Abstract
Free full text

Why? – Successful Pseudomonas aeruginosa clones with a focus on clone C
ABSTRACT
The environmental species Pseudomonas aeruginosa thrives in a variety of habitats. Within the epidemic population structure of P. aeruginosa, occassionally highly successful clones that are equally capable to succeed in the environment and the human host arise. Framed by a highly conserved core genome, individual members of successful clones are characterized by a high variability in their accessory genome. The abundance of successful clones might be funded in specific features of the core genome or, although not mutually exclusive, in the variability of the accessory genome. In clone C, one of the most predominant clones, the plasmid pKLC102 and the PACGI-1 genomic island are two ubiquitous accessory genetic elements. The conserved transmissible locus of protein quality control (TLPQC) at the border of PACGI-1 is a unique horizontally transferred compository element, which codes predominantly for stress-related cargo gene products such as involved in protein homeostasis. As a hallmark, most TLPQC xenologues possess a core genome equivalent. With elevated temperature tolerance as a characteristic of clone C strains, the unique P. aeruginosa and clone C specific disaggregase ClpG is a major contributor to tolerance. As other successful clones, such as PA14, do not encode the TLPQC locus, ubiquitous denominators of success, if existing, need to be identified.
INTRODUCTION
Pseudomonas aeruginosa is the prototype of an environmental bacterium the adaptability of which promotes selected fractions of the population to successfully occupy anthropized environments. To understand the genetic and physiological basis of the success of abundant clones, group of closely related strains, that thrive in environmental and clinical habitats, in contrast to less abundant clones with more restricted ecological niches, is of particular interest for population genetics. To unravel the genetic basis of ubiquity, adaptability and persistence of clones and its individual members is not only highly relevant from a basic science point of view such as to unravel the impact of individuality in a successful population, but also from a clinical point of view in order to prevent the emergence and spread of multidrug resistant clones. In this review, we describe the ecological and molecular characteristics of abundant P. aeruginosa clone C first consciously isolated from natural and clinical habitats in Germany and Canada in the 1980s. Unravelling in more detail the genetic background and physiology of this clone, not known to extensively bearing antimicrobial resistance markers, will shed light on survival strategies of microbial organisms.
The species Pseudomonas aeruginosa
The Gram-negative bacterium Pseudomonas aeruginosa is the type species of the genus Pseudomonas which consists today of almost 200 species (http://www.bacterio.net/-pseudomonas.html). With one polar flagellum, Pseudomonas aeruginosa, isolated the first time in 1882, was described and named by the botanist Walter Emil Friedrich August Migula (Migula 1894). The metabolic versatility and minimal growth requirements of P. aeruginosa characteristic for the species of the genus Pseudomonas in combination with robust isolation has led to the conventional view that P. aeruginosa is ubiquitous in nature accumulating preferentially in human-contaminated environments. Indeed P. aeruginosa is regularly isolated from oil-contaminated fields and sewage, but also swimming pools and household sinks (Grobe, Wingender and Truper 1995; Pirnay et al. 2005; Das and Mukherjee 2007). Recovery from distilled water and disinfectants such as triclosan contribute to its presence in the clinic (Lanini et al. 2011). Pseudomonasaeruginosa occurs in natural environments as diverse as natural freshwater water, the marine environment, plants, mushrooms and soil (Ojima et al. 2002; Khan et al. 2007; Kidd et al. 2012; Rutherford et al. 2018; Schroth et al. 2018). Due to unique products and its metabolic versatility P. aeruginosa also has gained interest to be used in biotechnological applications (Reetz and Jaeger 1998; Fenibo et al. 2019).
With eukaryotic hosts, P. aeruginosa shows a broad spectrum of interactions. In plants, the effect of the organism spans from growth promoting to being a plant pathogen (Rahme et al. 1995; Adesemoye, Obini and Ugoji 2008). Association of P. aeruginosa with an immunocompetent human being is usually infrequent and temporary with gastrointestinal and skin colonization (Cooke et al. 1970; Silvestre and Betlloch 1999; Dossel et al. 2012; Garcia et al. 2018). Approximately 5% of gastrointestinal carriage in humans points to an acquisition by produce or water in combination with an efficient colonization resistance by the gastrointestinal microbiome (Kerckhoffs et al. 2011). Superficial skin (hot tub folliculitis) and ear (otitis externa, also called swimmer's ear) infections with P. aeruginosa can be readily acquired in natural or anthropized environments with a high number of the organism (Ratnam et al. 1986; Ahlen, Mandal and Iversen 2001).
Upon the introduction of antibiotics, due to the innate and acquired resistance against antibiotics in combination with its nutritional minimalism, P. aeruginosa has developed into one of the most frequently hospital acquired (nosocomial) pathogens (Gould and Wise 1985). A local or systemic impairment of the innate or adaptive immune response such as lack of skin as an innate immune barrier in severe burn wounds, depletion of neutrophils in neutropenia, debiliated mucociliary clearance in cystic fibrosis and immune aging due to old age is usually the basis for the establishment of a successful infection with P. aeruginosa. As such, in a wide spectrum of infections including diabetic foot ulcer and ear infection P. aeruginosa is a frequent causative agent (Hatipoglu et al. 2014). With its notorious ability to form biofilms, P. aeruginosa infections are promoted by its colonization on artificial devices. Thus the prevalence of P. aeruginosa infections is especially prominent in catheter-associated urinary tract infection and ventilator associated pneumonia (VAP), two of the most common nosocomial acquired infection, but also in contact-lens associated keratitis in immunocompetent individuals (http://www.-antimicrobe.org/b112.asp; (Bouza et al. 2001; Chastre and Fagon 2002; Rello et al. 2006; Bjerklund Johansen et al. 2007; Willcox 2012)).
Prior to the introduction of genome-wide molecular techniques, the versatility of the genome, the broad spectrum of habitats and infections and the absence of unique characteristics such as virulence factors or serotypes associated with pathogenicity limited the epidemiology of P. aeruginosa allowing only a low discriminatory and inconclusive classification of isolates (Tümmler et al. 1991; Kidd et al. 2012; Parkins, Somayaji and Waters 2018). Furthermore, the molecular mechanisms of the environmental species P. aeruginosa to conduct this broad range of environmental, saphrophytic and clinical interactions remained enigmatic. The recent initiatives of genome wide typing approaches of large strain collections including whole genome sequencing, in combination with in depth investigations on the molecular analysis of gene products, begin to unravel the molecular and physiological details of such a versatility on the population and individual strain level.
