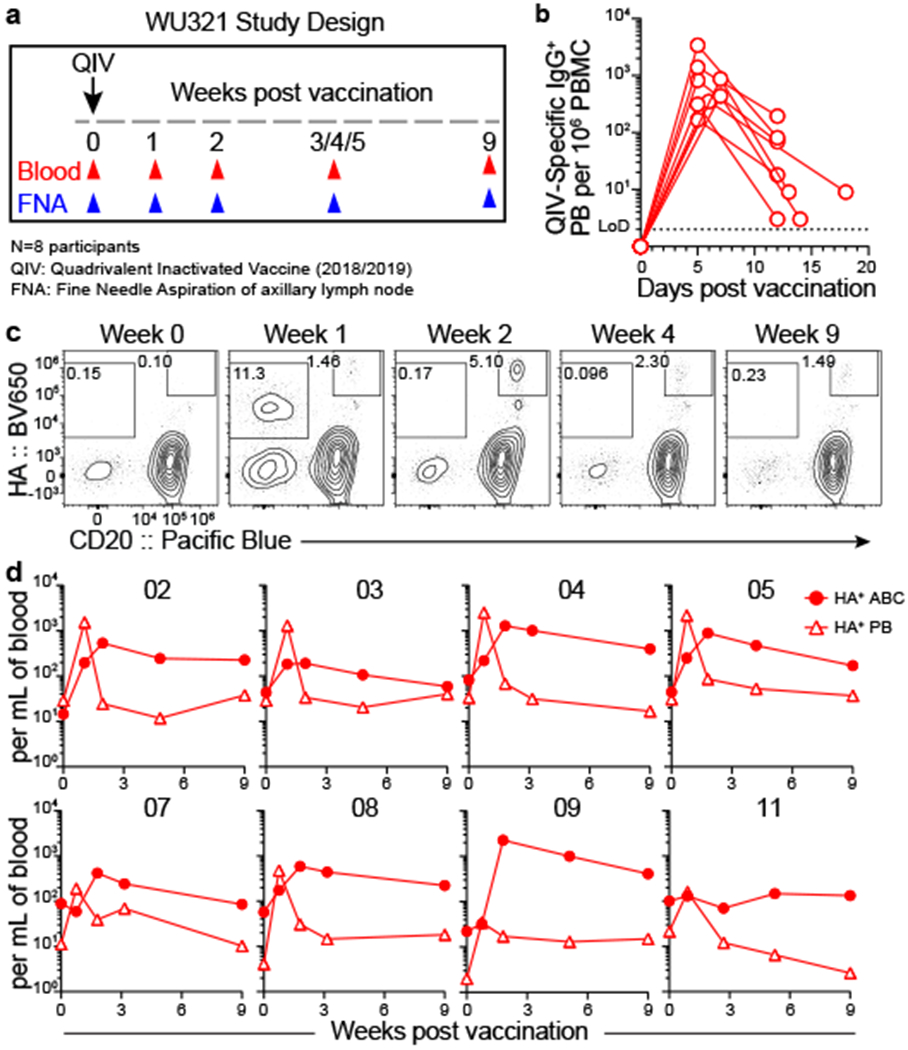
a) Study design. Eight healthy adults (ages 26–40) who had not been vaccinated against influenza for 3 years were enrolled and received quadrivalent inactivated vaccine (QIV) i.m. Blood and fine needle aspirates (FNAs) of ipsilateral axillary lymph nodes were collected pre-vaccination and at 1, 2, approximately 4, and 9 weeks after vaccination. b) ELISpot quantification of QIV-binding IgG-secreting plasmablasts (PBs) in blood at baseline, 1, and 2 weeks post-vaccination. c, d) Representative gating (c) and kinetics (d) of HA-binding activated B cells (ABCs, CD20+ HA+, closed circles) and PBs (CD20lo HA+, open triangles) in PBMC. Cells pre-gated IgD− CD19+ CD4− live singlet lymphocytes. Symbols at each timepoint in b and d represent one sample (n=8).
In humans, QIV is injected into the deltoid muscle of the upper arm. This area primarily drains into the lateral axillary LNs11,12. Both axilla of each participant were examined by ultrasound before vaccination to identify an accessible LN. Once identified, the hypoechoic LN cortex, which contains B cell follicles, was sampled by FNA (Fig. 2a), and QIV was administered in the ipsilateral muscle. The same LN was serially sampled, and its dimensions and cortical thickness were measured. LN cortical thickening following vaccination was observed in participants 04, 05, 09, and 11 (Fig. 2b, Extended Data Fig. 2a). The median number of live cells recovered per FNA was 8.9×105 (range, 2.3×104 to 6.9×106; Extended Data Fig. 2b). Flow cytometric analysis of FNA detected significantly higher frequencies of B cells and CD4+ T cells and lower frequencies of CD14+ monocytes or granulocytes than those in peripheral blood mononuclear cells (PBMCs) (Fig. 2c, Extended Data Fig. 1b). These trends were observed in FNA specimens from all participants except for participant 02 where the frequencies of CD14+ cells were consistently high, suggesting significant blood contamination (Extended Data Fig. 2c). FNA data from specimens in which fewer than 4×104 live cells were recovered or the CD14+ percentage exceeded 10% of CD45+ were excluded from further analyses, including all FNA specimens from participant 02.

a) Representative sonogram of FNA; note hyperechoic medulla “m” and needle sampling hypoechoic cortex “c” from top left. b) Representative sonograms of axillary LN before each FNA. c) Percentages of CD19+ CD4− B cells (left), CD14− CD4+ T cells (middle), and CD14+ CD4− monocytes or granulocytes (right) of CD45+ in paired PBMC (red) and FNA (blue) samples measured by flow cytometry. Lines represent means. Each symbol represents one sample (n=8). P-values from paired two-sided Student’s t - tests. d) Clustering via t-distributed stochastic neighbor embedding (tSNE) of all cells from whole PBMC (left) and FNA (right) scRNAseq samples pooled from all timepoints from participant 05. Each dot represents a cell, colored by phenotype as defined by the gene expression profile. Total numbers of cells are below clusters. e) Representative flow cytometry gating of CD20hi CD38int population in FNA. Cells pre-gated IgDlo CD19+ CD4− live singlet lymphocytes. f) Kinetics of total (closed circles) and HA+ (open circles) GC B cells in FNA, as defined by flow cytometry gates in e and Extended Data Fig. 2k. Symbols at each timepoint represent one sample (n=7). Daggers denote samples excluded from analysis due to low cell recovery or blood contamination.
Single cell RNA sequencing (scRNAseq) was performed on whole PBMCs and FNA samples from all timepoints for participant 05 to comprehensively examine their cellular compositions. Compared to PBMCs, B cells and CD4+ T cells comprised substantially larger populations in FNA (38.5% vs 5.8% for B cells and 42.3% vs 34.9% for CD4+ T cells), whereas monocytes and platelets were substantially lower (Fig. 2d, Extended Data Fig. 2d–h, Extended Data Table 1). Surface staining revealed a population of CD20hi CD38int IgDlo B cells that increased in frequency after vaccination in 5 of the 7 participants analysed (participants 03, 04, 05, 08, and potentially 11, whose baseline FNA specimen was among those excluded) (Fig. 2e). Intranuclear staining confirmed that this CD20hi CD38int IgDlo B cell population expressed Bcl6, indicating that it was comprised of GC B cells, whereas equivalently gated PBMCs did not (Extended Data Fig. 2i). The expression of additional markers on the CD20hi CD38int IgDlo population was consistent with a GC phenotype, including CD24lo, CD27int, CD71hi, CXCR5int, and Ki-67+ (Extended Data Fig. 2j). HA+ GC B cells were detected and increased in frequency after vaccination in participants 04, 05, and 11, along with HA+ IgDlo CD20− CD38+ PBs and IgDlo CD20+ CD38− ABCs, which also were detected in participants 07 and 09 (Fig. 2f, Extended Data Figs. 1c, 2k-m, and Extended Data Table 2).
Clonal overlap between PB and GC B cells
We clonally analysed PBs and GC B cells from participants 04, 05, and 11. We sorted single PBs isolated from 1-week post-vaccination PBMCs and expressed the corresponding immunoglobulin genes as recombinant monoclonal antibodies (mAbs, Extended Data Figs. 1d, ,3a3a)13,14. We generated 54, 206, and 74 clonally distinct mAbs from PBs, of which 4, 125, and 27 bound QIV, from participants 04, 05, and 11, respectively (Fig. 3a, Extended Data Fig. 3b). We also generated mAbs from single GC B cells sorted irrespective of their binding to HA probes from three timepoints for each of the three participants. We generated a total of 133, 153, and 87 clonally distinct mAbs, of which 42, 61, and 35 bound QIV by ELISA from participants 04, 05, and 11, respectively (Fig. 3a, Extended Data Figs. 1e, ,3c).3c). Clonal analysis using Vh gene information15,16 combined from the mAbs and repertoire data from bulk sorted PBs revealed substantial clonal overlap between B cell clones participating in the PB and GC responses, with 27%, 58%, and 7% of total GC sequences, and 88%, 78%, and 12% of the QIV-binding GC sequences clonally related to PBs for participants 04, 05, and 11, respectively (Fig. 3b, Extended Data Fig. 3d). The proportion of GC sequences clonally related to the early PB response over time was variable for the three participants; overlap increased, stayed fairly constant, and decreased for participants 04, 05, and 11, respectively (Extended Data Fig. 3e). Clones recruited to the early PB response were highly mutated, consistent with derivation from recalled memory B cells. Clones that participated in both GC and PB responses had similarly high levels of SHM, whereas GC clones not found in the early blood PB response carried significantly lower mutational burdens, suggesting a predominantly naïve B cell origin (Fig. 3c).