Genotyping of Pseudomonas aeruginosa strains
The classification of bacterial isolates on the strain level is relevant for ecology, epidemiology, taxonomy and biotechnology. Highly discriminatory genotyping methods for P. aeruginosa are either based on anonymous fingerprinting techniques like macrorestriction fragment pattern analysis or sequence-based typing approaches by multilocus sequence typing (MLST) and microarrays. Macrorestriction fragment pattern analysis has been made possible by the discovery of Schwartz and Cantor to separate kbp and Mbp long linear DNA fragments according to size in a gel matrix upon application of an alternately pulsed electric field (Schwartz and Cantor 1984). Generating barcode-like whole genome fingerprints created by rare cutting restriction enzymes such as SpeI is globally applicable to bacteria and hence is still the reference method for strain typing.
On the other hand, the P. aeruginosa MLST scheme utilizes nucleotide sequence data of internal fragments of seven housekeeping genes (https://pubmlst.org/paeruginosa/) (Kiewitz and Tummler 2000; Jolley, Bray and Maiden 2018) to scan the genetic diversity of the core genome by amplicon sequencing under high throughput. As a further development, a robust and rapid oligonucleotide microarray can type P. aeruginosa strains in both the conserved core and the flexible accessory genome (Wiehlmann et al. 2007). The microarray, hybridzed with the strain's DNA yields an electronically portable binary multimarker genotype that represents the core genome by single nucleotide polymorphisms (SNPs) and the accessory genome by markers of genomic islets and islands. A hexadecimal code summarizing the SNP genotypes assigns the strains to a clonal complex. Multimarker genotypes of 1448 strains are publicly available (Wiehlmann, Cramer and Tümmler 2015).
Examination of more than 550 P. aeruginosa isolates from environmental and clinical habitats by their macrorestriction SpeI fingerprints in the early 1990s identified more than 20% of the strains from various spatially and temporally separated habitats mainly from Germany to be variants of one major clone that since then is called clone C (Römling et al. 1994a,b). The hexadecimal code for clone C isolates reads C40A which matches in the MLST database with two rare (ST2691, ST2894) and two frequent MLST subtypes (ST17, ST845) the latter two accounting for more than 95% of clone C isolates.
Population biology of Pseudomonas aeruginosa
Cumulatively, recent whole genome sequencing projects have demonstrated that the population of the cosmopolitan P. aeruginosa grossly consists of one ExoS-positive and one ExoU-positive clade and three small groups of distant outliers (Stewart et al. 2014; Hilker et al. 2015; Freschi et al. 2019). Linkage groups, consecutive genes without recombination events, are just a few hundred base pairs in size indicating gene flow by recombination between clonal complexes (Dettman, Rodrigue and Kassen 2014; Hilker et al. 2015). These data support the conclusion of P. aeruginosa to mainly exhibit a non-clonal epidemic structure as previously drawn from polyphasic data sets (Pirnay et al. 2009).
To unravel the population structure of P. aeruginosa on the level of clones, several thousand isolates from more than 1500 independent habitats of diverse geographic origin have been investigated by microarray genotyping (Wiehlmann, Cramer and Tümmler 2015). This genotyping approach identified 323 different clone types. 109 clones made up for 82% of the population, whereby the 12 or 26 most frequent clones had absolute shares of 33.4% or 50%, respectively. On the other hand, 167 and 47 clone representatives were only found once or twice, respectively. In other words, the P. aeruginosa population is dominated by few epidemic clonal complexes (De Soyza et al. 2013). Overall, the most abundant genotype was the ExoS-positive clone C (C40A) (Römling et al. 1994a,b; Römling et al. 2005). Clone C was the most abundant genotype among the isolates from chronic human infections, the second most frequent clone in acute human and animal infections and the fourth most frequent clone among the isolates from the inanimate environment, i.e. soil and aquatic habitats (Wiehlmann, Cramer and Tümmler 2015).
aeruginosa population is dominated by few epidemic clonal complexes (De Soyza et al. 2013). Overall, the most abundant genotype was the ExoS-positive clone C (C40A) (Römling et al. 1994a,b; Römling et al. 2005). Clone C was the most abundant genotype among the isolates from chronic human infections, the second most frequent clone in acute human and animal infections and the fourth most frequent clone among the isolates from the inanimate environment, i.e. soil and aquatic habitats (Wiehlmann, Cramer and Tümmler 2015).
The P. aeruginosa community is more diverse in its clonal composition in soil and aquatic habitats than in the infected human host. Some P. aeruginosa clones like 149A, 081A or CBA3 (see Table 1) are common in the environment, but are rare or absent as causative agents of infections and thus behave like strains from the related Pseudomonas putida/Pseudomonas fluorescens group that are non-pathogenic for immunocompetent humans. The genetic repertoire to establish a niche in the mammalian host and/or to combat the host defense must be impaired in those clones. Consequently, in disease habitats the P. aeruginosa population narrows to proficient clones, which can colonize and persist in an animate host and thus, besides some generalists such as clone C (hexadecimal code C40A, Table 1) and PA14 (code D421), selects for minor clones to become dominant members of the populations in such atypical niches. A particular case is the lungs of cystic fibrosis patients (CF) where the microorganisms can be decade-long exposed to a hostile immune system and regular antimicrobial chemotherapy. These findings, made possible due to high resolution typing techniques and large strain collections, challenge the long standing dogma of environmental and clinical P. aeruginosa isolates being indistinguishable in their genetic properties and virulence factors.
Table 1.
Prevalence of the 15 most common environmental P. aeruginosa clones in human infections.
| Relative abundance [%]b | ||
|---|---|---|
| Clonea | Environment | Human infections |
| EA0A | 6.5 | 1.5 |
| B420 | 6.3 | 1.5 |
| C40A | 5.1 | 7.4 |
| 0812 | 4.4 | 1.6 |
| F46A | 4.0 | 0.6 |
| E429 | 3.5 | 2.2 |
| F429 | 3.3 | 2.1 |
| 0C2E | 2.6 | 3.8 |
| D421 | 2.1 | 4.3 |
| EC2A | 2.1 | 1.0 |
| 149A | 1.9 | < 0.1 |
| 081A | 1.9 | < 0.1 |
| CBA3 | 1.9 | ndc |
| 4C1A | 1.6 | < 0.1 |
| 6E1A | 1.4 | ndc |
In summary, the inanimate aquatic habitats harbor the largest pool of clones out of which subgroups spread to more specialized niches. The successful colonizers are either generalists like clone C found everywhere or minor clones that are endowed with clone-specific features to adapt to this peculiar niche. Clonal fitness is thereby subject to continuous genome evolution whereby in case of P. aeruginosa the horizontal transfer of mobile genetic elements is the most rapid and extensive process of strain diversification within clonal complexes (see below).
aeruginosa the horizontal transfer of mobile genetic elements is the most rapid and extensive process of strain diversification within clonal complexes (see below).
Pseudomonas aeruginosa clone C virulence
The range of infections caused by P. aeruginosa clone C strains seems to be as broad as the infection spectrum caused by the entire species. Clone C strains not only colonize the lung in individuals with different underlying etiology such as bronchiectasis (Hilliam et al. 2017), but also cause, for example, urinary tract (Tielen et al. 2011) and ear infections (Dinesh et al. 2003; Curran et al. 2004) and are found in the clinical environment (Bosshammer et al. 1995). Furthermore, clone C strains are widely distributed as they have been reported to infect CF patients on different continents (Römling et al. 1994b; Scott and Pitt 2004; Kidd et al. 2011; Fothergill, Walshaw and Winstanley 2012; Middleton et al. 2018; Parkins, Somayaji and Waters 2018).
Despite its prominent role in the global P. aeruginosa population and in acute and chronic infections, clone C has never been mentioned as highly virulent in comparison to other clones such as the PA14 clone D421 (STM253) (Rahme et al. 1995; He et al. 2004) or the international multidrug-resistant ST235 high-risk clone which has become the most frequent clone of severe acute infections in humans (Treepong et al. 2018). Like many other clonal lineages, the clone C genome harbors the genes for virulence factors and pathogenicity traits such as the type III secretion system effector proteins exotoxins S, T and Y or secreted proteases like LasA or LasB, although unconventional regulation of virulence factors by individual isolates has been demonstrated (Kamal et al. 2019). The presence of exotoxin S designates clone C as an invasive P. aeruginosa clone, in contrast to cytotoxic strains which mutually exclusive bear the patatin-like phospholipase exotoxin U as effector protein. Nevertheless, clone C strains successfully establish chronic, often life-long, infections in the airways of CF patients (Tümmler et al. 1991; Römling et al. 1994b). However, the chronic clone C carriers, as judged by semi-annually collected P. aeruginosa isolates from respiratory secretions, experience a rather mild course of their P. aeruginosa infection without a rapid decrease of lung function. Among the 29 individuals with CF who became chronically colonized with P. aeruginosa during the years 1984–1990 (Cramer et al. 2012), six of seven clone C carriers were still alive by June 2020. Conversely, 15 of the 22 carriers of other P. aeruginosa clones had passed away indicating that colonization by clone C was associated with a milder outcome of CF lung disease than chronic airway colonization with any other P. aeruginosa clone (P = 0.018, Fisher's exact test). This difference in the prognosis between clone C and non-clone C carriage is visualized as a Kaplan Meier plot (Fig. 1), which monitors the colonization time of CF airways with P. aeruginosa until death or lung transplantation by June 1st, 2020.
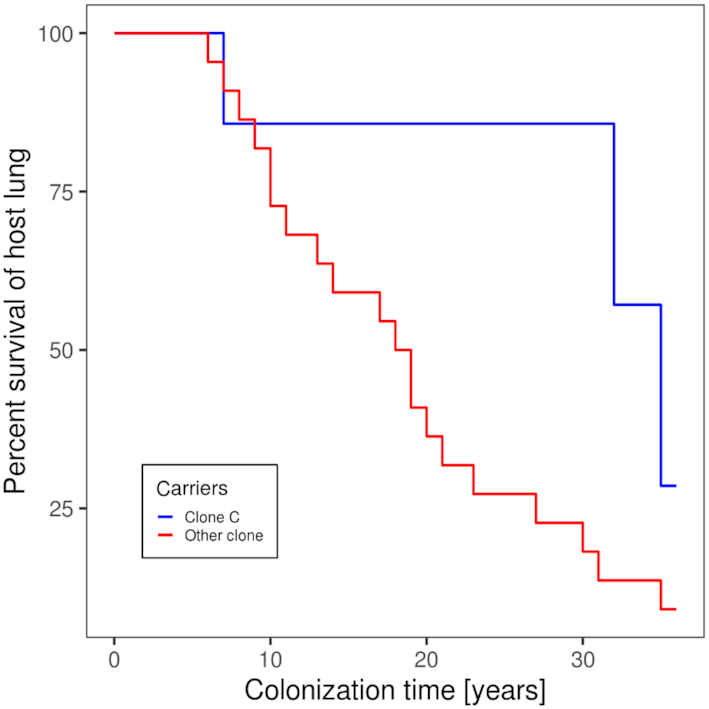
Duration of the chronic airway infection with P. aeruginosain individuals with cystic fibrosis carrying clone C (blue line) or any other clone (red line). The Kaplan–Meier plot shows individual length of colonization until lung transplantation or death because of respiratory insufficiency for the non-transplanted patients until June 1st, 2020. The data was extracted from the medical records of 29 CF patients regularly seen at the CF clinic Hannover who became chronically colonized with P. aeruginosa between 1984 and 1990.
Nevertheless, prima vista clone C cannot be described as less virulent. Representative strains of the 15 most common clones and five exclusively environmental clones have been compared in their virulence in three infection models (Hilker et al. 2015). In the murine airways with virulence monitored by lung function, ethology and inflammation (Munder and Tümmler 2014) (Fig. 2A), the clone C representative was lowly virulent ranked at positions 15 among 20 tested clones. In the plant infection model of lettuce leafs (Lactuca sativa var. longifolia) (Starkey and Rahme 2009) (Fig. 3), again the clone C representative was lowly virulent ranked at position 19 among 20 tested clones. Conversely, in the wax moth (Galleria melonella) larvae infection model, assessing the proportion of dead larvae (Pustelny et al. 2013; Kamal et al. 2019) (Fig. 2B), the clone C strain was the third most virulent strain. Likewise, the degree of virulence of clone C representatives in amoeba (Sandström and Römling, unpublished results) was close to that of a PA14 representative. Variable virulence phenotypes were also common for the other clones. These studies exemplarily demonstrated that the pathogenicity of P. aeruginosa is context-dependent with clone C demonstrating virulence especially in invertebrate hosts. However, even in one particular model different clonal isolates can show variable virulence properties indicating heterogeneity of the individuals (unpublished results). Nevertheless, the epidemiological evidence is rather strong that human infections with the most common clone C are more benign than those with the high-risk clones.