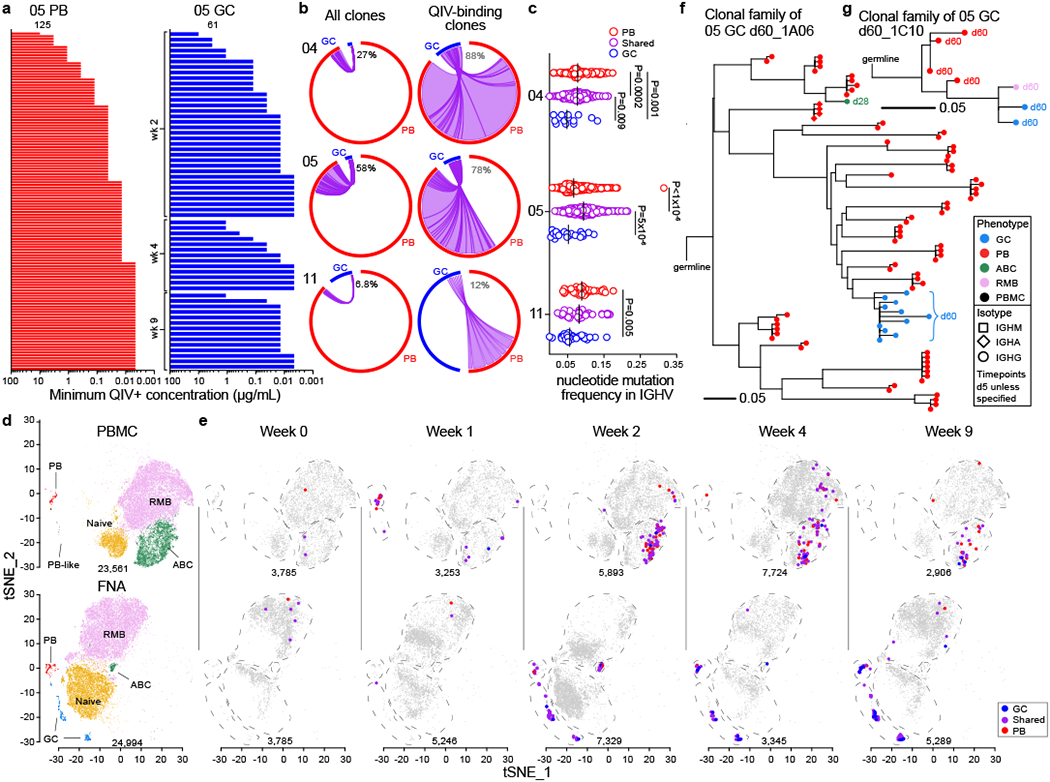
a) Minimum positive concentrations of clonally unique mAbs generated from singly sorted PBs from week 1 post-vaccination (left) and GC B cells at the indicated timepoints (right) from participant 05 as determined by QIV ELISA; positive binding defined as greater than 3× background. b) Clonal overlap of sequences from mAb cloning and bulk repertoire analysis between PBs sorted from PBMCs 1-week post-vaccination and GC B cells from all timepoints among total (left) and only QIV-binding (right) sequences. Chord width corresponds to clonal population size; numbers of sequences are in Extended Data Table 3. Percentages are of GC sequences overlapping with PBs. c) Immunoglobulin heavy chain variable region (IGHV) gene mutation frequency of sorted QIV-binding PBs and GC B cells that overlapped clonally (purple) or did not (red and blue). Vertical lines represent medians. Sequence counts were 14, 149, and 1000 (participant 04); 22, 1129, and 1034 (participant 05); 29, 43, and 57 (participant 11) for GC, shared, and PB, respectively. P-values from two-sided Dunn’s multiple comparisons test. d, e) Clustering via tSNE of B cells showing GC (blue), PB, (red), PB-like (brown), naïve (gold), ABC (green), and RMB (lavender) populations pooled from all timepoints (d) and QIV-binding clonal kinetics showing clones found in GC (blue), early PB (red), or both (purple) at the indicated timepoints (e). f, g) Dendrograms of clonal families of H1 HA–binding day 60 GC mAbs 1A06 (f) and 1C10 (g). Horizontal branch length represents the expected number of substitutions per codon in V -region genes, corresponding to the key in the lower left of each panel. Colored symbols represent sequences from cells isolated at day 5 unless otherwise specified, corresponding to the indicated phenotype and isotype.
We next tracked the clonal dynamics of vaccine-responding B cell clones using the scRNAseq data from participant 05. Gene expression–based clustering of B cells from whole FNA samples and from pooled MBC-enriched and whole PBMCs identified naïve, ABC, resting MBC (RMB), and PB populations in FNA and PBMC samples. GC B cells were identified as a distinct cluster only in the FNA samples (Fig. 3d, Extended Data Figs. 3g, ,f,f, ,4a4a–h). Isotype distribution among the clusters was consistent with ELISpot and flow cytometry analyses; PBs and GC B cells were predominantly IgG, naïve B cells expressed non-isotype switched receptors, and RMB expressed IgM, IgG, or IgA (Extended Data Fig. 4i). Sequences clonally related to QIV-binding mAbs were identified in PB and ABC compartments in 1-week post-vaccination PBMCs; thereafter, QIV-binding PBs declined rapidly and ABCs predominated. In FNA samples, QIV-binding GC B cells and PBs were observed after week 2, and QIV-binding RMB appeared at week 2–4 (Fig. 3e). The SHM frequency of clones recruited to the GC but not early PB response remained lower at all timepoints than that of clones found in both compartments (Extended Data Fig. 3h). We next constructed dendrograms of some responding B cell clonal lineages that were either highly mutated and extensively expanded (1A06, Fig. 3f) or minimally mutated and less expanded (1C10, Fig. 3g).
Mapping vaccine-induced PB and GC B cells
Influenza A viruses are categorized into two phylogenetic groups based on the HA sequence17. The QIV contains antigens that are derived from circulating H1N1 (group 1 HA) and H3N2 (group 2 HA) strains, in addition to strains from the two antigenically distinct influenza B lineages. Antibodies are considered broadly cross-reactive if they recoginze HAs from H1N1 or N3N2 strains that no longer circulate in humans. We analyzed the binding breadth of mAbs derived from PB and GC B cell using influenza virus protein microarrays (IVPMs)18,19 and found that both broadly cross-reactive and strain-specific clones were recruited to the GC and PB responses (Fig. 4a, Extended Data Fig. 5a–c). However, analysis of all QIV binding mAbs combined from participants 04, 05, and 11 indicated that the proportion of strain-specific mAbs among H1 HA and influenza B HA binders was significantly higher in clones recruited to the GC but not early PB response. Similar trends were observed for the individual participants (Fig. 4b, Extended Data Fig. 5d).
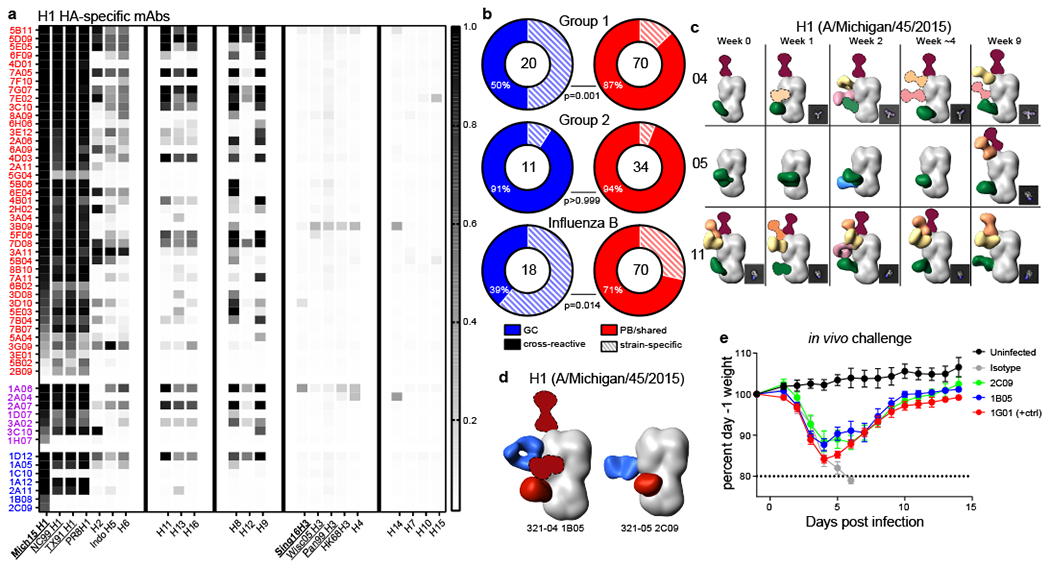
a) Binding of group 1–binding mAbs generated from singly sorted PBs and GC B cells that overlapped clonally (purple) or did not overlap (red and blue) for PB and GC B cells, respectively, from participant 05 using an influenza virus protein microarray (IVPM). Scale bar is median fluorescence intensity. Vaccine strains in bold type; underlined strains circulated in humans in participants’ lifetimes. b) Percentages of mAbs that bound two or more HA strains from participants 04, 05, and 11 from GC clones that did not participate in the early PB response (blue) and from PB and shared clones (red). P-values from Fisher’s exact test. The number of mAbs is indicated in the middle of the charts. c) Polyclonal epitopes of Fabs from plasma at indicated timepoints from participants 04, 05, and 11 with HA from A/Michigan/45/2015. Epitopes were determined by 3D reconstructions and/or 2D class averages (images to bottom right of 3D reconstructions). HA proteins shown in grey; Fabs shown in multiple colors. d) Monoclonal and polyclonal epitopes of immune complexes with HA from A/Michigan/45/2015 and Fabs generated from the indicated GC mAbs (blue) and plasma pAbs (red). Fabs with dashed outlines have predicted epitopes due to limited particle representation. e) Protection of GC mAbs 1B05 and 2C09 in a mouse influenza virus challenge model. Mice received 5 mg/kg of the indicated mAb intraperitoneally 1 day before intranasal challenge with A/California/04/2009 E3 (H1N1), and were weighed daily; 7 mice were used for 1B05 and 1G01, 6 for 2C09 and isotype control, and 5 for uninfected. Error bars indicate mean ±SEM.
We next examined the epitopes targeted by plasma anti-HA antibodies in participants 04, 05, and 11. We used electron microscopy polyclonal epitope mapping (EMPEM)20,21 of antigen binding fragments (Fabs) purified from the plasma of these participants to identify the binding landscape to the H1 and H3 HAs included in QIV. The polyclonal antibody (pAb) response diversified over time in participants 04, 05, and 11; new pAbs that appeared in the first four weeks after vaccination likely represented contributions from the early PB response, whereas later additions were likely also contributed by GC-derived longer-lived plasma cells (Fig. 4c, Extended Data Fig. 5e–g). We then mapped the epitopes targeted by some GC clones that did not participate in the early PB response by negative stain electron microscopy of Fab/HA complexes and found that the binding footprint of some Fabs overlapped with those found in plasma, whereas others targeted unique epitopes (Fig. 4d, Extended Data Fig. 5h–k). We identified two mAbs that targeted unique H1 HA epitopes and were protective in mice against lethal influenza challenge: 1B05 and 2C09 from participants 04 and 05, respectively (Fig. 4e). Notably, these and another mAb not detected in plasma, 1B08 from participant 05, were strain-specific and had low Vh mutation frequencies of 0.033, 0.026, and 0.026, respectively.
Discussion
This study sought to define the role of the GC reaction in human B cell responses to influenza virus vaccination. We found that a substantial proportion of vaccine-binding GC B cell clones were part of the early circulating PB response. These shared clones exhibited the two cardinal features of a recall B cell response to influenza: they possessed high levels of SHM, and they were broadly cross-reactive. By contrast, a subset of vaccine-induced GC B cells that were not detected among early circulating PBs had low SHM levels and were more likely to be strain-specific, indicating a probable naïve B cell origin. Studies of secondary GC responses in mice have found that naïve B cells and less somatically mutated MBCs are more efficiently recruited to GCs than MBCs that have undergone extensive GC selection22–25. One key difference is that in the current study, the vaccine antigens were likely different from the influenza antigens the participants had previously encountered, as none had been vaccinated for three years, whereas in the mouse studies, animals were boosted with the same antigen. Compared to homologous reimmunization, heterologous boosting may recruit MBCs to GC responses more efficiently due to decreased competition from preexisting antibodies.
To our knowledge, the current study provides the first direct evidence of vaccine-induced GC responses in humans, but it does have some limitations. We detected HA-binding GC B cells in only three participants despite readily detectable peripheral B cell responses in all eight participants. It is possible that the LNs sampled in the other five participants were not the primary draining LNs. It is also possible that vaccination did not elicit a GC response in these participants. A second limitation pertains to our clonal analyses of vaccine-responding B cells, in which we excluded those encoding mAbs that did not show detectable binding by QIV ELISA. Studies in mice26,27 and humans28 indicate that low-affinity B cells can be recruited to GCs despite encoding BCRs that, when expressed as mAbs, are below the ELISA detection threshold. This suggests that some very low-affinity clones that are in fact specific for QIV could have been excluded from our analyses, resulting in overestimation of the proportion of higher-affinity clones in the response. In a similar vein, the epitope analysis by EMPEM may have failed to detect some low-affinity and/or -frequency circulating pAbs, overestimating the contribution from more abundant, higher-affinity PB- and plasma cell-derived antibodies.
A significant fraction of vaccine-induced GC- and PB-derived mAbs were cross-reactive with previously circulating influenza virus strains. This response is reminiscent of the original antigenic sin (OAS) phenomenon, where antibody responses are preferentially directed against previously encountered influenza strains when a person is exposed to a contemporary strain4. We propose that suboptimal GC B cell responses after influenza vaccination result in a more pronounced OAS effect due to inefficient engagement of naïve B cells targeting novel epitopes. High levels of baseline antibodies against conserved influenza epitopes may interfere with the formation or continued maintenance of vaccination-induced GC responses in humans. If so, new vaccine formulations that promote robust GC reactions are more likely to induce a more diverse antibody response against circulating and emerging influenza virus strains.
Methods
Sample collection, preparation, and storage.
All studies were approved by the Institutional Review Board of Washington University in St. Louis. Written consent was obtained from all participants. Eight participants who had not been vaccinated against influenza for at least three years were enrolled, including 1 female and 7 males, aged 26–40 years old. Peripheral blood mononuclear cells (PBMCs) were isolated using Vacutainer CPT tubes (BD), the remaining RBCs were lysed with ammonium chloride lysis buffer (Lonza), and cells were immediately used or cryopreserved in 10% dimethylsulfoxide in FBS. Ultrasound guided fine-needle aspiration (FNA) of axillary lymph nodes was performed by a qualified physician’s assistant under the supervision of a radiologist. Lymph node dimensions and cortical thickness were measured before each FNA. For each FNA sample, 6 passes were made using 25 -gauge needles, each of which was flushed with 3 mL of RPMI 1640 supplemented with 10% FBS and 100 U/mL penicillin/streptomycin, followed by three 1 -mL rinses. Red blood cells were lysed with ammonium chloride buffer (Lonza), washed twice with PBS supplemented with 2% FBS and 2 mM EDTA, and immediately used or cryopreserved in 10% DMSO in FBS. Participants reported no adverse effects of phlebotomy, serial FNA, or vaccination.
Vaccine.
Flucelvax QIV influenza vaccine (2018/2019 season) was purchased from Seqirus.
Antigens.
For ELISpot, plates were coated with QIV, tetanus/diphtheria vaccine (Grifols), or recombinant hemagglutinin (HA) proteins derived from the pandemic H1N1 (A/Michigan/45/2015), H3N2 (A/Singapore/INFIMH-16-0019/2016), B/Yamagata/16/88-like lineage (B/Phuket/3073/2013), or B/Victoria/2/87-like lineage (B/Brisbane/60/2008) influenza viruses. HA proteins were expressed in a baculovirus expression system as described previously29. For flow cytometry staining, recombinant HA from A/Michigan/45/2015 (a.a.18–529), A/Singapore/INFIMH-16-0019/2016 (a.a.17–529), and B/Colorado/06/2017 (a.a. 18–546) expressed in 293F cells were purchased from Immune Technology and biotinylated using the EZ-Link Micro NHS-PEG4-Biotinylation Kit (Thermo Fisher); excess biotin was removed using 7-kDa Zeba desalting columns (Pierce).
ELISpot.
Direct ex-vivo ELISpot was performed to determine the number of total, vaccine-binding, or recombinant HA-binding IgM-secreting cells present in PBMC samples. ELISpot plates (Millipore) were coated overnight at 4°C with QIV (diluted 1:100), tetanus/diphtheria vaccine (diluted 1:50), and 10 µg/mL anti-human Ig (Jackson ImmunoResearch) and 3 µg/mL recombinant HA proteins. Plates were blocked the following morning for 90 min at 37°C with RPMI 1640 supplemented with 10% FBS. Dilutions of washed PBMCs were incubated for 18 h in RPMI supplemented with 10% FBS. After washing the plates, secreted antibodies were detected with anti-human IgM-biotin (Invitrogen, 1:2000) and streptavidin-HRP (Jackson ImmunoResearch), and plates were developed with AEC substrate (Sigma). Total, vaccine-binding, or recombinant HA-binding IgG and IgA-secreting cells were detected using IgG/IgA double -color ELISpot Kits (Cellular Technologies, Ltd.) according to the manufacturer’s instructions. ELISpot plates were analyzed using an ELISpot counter (Cellular Technologies Ltd.).
ELISA.
Assays were performed in 96-well plates (MaxiSorp; Thermo). Each well was coated with 100 µL of QIV (diluted 1:100), tetanus/diphtheria vaccine (diluted 1:50), or with 0.5 μg/mL of the recombinant HA antigens in PBS, and plates were incubated at 4 °C overnight. Plates were then blocked with 0.05% Tween20 and 10% FBS in PBS. Plasma or mAbs were serially diluted in blocking buffer and added to the plates. Plates were incubated for 90 min at room temperature and then washed 3 times with 0.05% Tween-20 in PBS. Goat anti-human IgG-HRP (Jackson ImmunoResearch, 1:2,500) was diluted in blocking buffer before adding to wells and incubating for 90 min at room temperature. Plates were washed 3 times with 0.05% Tween20 in PBS, and then washed 3 times with PBS before the addition of peroxidase substrate (SigmaFAST o-Phenylenediamine dihydrochloride, Sigma-Aldrich). Reactions were stopped by the addition of 1 M HCl. Optical density measurements were taken at 490 nm. The half-maximal binding dilution for plasma was calculated using nonlinear regression (Graphpad Prism v7). The minimum positive concentration for mAbs was defined as that with optical density at least 3-fold above background.
Hemagglutination inhibition (HAI).
Plasma and monoclonal antibody HAI titers were determined for the four constituent viruses of 18–19 QIV (A/Michigan/45/2015 [pH1N1], A/Singapore/INFIMH-16-0019/2016 [H3N2], B/Phuket/3073/2013 [Yamagata lineage], and B/Colorado/06/2017 [Victoria lineage] as described previously1. Briefly, plasma samples were treated with receptor -destroying enzyme (RDE; Denka Seiken) by adding 1 part plasma to 3 parts RDE and incubating at 37° C overnight. The following morning, RDE was inactivated by incubating the samples at 56° C for 1 h. Then, plasma samples or purified monoclonal antibodies were serially diluted with PBS in 96 -well u-bottom polystyrene plates, and 4 agglutinating doses (as determined by incubation with 1% turkey RBCs in the absence of plasma) of the appropriate inactivated virus were added to each well. After 30 min at room temperature, 50 μL of 0.5% turkey RBCs (Lampire) suspended in PBS was added to each well, and the plates were gently agitated. After an additional 30 min at room temperature, the titers were computed as the reciprocal of the final dilution for which non-agglutination was observed.
Influenza virus protein microarray (IVPM).
IVPMs were generated as described previously18,19. Briefly, recombinant influenza virus HAs were spotted onto expoxysilane-coated glass sides (Schott) using a Versa 100 automated liquid handler (Aurora Biomed). Each array included 13 HAs (Extended Data Table 5) diluted in 0.1% milk in PBS, and spotted in triplicate. Each slide contained 24 identical arrays. HAs were printed at a volume of 30 nL per spot with a concentration of 100 µg/mL. Slides were vacuum-packed and stored at –80°C until use. At the start of each assay, slides were removed from the –80°C freezer and allowed to warm to room temperature, then incubated in a humidity chamber at 95–98% relative humidity for 2 h. The slides were then inserted into 96-well microarray gaskets (Arrayit), dividing the slide into 24 separate arrays, which were blocked with 220 µL 3% milk in PBS containing 0.1% Tween 20 (PBS-T) for 2 h. The blocking solution was then removed, and mAbs diluted in 1% milk in PBS-T were incubated with arrays at a volume of 100 µL and a concentration of 5 µg/mL for 1 h. The arrays were washed 3 times with 220 µL PBS-T, and secondary antibody solution containing Cy5-labeled anti-human IgG secondary antibody (Abcam) diluted 1:3,000 in 1% milk in PBS-T was added to each well at a volume of 50 µL and incubated for 1 h. After removing the secondary antibody solution, arrays were washed 3 times with PBS-T and removed from their gaskets. The slides were rinsed with PBS-T, followed by a rinse with deionized water, and then dried with an air compressor. Arrays were imaged and analyzed with a Vidia microarray scanner (Indevr) using an exposure time of 1000 ms to measure spot median fluorescence.
Cell sorting and flow cytometry.
Staining for analysis and sorting was performed using fresh or cryo-preserved PBMCs or FNA single cell suspensions in 2% FBS and 2 mM EDTA in PBS (P2). For sorting, cells were stained for 30 min on ice with IgD-PerCP-Cy5.5 (IA6–2, 1:200), CD4-Alexa 700 (SK3, 1:400), CD20-APC-Fire750 (2H7, 1:100), and Zombie Aqua along with CD38-BV605 (HIT2, 1:100), CD71-FITC (CY1G4, 1:200), and CD19-PE (HIB19, 1:200) for PBs or CD19-BV421 (HIB19, 1:100), CD71-PE (CY1G4, 1:400), CXCR5-PE-Dazzle 594 (J252D4, 1:40), and CD38-PE-Cy7 (HIT2, 1:200) for GC B cells (all BioLegend). Cells were washed twice, and single PBs (live singlet CD19+ CD4− IgDlo CD38+ CD20− CD71+) and GC B cells (live singlet CD19+ CD4− IgDlo CD71+CD38int CD20+ CXCR5+) were sorted using a FACSAria II into 96-well plates containing 2 µL Lysis Buffer (Clontech) supplemented with 1 U/µL RNase inhibitor (NEB), or bulk sorted into buffer RLT Plus (Qiagen) and immediately frozen on dry ice.