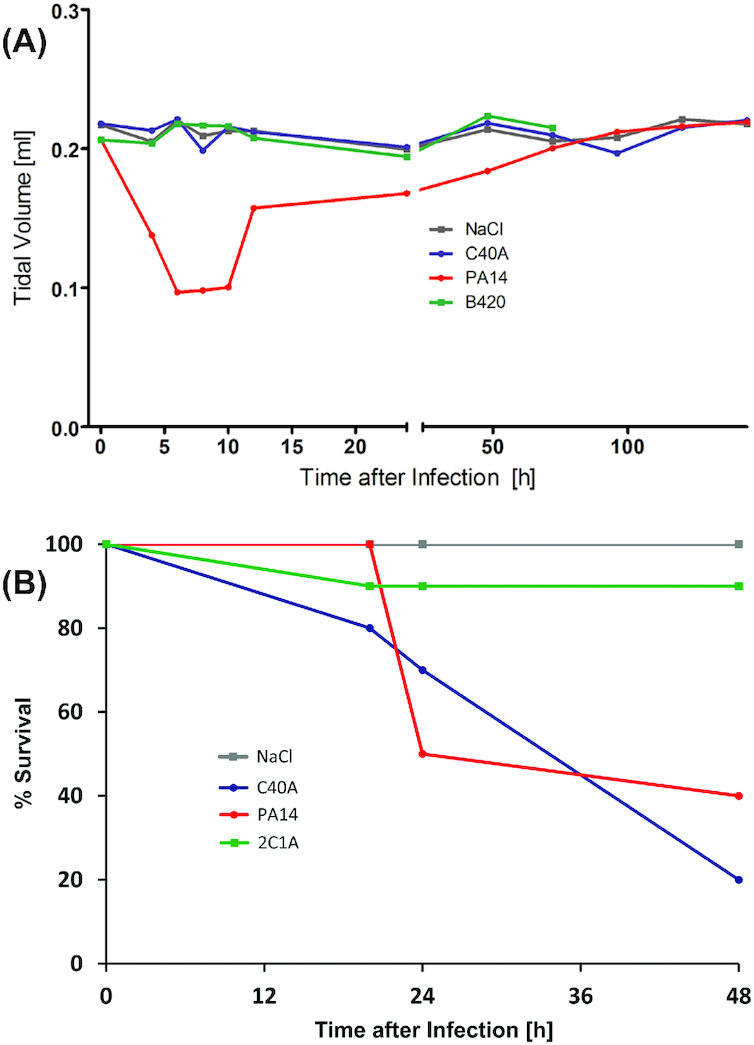
Virulence potential of clone C. The diagrams display the outcome of virulence tests for clone C (here termed C40A) in comparison to other clonal lineages (Hilker et al. 2015). The strains were tested in a murine airway infection model and in a wax moth (Galleria melonella) larvae infection model. For comparison of the severity of the mouse infection, lung pathology and cytokine responses were monitored and parameters such as body weight or rectal temperature were assessed. In addition, headout spirometry was performed on the infected mice. For negative controls, mice received NaCl solution. As an example for the different degrees of virulence in mouse infection, a diagram of displaying the development of tidal volumes are shown upon infection with clones C/C40A, B420 and PA14 (panel A). While clone PA14 inflicted a severe lung infection phenotype, clone C and B420 did not display much virulence potential in this assay. In contrast, clone C displayed high virulence in the G. melonella larvae infection model (panel B). While for some clones (such as 2C1A) most larvae could overcome the infection, clone C was found among the strains with the highest virulence in this invertebrate assay killing even higher proportions of the infected larvae than clone PA14.

Different degree of virulence in a salad leaf infection model. The examples display the different virulence of P. aeruginosa strains in a salad (Lactuca sativa var. longifolia) infection assay. MgCl2 solution containing 108 CFU of bacteria was instilled into the midrib of the lettuce leaf. Progress of infection was represented by the spread of a brownish rotten area to the different parts of the salad leafs. The panels show the spread of the infection 44 h after instillation of MgCl2 solution (negative control, panel A), clone C/C40A (panel B), clone PA14 (panel C) and B420 (panel D). The respective leaf appeared rather unaffected after instillation of clones C and PA14. Clone B420, however, which displayed very low virulence in the mouse infection model, caused a much more severe phenotype in this assay with rotting visible in wide areas of the lettuce leaf.
Intraclonal genomic sequence diversity
The median intraclonal sequence diversity among 58 clone C genomes at the single nucleotide level was determined to be 3.7 × 10−4 (Fischer et al. 2016). Remarkably, the sequence diversity of the core genome was just 8 × 10−6 (in comparison: 2 × 10−5 for clone PA14 (Fischer et al. 2016)), which is more than 100-fold lower than the sequence diversity among unrelated P. aeruginosa clones (Hilker et al. 2015). In other words, clone C strains differ in their core genome by just a few dozen SNPs from each other and are clearly distinguishable from unrelated clones. The few hot spots of mutations are phage- and plasmid-derived genes and genes encoding the heavy metal ion efflux protein CusA, the cyclic-di-GMP phosphodiesterase BifA and LasR, a key regulator of acyl homoserine lactone quorum sensing. Elements of the accessory genome, i.e. the genomic islands PAGI-2, PAGI-4 and pKLC102 and the regions of genome plasticity (RGPs) 5, 6, 10 and 26 (Fig. 4) (see below for a more extensive description), demonstrated the largest sequence diversity. These elements contain gene clusters, which are characteristic for certain classes of mobile elements. The orthologous conserved elements usually show a nucleotide identity below 98% and therefore typically display a significantly higher number of nucleotide exchanges among the genomes of clonal isolates compared to the core genome backbone genes.