For analysis, cells were stained for 30 min on ice with biotinylated recombinant HAs and PD-1-BB515 (EH12.1, BD Horizon, 1:100) diluted in P2, washed twice, then stained for 30 min on ice with IgA-FITC (M24A, Millipore, 1:500), CD45-PerCP (2D1, BD Bioscience, 1:25), IgG-BV480 (goat polyclonal, Jackson ImmunoResearch, 1:100), IgD-SB702 (IA6–2, Thermo, 1:50), CD38-BV421 (HIT2, 1:100), CD20-Pacific Blue (2H7, 1:400), CD27-BV510 (O323, 1:50), CD4-BV570 (OKT4, 1:50), CD24-BV605 (ML5, 1:100), streptavidin-BV650, CD19-BV750 (HIB19, 1:100), CXCR5-PE-Dazzle 594 (J252D4, 1:50), CD71-APC (CY1G4, 1:100), CD14-A700 (HCD14, 1:200), and IgM-APC-Cy7 (MHM-88, 1:400) (all BioLegend) diluted in Brilliant Staining buffer (BD Horizon). Cells were washed twice, then fixed and permeabilized for intranuclear staining for 1 h at 25°C with True Nuclear fixation buffer (BioLegend), washed twice with permeabilization/wash buffer, and stained for 30 min at 25°C with Bcl6-PE (7D1, 1:50) and Ki-67-PE-Cy7 (Ki-67, 1:400) (both BioLegend). Cells were washed twice with permeabilization/wash buffer and resuspended in P2 for acquisition on an Aurora using SpectroFlo v2.2 (Cytek). Flow cytometry data were analyzed using FlowJo v10 (Treestar).
Monoclonal antibody (mAb) generation.
Antibodies were cloned as described previously13. Briefly, VH, Vκ, and Vλ genes were amplified by reverse transcription-PCR and nested PCR reactions from singly sorted GC B cells and PBs using primer combinations specific for IgG, IgM/A, Igκ, and Igλ from previously described primer sets30 and then sequenced. To generate recombinant antibodies, restriction sites were incorporated via PCR with primers to the corresponding heavy and light chain V and J genes. The amplified VH, Vκ, and Vλ genes were cloned into IgG1 and Igκ expression vectors, respectively, as described previously1,30,31. Heavy and light chain plasmids were co-transfected into Expi293F cells (Gibco) for expression, and antibody was purified with protein A agarose (Invitrogen).
Bulk B cell receptor sequencing.
RNA was purified from whole PBMCs and sorted PBs from participants 04, 05, and 11 (Extended Data Table 3) using the RNeasy Micro Kit (Qiagen). Reverse transcription, unique molecular identifier (UMI) barcoding, cDNA amplification, and Illumina linker addition to B cell heavy chain transcripts were performed using the human NEBNext Immune Sequencing Kit (NEB) according to the manufacturer’s instructions. High-throughput 2×300 bp paired-end sequencing was performed on the Illumina MiSeq platform with a 30% PhiX spike-in according to the manufacturer’s recommendations, except that 325 and 275 cycles were performed for read 1 and 2, respectively.
Processing B cell receptor bulk sequencing reads.
Demultiplexed pair-end reads were preprocessed using pRESTO v0.5.10 [ref. 32] as follows. 1) Reads with a mean Phred quality score less than 20 were filtered. 2) Reads were aligned against template switch sequences and constant region primers, with a maximum mismatch rate of 0.5 and 0.2, respectively. 3) Reads were grouped based on unique molecular identifiers (UMIs) determined by the 17 nucleotides preceding the template switch site. 4) Separate consensus sequences were constructed for the forward and reverse reads within each UMI group, with a maximum error score of 0.1 and minimum constant region primer frequency of 0.6. If multiple constant region primers were associated with a particular UMI group, the majority primer was used. 5) Forward and reverse consensus sequence pairs were assembled by first attempting de novo assembly with a minimum overlap of 8 nucleotides and a maximum mismatch rate of 0.3. If unsuccessful, this was followed by reference-guided assembly using blastn v2.7.1 [ref 33] with a minimum identity of 0.5 and an E-value threshold of 1×10−5. 6) Isotypes were assigned by local alignment of the 3′ end of each consensus sequence to isotype-specific internal constant region sequences with a maximum mismatch rate of 0.3. Sequences with inconsistent isotype assignment and constant region primer alignment were removed. 7) Duplicate reads were collapsed into unique sequences, except for those spanning multiple biological samples and/or with different isotype assignments. Only sequences with at least two reads contributing to the UMI consensus sequence were used for further analyses. The template switch sequences, constant region primers, and isotype-specific internal constant region sequences that were used in these studies are available at: https://bitbucket.org/kleinstein/immcantation/src/master/protocols/AbSeq/.
Initial germline V(D)J gene annotation was performed using IgBLAST v1.14.0 [ref 34] with IMGT/GENE-DB release 201931–4 [ref 35]. IgBLAST output was processed using Change-O v0.4.3 [ref 36]. Additional quality control of these sequences was performed with the following requirements: aligned exclusively to heavy chain V and J genes; had Ns at fewer than 10% of V segment positions; had a minimum V segment coverage from nucleotide position 1 to 310 under the IMGT unique numbering scheme37; and had a junction length that was a multiple of 3, where junction was defined as from IMGT codon 104 encoding the conserved cysteine to codon 118 encoding phenylalanine or tryptophan.
Single cell RNAseq library preparation and sequencing.
Activated and memory B cells were enriched from PBMCs by first staining with IgD-PE and MojoSort anti-PE Nanobeads (BioLegend), and then processing with the EasySep Human B Cell Isolation Kit using the EasyEights magnet (Stemcell) to negatively enrich IgDlo B cells. Enriched IgDlo B cells, whole PBMCs, and whole FNA from each timepoint for participant 05 were processed using the following 10× Genomics kits: Chromium Single Cell 5′ Library and Gel Bead Kit v2 (PN-1000006); Chromium Single Cell A Chip Kit (PN-120236); Chromium Single Cell V(D)J Enrichment Kit; and Human, B cell (96rxns) (PN-1000016), and Chromium i7 Multiplex Kit (PN-120262). The cDNAs were prepared after GEM generation and barcoding, followed by GEM RT reaction and bead cleanup steps. Purified cDNA was amplified for 10–14 cycles before cleaning with SPRIselect beads. Then, samples were evaluated on a bioanalyser to determine cDNA concentration. BCR target enrichments were performed on full -length cDNA. GEX and enriched BCR libraries were prepared as recommended by the 10× Genomics Chromium Single Cell V(D)J Reagent Kit (v1 Chemistry) user guide, with appropriate modifications to the PCR cycles based on the calculated cDNA concentration. The cDNA libraries were sequenced on Novaseq S4 (Illumina), targeting a median sequencing depth of 50,000 and 5,000 read pairs per cell for gene expression and BCR libraries, respectively.
Processing 10× Genomics single-cell B cell receptor reads.
Demultiplexed pair-end FASTQ reads from participant 05 were preprocessed using the “cellranger vdj” command from 10× Genomics’ Cell Ranger v3.1.0 for alignment against the GRCh38 human reference v3.1.0 (refdata-cellranger-vdj-GRCh38-alts-ensembl-3.1.0). Initial germline V(D)J gene annotation was performed as described for bulk sequencing BCR reads. Additional quality control was performed, requiring sequences to be productively rearranged and have valid V and J gene annotations, consistent chain annotation (excluding sequences annotated with heavy chain V gene and light chain J gene), and a junction length that was a multiple of 3. The 429 heavy chain BCRs from the sample day 0 PBMC 2 were removed after they were found to have identical cell barcodes and sequences as BCRs from the sample day 5 PBMC 2. Only cells with exactly one heavy chain sequence paired with at least one light chain sequence were retained.
B cell receptor genotyping.
Processed BCR sequences from bulk sequencing (Extended Data Table 3) and, if available, single-cell sequencing (Extended Data Table 4), were combined with nested PCR and mAb sequences (Extended Data Table 3) for genotyping using TIgGER v0.3.1 [ref 38]. Individual genotypes, including novel V gene alleles missing from IMGT/GENE-DB, were computationally inferred and used to finalize V(D)J annotations. Non-productively rearranged sequences annotated as “non-functional” by IgBLAST were removed from further analysis.
Clonal lineage inference.
B cell clonal lineages were inferred based on productively rearranged heavy chain sequences using hierarchical clustering with single linkage15. First, sequences were partitioned based on common V and J gene annotations and junction region lengths. Within each partition, sequences whose junction regions were within 0.1 normalized Hamming distance from each other were clustered as clones. This distance threshold was determined by manual inspection in conjunction with kernel density estimates to identify the local minimum between the two modes of the within-participant bimodal distance-to-nearest distribution (Extended Data Fig. 3c). Following clonal clustering, full-length clonal consensus germline sequences were reconstructed for each clone with D-segment and N/P regions masked with N’s, resolving any ambiguous gene assignments by majority rule. Within each clone, duplicate IMGT-aligned V(D)J sequences from bulk sequencing were collapsed with the exception of duplicates derived from different B cell compartments or isotypes. Clonal rank abundance distributions for GC and early PB (Extended Data Fig. 3e) were produced using the “estimateAbundance” function from Alakazam v0.3.0 [ref 36].
Clonal overlap analysis.
Clonal overlap between week 1 PBs from blood and the GC B cells from later weeks was determined by the presence of sequences from both compartments in the same B cell clone. Visualization was achieved using the circlize package v0.4.8 [ref 39].
Calculation of SHM frequency.
Mutation frequency was calculated by counting the number of nucleotide mismatches from the germline sequence in the heavy chain variable segment leading up to the CDR3, while excluding the first 18 positions that could be error-prone due to the primers used for generating the mAb sequences. This calculation was performed using the calcObservedMutations function from SHazaM v0.1.11 [ref 36].
Construction of B cell lineage trees.
Phylogenetic trees for responding B cell clones were constructed using IgPhyML v1.1.0 [ref.40] with the HLP19 model41.
Processing of 10× Genomics single-cell 5′ gene expression data.
Demultiplexed pair-end FASTQ reads from participant 05 were first preprocessed on a by-sample basis using the “cellranger count” command from 10× Genomics’ Cell Ranger v3.1.0 for alignment against the GRCh38 human reference v3.0.0 (refdata-cellranger-GRCh38–3.0.0). To avoid a batch effect introduced by sequencing depth, the “cellranger aggr” command was used to subsample from each sample so that all samples had the same effective sequencing depth, which was measured in terms of the number of reads confidently mapped to the transcriptome or assigned to the feature IDs per cell. The feature biotypes were retrieved using biomaRt v2.42.0 [ref 42] from Ensembl release 93 [ref 43]. Additional quality control was performed as follows. 1) To remove presumably lysed cells, cells with mitochondrial content greater than 12.5% of all transcripts were removed. 2) To remove likely doublets, cells with more than 7,000 features or 70,000 total UMIs were removed. 3) To remove cells with no detectable expression of common housekeeping genes, cells with no transcript for any of ACTB, GAPDH, B2M, HSP90AB1, GUSB, PPIH, PGK1, TBP, TFRC, SDHA, and LDHA were removed44. 4) The feature matrix was subset, based on their biotypes, to protein-coding, immunoglobulin, and T cell receptor genes that were expressed in at least 0.05% of the cells in any sample. The resultant feature matrix contained 15,479 genes. 5) Cells with detectable expression of fewer than 200 genes were removed. After quality control, there were a total of 130,757 cells from 21 samples (Extended Data Table 4).
Single-cell gene expression analysis.
Single-cell gene expression analysis was performed using Seurat v3.1.1 [ref 45]. UMI counts measuring gene expression were log-normalized. The top 2,000 highly variable genes (HVGs) were identified using the “FindVariableFeatures” function with the “vst” method. A set of 317 immune-related, “immunoStates” marker genes46 were added to the HVG list, whereas immunoglobulin and T cell receptor genes were removed. The data were scaled and centered, and principal component analysis (PCA) was performed based on HVG expression. The PCA-guided t-distributed stochastic neighbor embedding (tSNE) plot was generated using the top 20 principal components (PCs).
Overall clusters (Extended Data Table 1) were identified using the “FindClusters” function with resolution 0.07 (Extended Data Fig. 2b). Cluster identities were assigned by examining the expression of a set of marker genes for different cell types (Extended Data Fig. 2c–e): MS4A1, CD19, and CD79A for B cells; CD3D, CD3E, CD3G, IL7R, and CD4 or CD8A for CD4+ or CD8+ T cells, respectively; GZMB, GNLY, NKG7, and NCAM1 for natural killer (NK) cells; CD14, LYZ, CST3, and MS4A7 for monocytes; IL3RA and CLEC4C for plasmacytoid dendritic cells (pDCs); and PPBP for platelets.
B cells from the overall B cell cluster were further clustered to identify B cell subsets (Extended Data Table 1) using the “FindClusters” function with resolution 0.2 (Extended Data Fig. 4a). Cluster identities were assigned by examining the expression of a set of marker genes for different B cell subsets and the availability of BCRs (Extended Data Fig. 4b–g). The following marker genes were examined: BCL6, RGS13, MEF2B, STMN1, ELL3, and SERPINA9 for GC B cells; XBP1, IRF4, SEC11C, FKBP11, JCHAIN, and PRDM1 for PBs; TCL1A, IL4R, CCR7, IGHM, and IGHD for naïve B cells; TBX21, FCRL5, ITGAX, NKG7, ZEB2, and the lack of CR2 for activated B cells (ABCs); and TNFRSF13B, CD27, and CD24 for resting memory B (RMB) cells. Although clusters 5 and 9 both clustered with B cells during overall clustering, cluster 5 was labelled “B & T” as its cells tended to have both BCRs (Extended Data Fig. 4c) and high expression levels of CD2 and CD3E (Extended Data Fig. 4e), whereas cluster 9 was labelled “Unassigned” as its cells tended to have no BCR (Extended Data Fig. 4c) and high expression levels of CD2 and CD3E (Extended Data Fig. 4e). Clusters 5 and 9 were excluded from the final B cell clustering (Extended Data Fig. 4f). Cells that were found in the GC cluster but came from the blood samples were labelled “PB-like” (Extended Data Fig. 4f–g). To further confirm the identities of these B cell subsets, we assessed their corresponding heavy chain BCRs for SHM frequency (Extended Data Fig. 4h) and isotype usage (Extended Data Fig. 4i), which were consistent with expected values.
EMPEM sample preparation and complexing.
Polyclonal antibodies were prepared similarly to Bianchi and Turner et al20. For each time point after vaccination, 2 mL plasma samples were heat-inactivated at 55°C for 30 min and incubated on protein G resin (GE Healthcare) for 72 h. Protein G/sera samples were washed three times with PBS, IgG was eluted off of protein G resin with 0.1 M glycine pH 2.5 buffer and neutralized with 1M Tris-HCl pH 8 buffer, and IgG was buffer -exchanged with PBS by centrifugation with 30 kDa concentrators (Amicon). For digestion, 4 mg IgG were buffer -exchanged with freshly prepared digestion buffer (20 mM sodium phosphate, 10 mM EDTA, 20 mM cysteine, pH 7.4) and incubated with immobilized papain (Fisher) for 20 h at 37°C. Digestion products were separated from immobilized papain using Pierce spin columns (Fisher), and buffer -exchanged by centrifugation with 10 kDa concentrators (Amicon). IgG and Fab/Fc were separated by size exclusion chromatography using a Superdex 200 increase 10/300 column (GE Healthcare). To generate polyclonal immune complexes, 20 µg HA from A/Singapore/INFIMH-16-0019/2016 (H3N2) and A/Michigan/51/2015 (H1N1) were incubated with 1 mg Fab for 16 h at room temperature. Immune complexes were purified by size exclusion chromatography using a Superose 6 increase 10/300 column (GE Healthcare). Purified immune complexes were concentrated to 50 µL for negative stain electron microscopy analysis.
mAb complexing.
mAbs expressed as Fab were complexed with HA (A/Singapore/INFIMH-16-0019/2016 (H3N2) and A/Michigan/51/2015 (H1N1)) in a 3:1 molar ratio of Fab to HA for 1 h at room temperature or 16 h at 4°C.
Negative stain electron microscopy.
For negative stain analysis, 400 mesh copper grids (Electron Microscopy Sciences, EMS) were coated with collodion 2% in amyl acetate (EMS), deposited with carbon using a Cressington carbon evaporator, and plasma cleaned (EMS). Immune complexes were diluted to 30 µg/mL, loaded onto prepared grids, and stained with 2% w/v uranyl formate. Samples were imaged on three Tecnai transmission electron microscopes (FEI): Spirit T12 with a CMOS 4k camera (TVIPS), T20 with an Eagle CCD 4k camera (FEI), and Talos 200C with a Falcon II direct electron detector and a CETA 4k camera (FEI). Micrographs were obtained using Leginon47, 100,000–300,000 particles were picked and stacked using Appion48, and data were processed using 2D and 3D classification and reconstruction in Relion49,50. Fab densities were segmented and resampled onto reference trimers (PDB IDs 4M4Y for group 1 HA and 3M5G for group 2 HA), and final images were made using UCSF Chimera51 and Photoshop (Adobe). Polyclonal Fab specificities were determined using 3D reconstructions and 2D class averages. Some specificities did not reconstruct fully due to limited angular sampling caused by orientation bias, sub-stoichiometric binding of Fabs, and/or low abundance of these immune complexes even within the large datasets collected. However, in addition to suggestive 3D density, our large internal database of images of HA-Fab complexes provide references to extrapolate the 2D images onto 3D models of HA using the same procedure we recently reported for interpreting neuraminidase-Fab complexes52.
In vivo challenge.
Animal experiments were approved by approved by the Washington University IACUC. Mice were housed under specific-pathogen free conditions at 55±10% relative humidity with a 12h day/night cycle. Female 6–8-week-old C57BL/6 mice (5–7 mice/group) received 5 mg/kg mAb 321–04 1B05, 321–05 2C09, the broadly protective mAb 1G01 [ref 14] as a positive control, or irrelevant human IgG control mAb in 100 μL PBS via intraperitoneal administration. One day later, the mice were anesthetized with isofluorane and challenged intranasally with 104 TCID50 A/California/04/2009 E3 (H1N1). Weight was measured daily for 14 days; mice were euthanized if weight loss exceeded 20% of original weight.
Extended Data
Extended Data Figure 1.
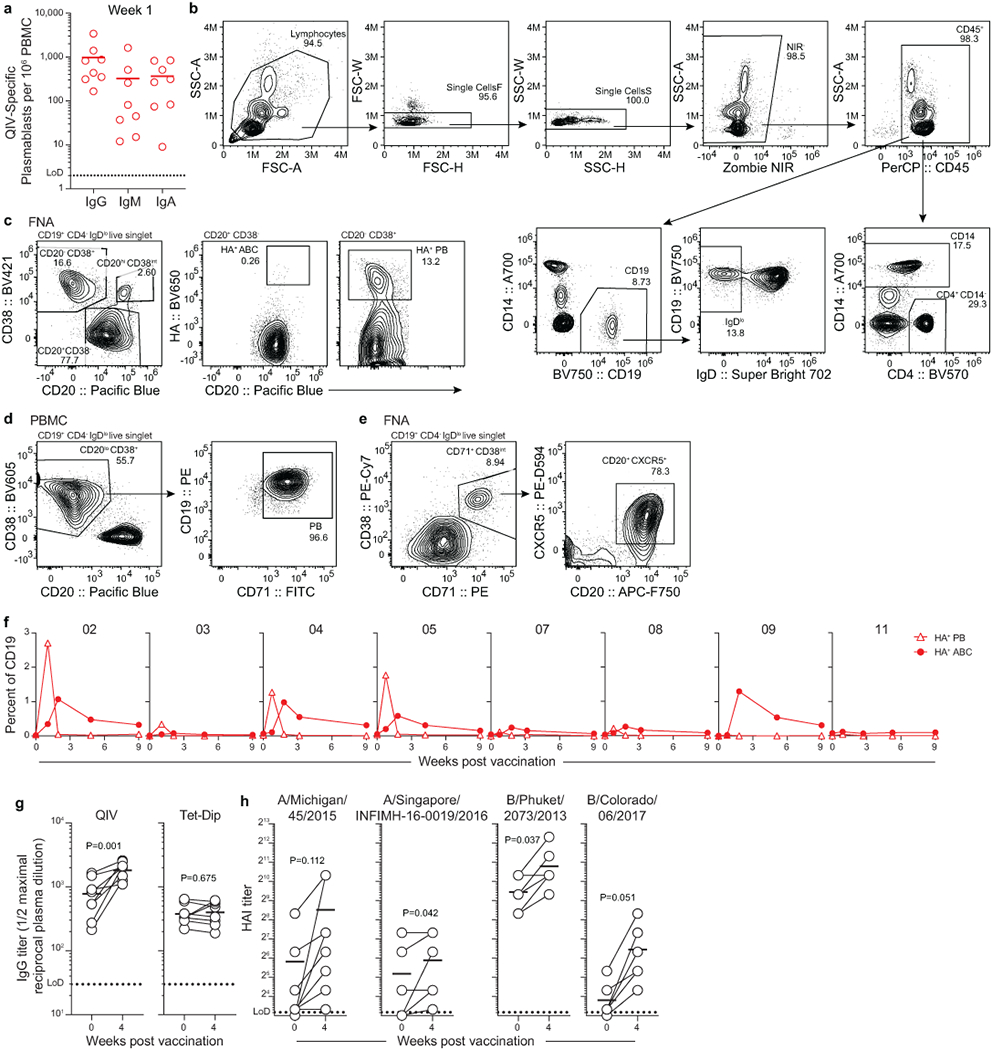
a) ELISpot quantification of QIV-binding IgG-, IgM-, and IgA-secreting QIV-binding PBs 1 -week post-vaccination. Each symbol represents one participant (n=8). b-e) Flow cytometry (b, c) and sorting (d, e) gating strategies for PBMC (b, d) and FNA (c, e). Population counts per mL of blood and frequencies are presented in f, below and in Figs. 1d, ,2f, and2f, and Extended Data Fig. 2c, ,m.m. f) Kinetics of HA-binding PBs (CD20lo HA+, open triangles) and activated B cells (ABCs, CD20+ HA+, closed circles) in PBMCs, gated as in Fig. 1c, pre-gated IgDlo CD19+ CD4− live singlet lymphocytes as in (b). Symbols at each timepoint represent one sample (n=8). g, h) IgG plasma antibody titers against QIV and Tetanus/Diptheria vaccine measured by ELISA (g) and hemagglutination inhibition titers against QIV constituent viruses (h) pre- and 4-weeks post-vaccination. Symbols at each timepoint represent one sample (n = 8). Horizontal lines in a, g, and h represent means. Dotted lines represent limit of detection. P-values from paired two-sided Student’s t -tests.