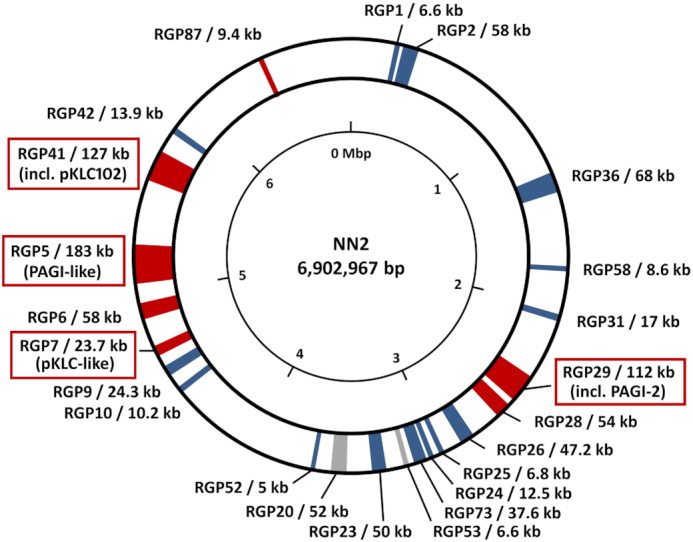
Accessory DNA elements in the genome of the clone C isolate NN2. Regions of the NN2 genome containing accessory DNA are indicated by coloured segments according to conservation in other P. aeruginosa genomes. Grey segments indicate accessory DNA occurring in other clone C isolates as well as in other clonal lineages while clone C specific accessory elements are shown in blue. Red segments indicate accessory elements which are fully conserved only in NN2 and closely related isolates but are absent or only partially conserved in other clone C genomes. The accessory elements are tagged by the so-called ‘region of genome plasticity’ (RGP) assignment defining the flanking core genome parts (Mathee et al. 2008, Klockgether et al. 2011) and the size of the accessory DNA inserted at the respective locus. Tags of RGPs harbouring PAGI-2- or pKLC102-like genomic island are marked by red boxes. Accessory DNA blocks < 5 kbp are not shown in this figure. The 23 displayed accessory elements of isolate NN2 make up for approx. 990 kbp of DNA in total, equivalent to 14% of its chromosomal DNA. The majority of the smaller elements is generally conserved in other clone C strains or even in other clonal lineages while most large accessory elements seem to be specific for the reference or for subsets of clone C isolates. Among them, the largest elements belong to the PAGI-/pKLC-like island family (Klockgether et al. 2007) of which three representatives > 100 kbp (RGP29, RGP5, RGP41) and a fourth fragmentary element (RGP7) are present in the NN2 genome.
Consistent with the low intraclonal sequence diversity of the core genome, the length of syntenic segments with 100% sequence identity had a median size of 99 kb between pairs of clone C strains (Fischer et al. 2016). Thus the length of 100% pairwise conserved sequences is 1000-fold longer than between unrelated clones (Hilker et al. 2015). The chromosomal frame of the core genome is thus conserved among clone C members and only in a few cases disrupted by larger deletions (Fig. 5). However, rapid evolution of the clone C strains’ genome can occur, for example, in the CF lung. Hypermutators, which are impaired in DNA repair or replication fidelity genes and thus possess an up to 1000-fold higher mutation rate, arise in clone C strains during CF lung colonisation (Oliver et al. 2000; Kresse et al. 2003; Mena et al. 2008). As another mechanism of diversification, we observed the expansion of the insertion sequence ISPa20 in the C13 clone C sublineage in one patient (Kresse, Blöcker and Römling 2006). Thirdly, large chromosomal inversions around the origin of replication that conventionally accompany bacterial speciation, creating a CF adapted phenotype have been observed in CF isolates (Römling, Schmidt and Tümmler 1997a; Kresse et al. 2003).
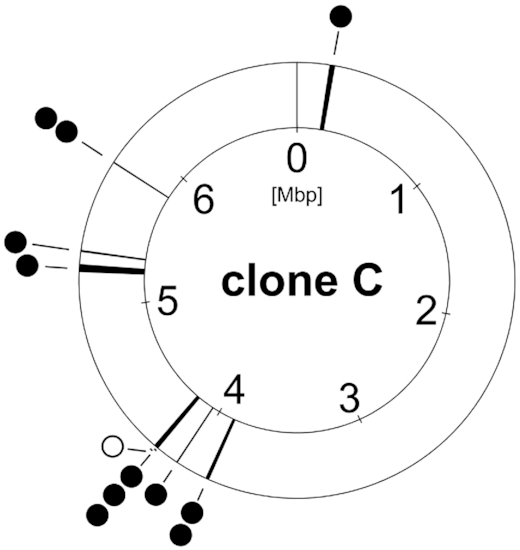
Clone C core genome deletions. Deletions in the core genome found in the clone C strain panel from different habitats (full circle =
=  chronic infection, open circle
chronic infection, open circle =
=  acute infection). Environmental isolates showed no deletions within the core genome.
acute infection). Environmental isolates showed no deletions within the core genome.
Using the syntenic segment length a parameter to assess the relatedness of clone C strains, the majority of strains form a star-like structure of closely related independent singletons of just one strain in a split tree (Fig. 6; (Poigbo, Wolf and Koonin 2012)). A few clone C strains are distant outliers. The preponderance of singletons suggests that most isolates of clone C diverged from a common ancestor by few independent events.
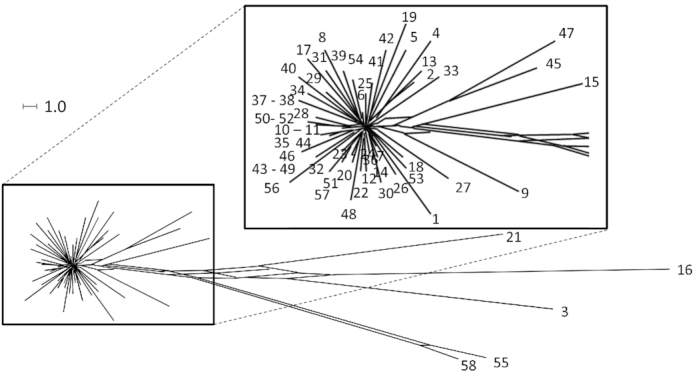
Clone C strain panel dendrogramm. SNP-based phylogenetic trees of the clonal complex C. Only polymorphisms of the core genome were used because mobile elements show a higher variance and are acquired through horizontal gene transfer. The star-like structure visualizes the variability of the core genome by independent de novo mutations. Some outlier strains form a distinct group and will become independent clonal lineages.