Extended Data Figure 2.

a) Cortical thickness measurements of axillary LNs before each FNA collection. b) FNA cell yields for each participant at the indicated timepoint. Symbols at each timepoint represent one sample (n=7). c) Participant 02 percentages of CD19+ CD4− B cells (left), CD14− CD4+ T cells (middle), and CD14+ CD4− monocytes or granulocytes (right) of CD45+ in PBMC (red) and FNA (blue) from one set of paired samples, representative of 6 FNA samples. d) Unsupervised clustering via tSNE based on scRNAseq gene expression of all cells pooled from all samples and timepoints from participant 05. Each dot represents a cell, colored by phenotype as defined by gene expression profile. e) Dot plot showing the average log-normalized expression of a set of marker genes and the fraction of cells expressing the genes in each unsupervised cluster. f, g) Annotated tSNE clusters of all cells from all scRNA-seq samples (f) and IgDlo enriched B cells from PBMC scRNA-seq samples (g) pooled from all time points from participant 05. Total number of cells is below clusters. h) Dot plot for annotated clusters. i) Representative flow cytometry gating of Bcl6 expression within CD20hi CD38int in PBMC and FNA. Cells pre-gated IgDlo CD19+ CD4− live singlet lymphocytes. j) Representative histographs (upper) and median fluorescence intensity (lower) of the indicated markers on GC B cells (IgDlo CD20hi CD38int) compared to PBs (IgDlo CD20− CD38+), memory B cells (IgDlo CD27+ CD38−), and naïve B cells (IgD+ CD27−). All populations pre-gated CD19+ CD4− live singlet lymphocytes. MFIs from 2- or 4-week FNA samples from participants 04, 05, 07, 08, 09, and 11. Lines represent medians. k) Representative gating of HA+ GC B cells. Cells pre-gated CD20hi CD38int IgDlo CD19+ CD4− live singlet lymphocytes. l) Kinetics of HA-binding percent of GC B cells measured by flow cytometry in participants 04, 05, and 11. m) Kinetics of HA+ CD38+ CD20lo PBs (open triangles) and HA+ CD38− CD20+ ABCs (closed circles) in FNA, as gated in Extended Data Fig. 1c. Symbols at each timepoint represent one sample (n=7). Daggers denote samples excluded from analysis due to low cell recovery or blood contamination.
Extended Data Figure 3.
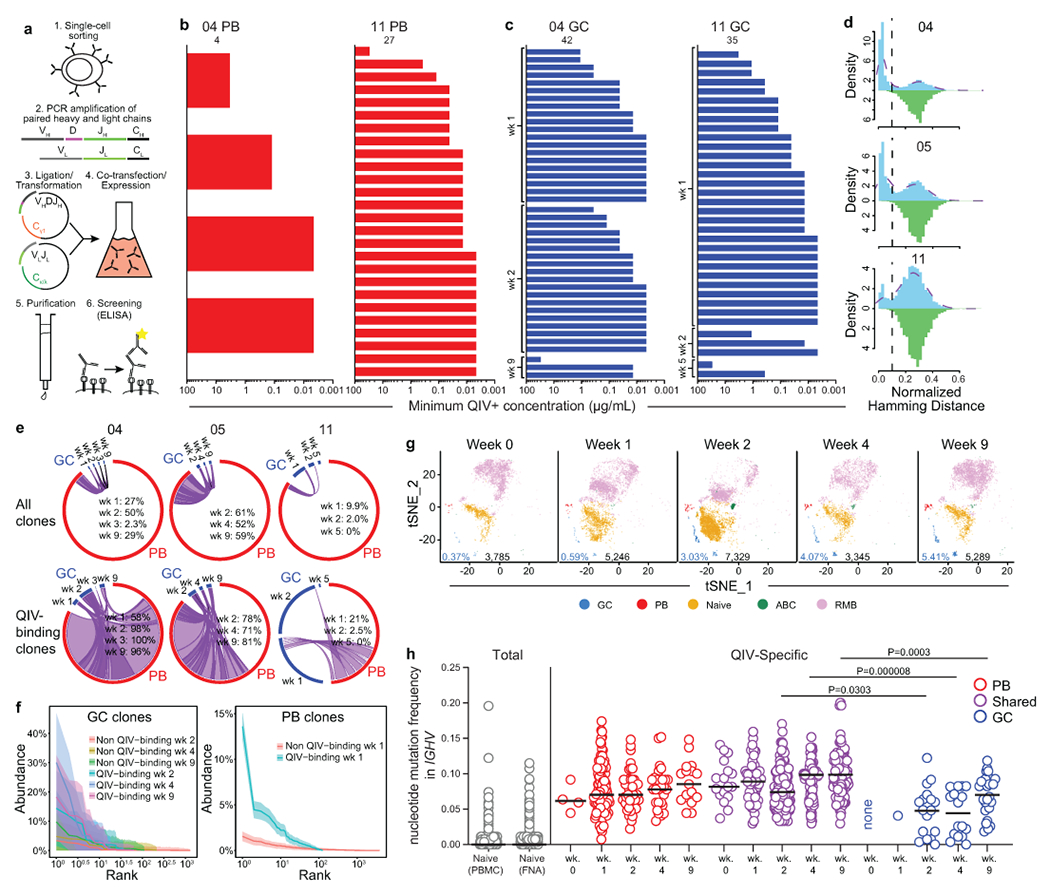
a) Schematic of single cell mAb cloning and expression. Paired heavy and light chain genes were amplified from singly sorted PBs or GC B cells. Variable portions of heavy chains were cloned into a Cγ1 expression vector and variable portions of κ and λ light chains were cloned into respective expression vectors. Paired heavy and light chain expression vectors were co-transfected into 293F cells, and mAbs were purified from culture supernatant by protein A affinity chromatography, then screened for QIV specificity by ELISA. b) Minimum positive concentrations of clonally unique mAbs generated from singly sorted PBs as determined by QIV ELISA; positive binding defined as greater than 3× background. c) Distance-to-nearest-neighbor plots for choosing a distance threshold for inferring clones via hierarchical clustering. After partitioning sequences based on common V and J genes and junction length, the nucleotide Hamming distance of a junction to its nearest non-identical neighbor from the same participant within its partition was calculated and normalized by junction length (blue histogram). For reference, the distance to the nearest non-identical neighbor from other participants was calculated (green histogram). A clustering threshold of 0.1 (dashed black line) was chosen via manual inspection and kernel density estimate (dashed purple line) to separate the two modes of the within-participant distance distribution representing, respectively, sequences that were likely clonally related and unrelated. d) Clonal overlap of sequences from mAb cloning and bulk repertoire analysis between PBs sorted from PBMCs 1-week post-vaccination and GC B cells from the indicated timepoint among total (top) and only QIV-binding (bottom) sequences. Purple chords link overlapping GC and PB clones; black chords link GC clones found at multiple timepoints that did not participate in the early PB response. Chord width corresponds to clonal population size. Percentages are of GC sequences overlapping with PBs. e) Clonal rank-abundance distributions of GC B cells from indicated timepoints (left) and of early blood PBs (right). The number of GC B cells or early blood PBs in a clone as a percentage of the total GC or early blood PB repertoire (y-axis) is plotted against the abundance rank of that clone (x-axis). Solid lines represent the estimated clonal abundance curves, with shaded bands representing the 95% confidence intervals from 200 bootstraps. g) tSNE clusters of B cells from FNA scRNAseq samples from participant 05. Each dot represents a cell, colored by phenotype as defined by gene expression profile. Total numbers of cells are given below clusters. GC percentages are indicated in blue. h) IGHV mutation frequency of naïve B cells pooled from all timepoints (left) and the indicated populations at the indicated timepoint (right) from scRNAseq of whole and memory B cell-enriched PBMC and FNA samples from participant 05. Horizontal lines represent medians. P-values from two-sided Dunn’s multiple comparisons test.
Extended Data Figure 4.

a) tSNE plot showing unsupervised clusters based on scRNAseq gene expression of cells in the “B cell” cluster of Extended Data Fig. 2e, pooled from all samples and timepoints from participant 05. b) Dot plot showing the average log-normalized expression of a set of marker genes and the fraction of cells expressing the genes in each unsupervised cluster. c) tSNE plot showing BCR availability. d) tSNE plot showing interim annotated clusters. e) Dot plot for interim annotated clusters. f) tSNE plot showing final annotated clusters. g) Dot plot for final annotated clusters. h) tSNE plot showing IHGV mutation frequency in BCRs. Total numbers of cells are given below clusters. i) Bar plots showing isotype usage in annotated B cell clusters. Numbers of cells per cluster are in Extended Data Table 1.
Extended Data Figure 5.
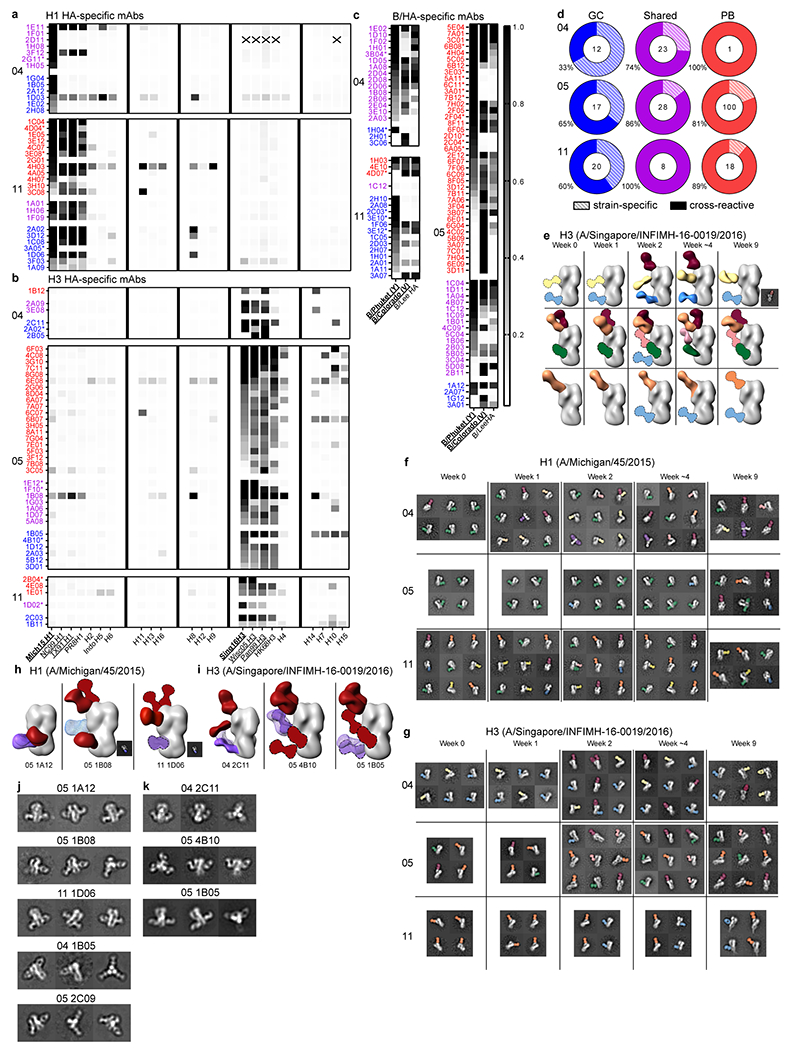
a–c) IVPM binding of H1- (a), H3- (b), and influenza B/HA- (c) binding mAbs generated from singly sorted PBs and GC B cells that overlapped clonally (purple) or did not overlap (red and blue) from the indicated participant. Scale bar represents median fluorescence intensity. Asterisks denote HAI+ mAbs. Vaccine strains in bold type; underlined strains circulated in humans in participants’ lifetimes. d) Percentages of mAbs that bound two or more HA strains from participants 04, 05, and 11 from GC clones that did not participate in the early PB response (blue), clones that participated in both GC and early PB responses (purple), and from PB clones not found in GCs (red). Numbers of mAbs are indicated in the middle of the charts. e) Polyclonal epitopes of Fabs from plasma at indicated timepoints from participants 04, 05, and 11 with HA from A/Singapore/INFIMH-16-0019/2016. Epitopes were determined by 3D reconstructions and/or 2D class averages (images to bottom right of 3D reconstructions). HA proteins shown in grey; Fabs shown in multiple colors; Fabs with dashed outlines have predicted epitopes due to limited particle representation. f, g) Example 2D class averages of immune complexes from participants 04, 05, and 11 plasma with HA from A/Michigan/45/2015 (f) and A/Singapore/INFIMH-16-0019/2016 (g). h, i) Monoclonal and polyclonal epitopes of immune complexes with HA from A/Michigan/45/2015 (h) or A/Singapore/INFIMH-16-0019/2016 (i) and Fabs generated from indicated GC mAbs in blue or purple mesh and plasma pAbs in red. Fabs with dashed outlines have predicted epitopes due to limited particle representation. j, k) Example 2D class averages of immune complexes from the indicated mAb with HA from A/Michigan/45/2015 (j) and A/Singapore/INFIMH-16-0019/2016 (k).
Extended Data Table 1.
Cell counts in overall and B cell clusters based on single-cell gene expression (Participant 05)
| Overall cluster | PBMC | Memory B (PBMC) | FNA |
|---|---|---|---|
| B | 2078 (5.8%) | 21855 (89.6%) | 27223 (38.5%) |
| CD4+ T | 12408 (34.9%) | 734 (3.0%) | 29914 (42.3%) |
| CD8+ T | 10138 (28.5%) | 728 (3.0%) | 11693 (16.5%) |
| NK | 4886 (13.7%) | 291 (1.2%) | 965 (1.4%) |
| Monocyte | 5418 (15.2%) | 678 (2.8%) | 837 (1.2%) |
| pDC | 178 (0.5%) | 29 (0.1%) | 159 (0.2%) |
| Platelet | 473 (1.3%) | 67 (0.3%) | 5 (0.0%) |
| B cell cluster | |||
| GC | 0 (0.0%) | 0 (0.0%) | 689 (2.8%) |
| PB | 136 (7.3%) | 47 (0.2%) | 358 (1.4%) |
| PB-like | 20 (1.2%) | 22 (0.1%) | 0 (0.0%) |
| Naïve | 1200 (64.7%) | 1709 (7.9%) | 8647 (34.6%) |
| ABC | 115 (6.2%) | 4950 (22.8%) | 539 (1.4%) |
| RMB | 384 (20.7%) | 14978 (69.0%) | 14941 (59.8%) |
Extended Data Table 2.
Cell counts of FNA populations based on flow cytometry
| Participant | week | Live cells | GC B cells | HA+ GC B cells | HA+ PB | HA+ ABC |
|---|---|---|---|---|---|---|
| 321-03 | 0 | 1.14E+06 | 627 | 0 | 0 | 0 |
| 321-03 | 1 | 5.25E+06 | 0 | 0 | 20 | 0 |
| 321-03 | 2 | 1.84E+06 | 15929 | 0 | 19 | 0 |
| 321-03 | 4 | 2.35E+06 | 493 | 0 | 0 | 0 |
| 321-03 | 9 | 3.48E+06 | 515 | 0 | 0 | 0 |
| 321-04 | 0 | 6.60E+05 | 7407 | 0 | 0 | 0 |
| 321-04 | 1 | 5.25E+06 | 19842 | 327 | 20006 | 4765 |
| 321-04 | 2 | 1.05E+06 | 22295 | 2296 | 194 | 375 |
| 321-04 | 3 | 5.48E+05 | 1996 | 10 | 0 | 411 |
| 321-04 | 9 | 1.00E+06 | 35749 | 504 | 18 | 696 |
| 321-05 | 0 | 1.02E+06 | 859 | 0 | 0 | 0 |
| 321-05 | 1 | 7.60E+05 | 826 | 0 | 98 | 373 |
| 321-05 | 2 | 6.88E+06 | 31453 | 4954 | 1995 | 13787 |
| 321-05 | 4 | 3.95E+06 | 49434 | 9726 | 2210 | 3135 |
| 321-05 | 9 | 1.13E+06 | 28017 | 6592 | 1615 | 1580 |
| 321-07 | 0 | 1.55E+06 | 1862 | 0 | 0 | 0 |
| 321-07 | 1 | 1.92E+06 | 9288 | 0 | 20857 | 3224 |
| 321-07 | 2 | 2.68E+05 | 91 | 18 | 9 | 209 |
| 321-07 | 4 | 1.98E+06 | 9754 | 19 | 149 | 1488 |
| 321-07 | 9 | 1.93E+05 | 67 | 17 | 17 | 452 |
| 321-08 | 0 | 5.33E+05 | 6 | 0 | 0 | 28 |
| 321-08 | 1 | 5.67E+05 | 3883 | 0 | 101 | 124 |
| 321-08 | 2 | 1.01E+06 | 8559 | 0 | 39 | 371 |
| 321-08 | 3 | 2.61E+05 | 292 | 0 | 13 | 0 |
| 321-08 | 9 | 1.84E+06 | 9305 | 0 | 0 | 455 |
| *321-09 | 0 | 1.56E+05 | ||||
| 321-09 | 1 | 5.80E+06 | 4915 | 8 | 9074 | 3556 |
| *321-09 | 2 | 2.69E+04 | ||||
| 321-09 | 4 | 1.67E+05 | 70 | 0 | 0 | 44 |
| 321-09 | 9 | 7.74E+05 | 1055 | 0 | 0 | 53 |
| *321-11 | 0 | 2.29E+04 | ||||
| 321-11 | 1 | 1.58E+06 | 4817 | 458 | 4522 | 6215 |
| 321-11 | 2 | 7.07E+05 | 1040 | 870 | 28 | 601 |
| 321-11 | 4 | 3.32E+05 | 379 | 13 | 13 | 105 |
| 321-11 | 9 | 1.66E+05 | 48 | 0 | 6 | 54 |
Extended Data Table 3.
Processing of bulk sequencing BCR reads and number of nested PCR sequences and clonally distinct monoclonal antibody sequences
| Participant | Sample | Timepoint | Compartment | Cell count | Sequence count | |||
|---|---|---|---|---|---|---|---|---|
| Input | Preprocessed | Post-QC | Unique VDJ | |||||
| 321-04 | 7 | d5 | Plasmablast | 101555 | 2433663 | 73442 | 70096 | 31219 |
| 321-04 | 10 | d0 | PBMC | 515000 | 2605448 | 99076 | 80997 | 33651 |
| 321-05 | 8 | d5 | Plasmablast | 8007 | 2495273 | 63677 | 50292 | 9898 |
| 321-05 | 11 | d0 | PBMC | 299000 | 2126862 | 75363 | 68028 | 33853 |
| 321-11 | 9 | d6 | Plasmablast | 2007 | 2771332 | 11526 | 8928 | 2076 |
| 321-11 | 12 | d0 | PBMC | 481000 | 2719946 | 114850 | 102198 | 83059 |
| Participant | Nested PCR
sequences | QIV+ clonally
distinct mAb sequences | ||
|---|---|---|---|---|
| Plasmablast | Germinal center | Plasmablast | Germinal center | |
| 321-04 | 187 | 448 | 4 | 42 |
| 321-05 | 226 | 187 | 125 | 67 |
| 321-11 | 107 | 158 | 27 | 35 |
Extended Data Table 4.
Processing of 10x Genomics single-cell BCR and 5’ gene expression data (Participant 05)
| Sample | BCR | 5’ gene
expression | ||||
|---|---|---|---|---|---|---|
| Pre-QC number of cells | Post-QC number of cells | Pre-QC number of cells | Post-QC number of cells | Median number of UMIs per cell | Median number of genes per cell | |
| d0 PBMC 2 | 794 | 296 | 8165 | 7961 | 4102 | 1357 |
| d5 PBMC 2 | 971 | 896 | 7242 | 7106 | 4309 | 1378 |
| d12 PBMC 2 | 437 | 402 | 6795 | 6712 | 4182 | 1407 |
| d28 PBMC 2 | 423 | 406 | 6435 | 6317 | 4416 | 1493 |
| d60 PBMC | 605 | 556 | 7594 | 7483 | 4110 | 1392 |
| d0 memory B (PBMC) | 3552 | 3261 | 3998 | 3899 | 4548 | 1322 |
| d5 memory B (PBMC) | 3141 | 2768 | 3365 | 3265 | 4328 | 1231 |
| d12 memory B (PBMC) | 5859 | 5244 | 6587 | 6406 | 4317 | 1266 |
| d28 memory B (PBMC) | 7780 | 6909 | 7865 | 7755 | 4426 | 1315 |
| d60 memory B (PBMC) | 2817 | 2592 | 3143 | 3057 | 4587 | 1397 |
| d0 FNA | 2138 | 1951 | 6702 | 6429 | 3738 | 1414 |
| d0 FNA 2 | 2223 | 2052 | 6167 | 5882 | 3670 | 1407 |
| d5 FNA | 3399 | 3122 | 8840 | 8426 | 4075 | 1557 |
| d5 FNA 2 | 2659 | 2439 | 6959 | 6657 | 4039 | 1557 |
| d12 FNA | 3408 | 3128 | 7491 | 7178 | 4066 | 1514 |
| d12 FNA 2 | 2537 | 2315 | 5932 | 5661 | 3969 | 1486 |
| d12 FNA 3 | 3157 | 2902 | 7322 | 6992 | 4004 | 1496 |
| d28 FNA 2 | 2005 | 1809 | 6604 | 6274 | 3878 | 1541 |
| d28 FNA 3 | 2071 | 1869 | 6491 | 6173 | 3854 | 1539 |
| d60 FNA | 3033 | 2790 | 5799 | 5540 | 3893 | 1436 |
| d60 FNA 2 | 3146 | 2832 | 5865 | 5584 | 3900 | 1440 |
Extended Data Table 5.