The NN2 clinical clone C reference genome
The cystic fibrosis (CF) isolate NN2 was selected as the clinical reference strain for clone C. NN2 is the first P. aeruginosa clone C isolate from a P. aeruginosa naïve subject with CF. The 30-year-long genomic microevolution of the NN lineage was resolved in its CF host until lung transplantation (Cramer et al. 2011). The 6902,967 bp large NN2 genome encodes 6601 open reading frames (ORFs), 62 transfer RNAs, 13 ribosomal RNAs and 1 transfer-messenger RNA. Strain NN2 shares 5455 genes with P. aeruginosa PAO1. Major phenotype-stratifying differences in coding sequence between NN2 and PAO1 were noted in 39 loci including the adherence sensor ladS (Broder, Jaeger and Jenal 2016), genes of pyocyanin and phenazine production (ptsP) (Xu et al. 2005), protein secretion (ftsY) (Ma et al. 2003), chemotaxis (cheR) (Sheng et al. 2019) and biofilm formation (wspR) (Huangyutitham, Guvener and Harwood 2013). Strain NN2 harbours a repertoire of 47 inserted elements in its accessory genome. Of the 1246 non-PAO1 ORFs numerous genes may confer specific fitness traits to clone C isolates such as DNA repair genes or heavy metal resistance determinants (see the next sections for a more extensive description of the accessory genome).
5′ untranslated regions (5′-UTRs) are major regulatory components at the mRNA level. Specific secondary RNA structures (aptamers) constitute binding sites for effector molecules, which are part of expression platforms to regulate downstream genes. The 5′UTRs of expressed genes in the NN2 genome were examined by RNA-sequencing (own unpublished data; (D'Arrigo et al. 2016)). With 70 nucleotides, the median length of 5′UTRs is similar in NN2 as reported for strain PA14 (Wurtzel et al. 2012). However, short 5′-UTRs of 10 to 20 nucleotides in length are more common in the NN2 clone C genome. Those transcripts with short 5’UTRs or even leaderless transcripts lack aptamer-based regulation. When growing under nutrient rich conditions in a fermenter, the 73 NN2 ORFs with the shortest 5’-UTR were expressed at significantly lower mRNA transcript level (P < 5 × 10−5) compared to the corresponding orthologs with longer 5’-UTRs by the reference strain PA14 (Dotsch et al. 2012).
In P. aeruginosa, the role of the 5′-UTRs has been investigated for few loci, including genes involved in virulence (lasB) (Fukushima et al. 1997; Brumlik and Storey 1998), quorum sensing (rhlA and lasI) (Grosso-Becerra et al. 2014), quinolone signaling (pqsABCDE) (Brouwer et al. 2014) and phenazine synthesis (Li et al. 2011). For lasI, a 5′-UTR of only 11 bp was detected in the clone C strain NN2 thus lacking the ROSE family RNA thermometer motif described for the 5′-UTR of lasI in the reference strain PAO1. As this motif mediates thermoregulation by binding of heat shock proteins (Grosso-Becerra et al. 2014), the NN2 should lack this type of temperature regulation for the central quorum sensing autoinducer synthase gene lasI.
The accessory genome of Pseudomonas aeruginosa clone C isolates
A P. aeruginosa genome typically consists of a single circular chromosome and, in some cases, episomal plasmids. The major part of the circular chromosomes represents the highly conserved ‘core genome’ found in all strains of the species with nucleotide identities > 99%. At various positions, however, DNA blocks specific for subgroups of strains or even single isolates are inserted. The specific DNA blocks typically contain genes derived from phages, plasmids, transposons, insertion elements or other DNA mobility elements such as integrase/transposase genes, DNA helicases, nucleases or genes encoding components of a DNA transfer machinery. Such elements are therefore considered formerly mobile DNA elements acquired by horizontal gene transfer and integrated into the host genome. These DNA blocks described as accessory elements can have an individual size of less than 1000 bp, but can also be as large as > 200 kbp. Genome comparison shows that P. aeruginosa genomes typically harbour several dozen accessory elements, which together represent the accessory genome of an isolate. The total size of the accessory genome elements is variable, but usually accounts for more than 10% of the overall genomic DNA of a P. aeruginosa strain (Klockgether et al. 2011; Freschi et al. 2019).
A special type of accessory elements—PAGI-2/pKLC102 like genomic islands
Initial assessment of the accessory genome of clone C strains by physical mapping revealed a common pKLC102 plasmid in a collection of 21 isolates and the presence of various segments of non-PAO DNA often only present in a subgroup or a single strain (Schmidt, Tümmler and Römling 1996; Römling, Schmidt and Tümmler 1997b). Very large specific DNA elements (> 100 kbp) termed PAGI-2/pKLC102-like islands were predominantly detected at three distinct genomic regions inserted at tRNA genes (Larbig, Kiewitz and Tümmler 2002; Klockgether et al. 2004). These genomic islands, which apply a phage like integration mechanism (Kiewitz et al. 2000; Burrus et al. 2002; Larbig, Kiewitz and Tümmler 2002), also display features of conjugative plasmids. Up to 60 ORFs representing a conserved ‘backbone’ of genes involved in DNA organization and transfer are shared among such islands with nucleotide identity values of 70–100%. The islands also contain blocks of unrelated ‘cargo’ DNA, which confer individual features to the host strains (Klockgether et al. 2008). Due to this combination of ‘backbone’ and ‘cargo’ genes, PAGI-2/pKLC102-like islands display a ‘semi-conserved’ composition. Among the conserved backbone genes, many code for yet unknown functions. However, genes with similarity to type IV secretion system components indicate formation of a DNA transfer machinery (Kung, Ozer and Hauser 2010), while other genes code for products involved in the integration and/or excision of the islands such as parA or parB-like chromosome partitioning or integrase genes.
Annotation of the cargo genes has revealed unconventional physiological traits encoded by the individual islands beyond conventional pathogenicity islands. For example, PAGI-2 of the clinical isolate NN2 harbours genes involved in energy metabolism, such as components of the disulphide bond (dsb) formation system, cytochrome C biogenesis and oxidase proteins; and determinants of heavy metal resistance. PAGI-3 from the environmental strain SG17M has among its cargo putative pnt genes encoding nicotinamidenucleotide transhydrogenase proteins and predicted glutamine synthase genes (Larbig, Kiewitz and Tümmler 2002; Lee et al. 2014). Cargo genes thus endow the host strain with specific individual metabolic and resistance traits that allows the colonisation of otherwise inaccessible habitats or confer advantages in competition with other strains or species upon colonising a new habitat. In addition, as exemplified with the clone C specific TLPQC island (see below), competitive advantages might not contribute to new traits exclusively encoded in genomic islands. The acquisition of metabolic gene clusters homologous or functionally similar to core genome genes might provide the host strain with extended opportunities such as regulation of carbon and energy metabolism, which provides metabolic fine-tuning and flexibility to adapt to changing environmental conditions. For example, the PAGI-2 genes in the clinical isolate NN2 mentioned above could aid in the protection against oxidative stress as experienced in the CF lung habitat. Similarly, pnt genes have been shown to be required for optimal growth and tolerance against ethanol (Kamarainen et al. 2017; Long et al. 2018; Liu et al. 2019) and can potentially be involved in the protection against oxidative stress.