HA Strains for IVPM
| Strain Name (Subtype) | Abbreviation |
|---|---|
| A/Michigan/45/2015 (H1) | Mich15 H1 |
| A/New Caledonia/20/1999 (H1) | NC99 H1 |
| A/Texas/36/1991 (H1) | TX91 H1 |
| A/Puerto Rico/8/1934 (H1) | PR8 H1 |
| A/Japan/305/1957 (H2) | H2 |
| A/Indonesia/05/2005 (H5) | Indo H5 |
| A/mallard/Sweden/81/2002 (H6) | H6 |
| A/shoveler/Netherlands/18/1999 (H11) | H11 |
| A/black headed gull/Sweden/1/1999 (H13) | H13 |
| A/black headed gull/Sweden/5/1999 (H16) | H16 |
| A/mallard/Sweden/24/2002 (H8) | H8 |
| A/mallard/Interior Alaska/7MP0167/2007 (H12) | H12 |
| A/guinea fowl/Hong Kong/WF10/1999 (H9) | H9 |
| A/Singapore/INFIMH-16-0019/2016 (H3) | Sing16H3 |
| A/Wisconsin/67/2005 (H3) | Wisc05 H3 |
| A/Panama/2007/1999 (H3) | Pan99 H3 |
| A/Hong Kong/1/1968 (H3) | HK68 H3 |
| A/red knot/Delaware/541/1988 (H4) | H4 |
| A/mallard/Gurjev/263/1982 (H14) | H14 |
| A/chicken/BC/CN-6/2004 (H7) | H7 |
| A/mallard/Interior Alaska/10BM01929/2010 (H10) | H10 |
| A/shearwater/West Australia/2576/1979 (H15) | H15 |
| B/Lee/1940 (B-HA) | B/Lee HA |
| B/Colorado/06/2017 (B-HA Victoria lineage) | B/Colorado (V) |
| B/Phuket/3073/2013 (B-HA Yamagata lineage) | B/Phuket (Y) |
Acknowledgments
We thank Ken Hoehn for discussion on phylogenetic analysis, Adrianus Boon and Houda Harastani for providing A/California/04/2009 E3 (H1N1) virus; Erica Lantelme for facilitating sorting; Lisa Kessels, Michael Royal, and the staff of the Infectious Diseases Clinical Research Unit at Washington University School of Medicine for assistance with vaccination and sample collection; the Yale Center for Research Computing for use of high-performance computing infrastructure, the Genome Technology Access Center (GTAC) in the Department of Genetics at Washington University School of Medicine and the Yale Center for Genome Analysis for help with genomic analysis. GTAC is partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant# UL1 TR000448 from the NCRR.
The authors declare the following competing interests: A.H.E. is a consultant for InBios and Fimbrion Therapeutics. The Ellebedy laboratory received funding under sponsored research agreements from Emergent BioSolutions. All other authors declare no competing interests.
The Ellebedy laboratory was supported by NIAID grants R21 AI139813, U01 AI141990, and NIAID Centers of Excellence for Influenza Research and Surveillance (CEIRS) contract HHSN272201400006C. The Kleinstein laboratory was supported by NIH grant R01AI104739 and HIPC-CEIRS grant U19AI089992. The Ward laboratory was supported by Collaborative Influenza Vaccine Innovation Centers contract 75N93019C00051-0-9999-1. The Krammer laboratory was supported by NIAID CEIRS contract HHSN272201400008C and NIAID grants AI117287 and AI128821. J.S.T. was supported by NIAID 5T32CA009547. J.H. was supported by NIAID 2 T32 AI007244-36.
The WU321 study was reviewed and approved by the Washington University Institutional Review Board (approval no. 201808171). The manuscript was edited by the Scientific Editing Service of the Institute of Clinical and Translational Sciences at Washington University, which is supported by an NIH Clinical and Translational Science Award (UL1 TR002345).
Footnotes
Data availability statement
Raw fastq files and associated RNA sequencing have been uploaded to the NCBI Gene Expression Omnibus (GEO) database under identifiers SRP251458 and GSE148633. Antibody sequences are deposited on GenBank under the following accession numbers: MT200037-MT200636, KEB[P-Z]00000000, KEC[A-Z]00000000, KED[A-Z]00000000, and KEE[A-F]00000000, available from GenBank/EMBL/DDBJ. The template switch sequences, constant region primers, and isotype-specific internal constant region sequences that were used in these studies are available at: https://bitbucket.org/kleinstein/immcantation/src/master/protocols/AbSeq/. 3D reconstructions are deposited to the Electron Microscopy Data Bank (EMDB) under the following accession numbers: EMD-21703, EMD-21710, EMD-21733, EMD-21734, EMD-21737, EMD-21765, EMD-21809, EMD-21768, EMD-21776, EMD-21778, EMD-21779, EMD-21780, EMD-21782, EMD-21783 (participant 4 polyclonal immune complexes); EMD-21739, EMD-21741, EMD-21745, EMD-21746, EMD-21750, EMD-21784, EMD-21785, EMD-21786, EMD-21787, EMD-21788, EMD-21792, EMD-21793, EMD-21797, EMD-21798, EMD-21799, EMD-21800, EMD-21802 (participant 5 polyclonal immune complexes); EMD-21714, EMD-21715, EMD-21717, EMD-21719, EMD-21722, EMD-21723, EMD-21726, EMD-21729, EMD-21730, EMD-21731, EMD-21751, EMD-21752, EMD-21754, EMD-21803, EMD-21804, EMD-21806, EMD-21808 (participant 11 polyclonal immune complexes); EMD-21757, EMD-21758, EMD-21759, EMD-21760, EMD-21761, EMD-21762, EMD-21763 (monoclonal immune complexes). Other relevant data are available from the corresponding author upon reasonable request. Source data are provided with this paper.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/s41586-020-2711-0
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7566073
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Vaccination against rapidly evolving pathogens and the entanglements of memory.
Nat Immunol, 25(11):2015-2023, 09 Oct 2024
Cited by: 0 articles | PMID: 39384979
Review
Meeting Report From "Correlates of Protection for Next Generation Influenza Vaccines: Lessons Learned From the COVID-19 Pandemic".
Influenza Other Respir Viruses, 18(10):e13314, 01 Oct 2024
Cited by: 0 articles | PMID: 39380156 | PMCID: PMC11461279
High-throughput sequencing-based neutralization assay reveals how repeated vaccinations impact titers to recent human H1N1 influenza strains.
J Virol, 98(10):e0068924, 24 Sep 2024
Cited by: 4 articles | PMID: 39315814 | PMCID: PMC11494878
Robust and persistent B-cell responses following SARS-CoV-2 vaccine determine protection from SARS-CoV-2 infection.
Front Immunol, 15:1445653, 17 Sep 2024
Cited by: 1 article | PMID: 39355249 | PMCID: PMC11442242
Immune imprinting: The persisting influence of the first antigenic encounter with rapidly evolving viruses.
Hum Vaccin Immunother, 20(1):2384192, 16 Aug 2024
Cited by: 0 articles | PMID: 39149872 | PMCID: PMC11328881
Review Free full text in Europe PMC
Go to all (144) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Electron Microscopy Data Bank (Showing 10 of 10)
- (7 citations) Electron Microscopy Data Bank - EMD-2175
- (7 citations) Electron Microscopy Data Bank - EMD-2180
- (6 citations) Electron Microscopy Data Bank - EMD-2173
- (6 citations) Electron Microscopy Data Bank - EMD-2176
- (5 citations) Electron Microscopy Data Bank - EMD-2171
- (5 citations) Electron Microscopy Data Bank - EMD-2179
- (4 citations) Electron Microscopy Data Bank - EMD-2172
- (3 citations) Electron Microscopy Data Bank - EMD-2174
- (3 citations) Electron Microscopy Data Bank - EMD-2177
- (1 citation) Electron Microscopy Data Bank - EMD-2170
Show less
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE148633
Nucleotide Sequences (2)
- (1 citation) ENA - MT200037
- (1 citation) ENA - MT200636
Protein structures in PDBe (2)
-
(1 citation)
PDBe - 3M5GView structure
-
(1 citation)
PDBe - 4M4YView structure
Electron Microscopy Data Bank (EMDB) at PDBe (Showing 10 of 65)
- https://www.ebi.ac.uk/emdb/EMD-21710
- https://www.ebi.ac.uk/emdb/EMD-21703
- https://www.ebi.ac.uk/emdb/EMD-21801
- https://www.ebi.ac.uk/emdb/EMD-21809
- https://www.ebi.ac.uk/emdb/EMD-21714
- https://www.ebi.ac.uk/emdb/EMD-21715
- https://www.ebi.ac.uk/emdb/EMD-21717
- https://www.ebi.ac.uk/emdb/EMD-21719
- https://www.ebi.ac.uk/emdb/EMD-21722
- https://www.ebi.ac.uk/emdb/EMD-21758
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses.
Nature, 596(7870):109-113, 28 Jun 2021
Cited by: 472 articles | PMID: 34182569 | PMCID: PMC8935394
Human B cell lineages associated with germinal centers following influenza vaccination are measurably evolving.
Elife, 10:e70873, 17 Nov 2021
Cited by: 26 articles | PMID: 34787567 | PMCID: PMC8741214
Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells.
Proc Natl Acad Sci U S A, 109(23):9047-9052, 21 May 2012
Cited by: 298 articles | PMID: 22615367 | PMCID: PMC3384143
Tfh cell response in influenza vaccines in humans: what is visible and what is invisible.
Curr Opin Immunol, 59:9-14, 25 Mar 2019
Cited by: 19 articles | PMID: 30921542 | PMCID: PMC6761038
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCATS NIH HHS (2)
Grant ID: UL1 TR002345
Grant ID: UL1 TR000448
NCI NIH HHS (2)
Grant ID: P30 CA091842
Grant ID: T32 CA009547
NIAID NIH HHS (16)
Grant ID: 75N93019C00051
Grant ID: R56 AI117287
Grant ID: HHSN272201400006C
Grant ID: R01 AI104732
Grant ID: R01 AI104739
Grant ID: U19 AI089992
Grant ID: R01 AI117287
Grant ID: AI117287
Grant ID: 2 T32 AI007244-36
Grant ID: AI128821
Grant ID: HHSN272201400008C
Grant ID: R21 AI139813
Grant ID: R01 AI128821
Grant ID: T32 AI007244
Grant ID: 5T32CA009547
Grant ID: U01 AI141990