Semi-conserved PAGI-2 or pKLC102-like islands are frequently present in clone C genomes. About 58 genome-sequenced isolates of a clone C collection harbor at least one island, while 32 (55%) of the isolates possess two or even more islands (Fischer et al. 2016). Although with lower frequency, PAGI- or pKLC102-like island are also present in P. aeruginosa strains from other clonal lineages. Prominent examples are the pathogenicity island PAPI-1 in reference strain PA14 with genes contributing to plant and mouse virulence (He et al. 2004) and the ExoU-island A (Kulasekara et al. 2006). PAGI-like islands were also detected in other Pseudomonads and in other genera, mainly beta- or gamma-proteobacteria that had been classified in the pre-genomic era as ‘honorary pseudomonads’. These islands might have emerged from an ancestral mobile element, which allowed the uptake/exchange of DNA via horizontal gene transfer between different species and genera. For instance, an identical copy of the PAGI-2 island from a German clinical clone C isolate (Larbig, Kiewitz and Tümmler 2002) was found in a Cupriavidus metallidurans isolate from a metal-contaminated environment in Belgium (Mergeay et al. 2003). Another example is the clc element, which was transferred from P. knackmussii B13 to P. putida F1 (Ravatn, Zehnder and van der Meer 1998) and P. aeruginosa PAO1 (Gaillard et al. 2008).
Pseudomonas aeruginosa plasmid pKLC102 (Kiewitz et al. 2000; Klockgether et al. 2008), the clc island and their derivatives are integrative and conjugative elements (ICE). The pKLC102 element is present in all clone C strains. Mobilisation from the host chromosome and formation of a circular element with copy numbers up to 30 per host chromosome occurs for pKLC102 in P. aeruginosa strain SG17M (Klockgether et al. 2007) and consequently made up 10% of the mRNA content (Klockgether et al. 2008). Upon mobilisation, the islands were precisely excised from the host chromosome without affecting the surrounding core genome DNA. The retained potential for autonomous replication is consistent with a postulated replication origin (oriV) within pKLC102. Also, the clc island can excise from the chromosome and form a circular intermediate in which both ends are connected (Sentchilo, Zehnder and van der Meer 2003). Circular isoforms could not be detected in clone C strains for PAGI-2 and PAGI-3 islands under laboratory conditions (Klockgether et al. 2007), but chromosomal excision can occur at low frequency in sequential isolates. Loss of PAGI-2 from the chromosome was observed in serial clone C isolates that had been retrieved from the airways of a CF patient in half-year intervals (Fig. 7). A comparably precise excision as seen for the excision/mobilisation of pKLC102 can be postulated.
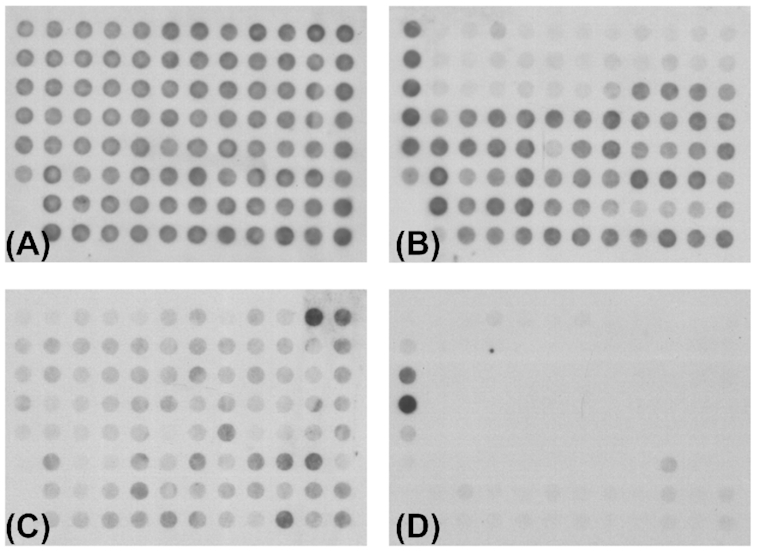
Loss of PAGI-type islands in sequential clone C islands from cystic fibrosis airways. The presence of PAGI-2 or partially related islands in P. aeruginosa genomes was tested with hybridisation of macroarrays representing the ORFs located in PAGI-2- Upper row: Hybridisation patterns of clone C strains SG1 (A) and SG3 (B), isolated at the onset of a chronic P. aeruginosa infection or three months later, respectively. Lower row: Hybridisation patterns for clone C strains NN18 (C) and NN86 (D), isolated three or 17 years, respectively, after the onset of a chronic P. aeruginosa infection. The patterns for both pairs demonstrate the loss of an island from the host genome in the later isolate while it was still present in the earlier isolate. In case of SG3, the PAGI-2 island itself was lost. Another semi-conserved element is still present but the absence of signals in the upper rows, representing the PAGI-2 specific ORFs, indicate the loss of this island. The weaker hybridisation pattern for NN18 indicates the presence of a related PAGI-like island. This island is apparently absent in the later isolate NN86 as the corresponding hybridisation result displayed only two prominent control spots but no clearly positive PAGI-2 specific signals. This figure and the corresponding results were originally published in Klockgether et al., J Bacteriol 2007, Vol 189(6), p. 2443–2459. The reuse of this figure was kindly permitted by the Copyright Holders (Copyright ©2007, American Society for Microbiology).
Individual clone C genomes may harbor several PAGI-like islands (Klockgether et al. 2007; Wiehlmann et al. 2007). For example, the sequenced clinical isolate NN2 carries pKLC102, PAGI-2 and a hybrid of two PAGI-like elements (Fischer et al. 2016) and the aquatic isolate SG17M pKLC102, PAGI-3 and PACGI-1 (Lee et al. 2015). Interestingly, pKLC102 and PACGI-1, or variants thereof, were detected in all clone C genomes analysed so far. In NN2, the ‘hybrid’ island is highly similar to the SG17M PACGI-1 with approx. 73 kb of conserved DNA containing not only the typical ‘backbone genes’, but also ‘cargo’ genes such as TLPQC-1 genes assigned to protein homeostasis (Lee et al. 2015). These closely related islands were both inserted at tRNAGly genes, but are located in different genomic areas: in SG17M PACGI-1 is found at RGP27, while in NN2 the ‘hybrid’ island with the counterpart is found at RGP5. So, apparently both the clinical and the environmental isolate, or a common precursor strain, took up a similar accessory element. The element can be either integrated at different sites into the chromosome a priori, or after horizontal transfer into an ancestor clone C stain changed its location by subsequent transposition events. If chromosomally integrated, pKLC102 is alternately inserted at RGP41 or RGP7 into a tRNALys gene.
After integration into the host genome PAGI-like islands can rapidly diversify by nucleotide substitutions, insertions or deletions. Secondary insertions of IS elements or transposons generate a mosaic-like architecture of the islands. For example, a transposon and remnants of pKLC102 were assembled to genome island PAGI-4 in strain NN2. Due to these secondary events pKLC102 sequences may become irreversibly fixed in the chromosome (Romling et al. 1997) as has been seen in a subgroup of clone C strains from CF airways which integrate a hybrid of class I integron, IS elements and aminoglycoside resistance gene cassette called TNCP23 into their pKLC102 sequence (Klockgether et al. 2004).
These secondary insertions into PAGI-like islands triggered large chromosomal inversions in some clinical CF clone C strains (Römling, Schmidt and Tümmler 1997a). The inversion breakpoints were mapped to an IS element at the border of the TNCP23 element in the pKLC102 sequence (Kresse et al. 2003). Copies of the IS element were identified at both recombination breakpoints indicating that the duplication of the IS element and its subsequent integration at a CF-relevant genomic locus might have initiated the inversion of several Mbp of DNA. There does not seem to be a specificity in the IS elements that can provide the basis for large chromosomal inversions (Kresse, Blöcker and Römling 2006). However, the duplication of an IS element is not mandatory to generate an inversion. In another strain, we have localized the inversion breakpoints in conserved DNA blocks of two PAGI-2 like islands, but no IS element or equivalent cover the breakpoint loci (unpublished data).
Accessory elements of Pseudomonas aeruginosa clone C clinical reference strain NN2
Elements of the accessory genome other than the PAGI-like islands are typically smaller in size. Many of them contain only few ORFs. For example, of the 47 accessory elements in the NN2 clone C genome that distinguish it from P. aeruginosa PAO, 24 elements are smaller than 5 kbp (Table 2). Forty-five of the 47 blocks were found in one of the 89 so-called ‘regions of genome plasticity’ (RGPs), loci already defined as candidates for harbouring accessory elements upon genome comparisons of P. aeruginosa strains (Mathee et al. 2008; Klockgether et al. 2011). Whereas the larger PAGI-like islands are specific for a strain or subgroup of clone C strains, the majority of the other elements is shared among all tested clone C strains and likely constitutes the clone-specific signature of the accessory genome. Of the latter group, the element inserted at RGP24 harbours genes that are annotated as CRISPR-related cas and csy genes that are part of a CRISPR/Cas system in clone C strains similar to the one described for the P. aeruginosa reference strain PA14.
Table 2.
Accessory elements detected in the genome sequence of clone C strain NN2.
| Region1 | Size [kbp] | No. of ORFs | Comment |
|---|---|---|---|
| RGP46 | 1.6 | 3 | |
| RGP1 | 6.6 | 6 | |
| RGP2 | 58 | 39 | with type I restriction modification system genes |
| RGP66 | 3.4 | 4 | phage resistance and type I restriction modification genes |
| RGP3 | 2.8 | 4 | |
| RGP4 | 4.1 | 6 | |
| RGP5 | 183 | 175 | combination of two PAGI-2-like integrated elements2 |
| RGP6 | 58 | 64 | with trb conjugative transfer gene cluster |
| RGP7 | 23.7 | 24 | PAGI-4, composed of pKLC102 fragment and transposon DNA |
| RGP9 | 24.3 | 22 | flagella glycosylation genes3 |
| RGP47 | 2.5 | 2 | with gene encoding for S-type pyocin |
| RGP10 | 10.2 | 16 | |
| RGP11 | 1.8 | 2 | |
| RGP48 | 2.8 | 4 | |
| RGP13 | 2.3 | 2 | |
| RGP15 | 2.7 | 3 | |
| RGP76 | 1.6 | 2 | |
| RGP52 | 5 | 4 | |
| RGP20 | 52 | 51 | conserved in many clonal linages, also present in strain PAO1 |
| RGP22 | 2.9 | 5 | |
| RGP23 | 50 | 44 | PAGI-14, present in many clonal lineages but absent in PAO1 |
| RGP53 | 6.6 | 4 | |
| RGP73 | 37.6 | 10 | pyoverdine biosynthesis genes3 |
| RGP24 | 12.5 | 8 | with CRISPR-related cas and csy genes |
| PA2425/28 | 2.1 | 2 | pyoverdine biosynthesis genes pvdS and pvdY |
| RGP25 | 6.8 | 4 | |
| RGP26 | 47.2 | 54 | with trb conjugative transfer gene cluster |
| RGP71 | 1.6 | 1 | |
| RGP28 | 54 | 44 | with phage like genes |
| RGP43 | 2.2 | 4 | |
| RGP56 | 3.2 | 3 | |
| RGP29 | 112 | 126 | PAGI-2 plus additional 7 kbp accessory element |
| RGP31 | 17 | 17 | LPS biosynthesis genes (O-antigen, defining serotype)3 |
| RGP58 | 8.6 | 6 | |
| PA3576/78 | 2.3 | 3 | |
| RGP36 | 68 | 59 | with trb conjugative transfer gene cluster |
| RGP89 | 0.6 | 1 | |
| RGP68 | 2.4 | 3 | with exoS and ExoS chaperone gene |
| PA4092/93 | 3.2 | 3 | |
| RGP44 | 2 | ||
| RGP39 | 4.2 | 5 | |
| RGP60 | 0.7 | 1 | major pilin gene pilA3 |
| RGP41 | 127 | 131 | integrated element pKLC102, with add. 23 kbp integron |
| RGP42 | 13.9 | 18 | with phage like genes |
| PA5085/90 | 4.9 | 4 | |
| RGP87 | 9.3 | 10 | |
| RGP80 | 4.2 | 5 |
If the accessory DNA was located in an already defined region of genome plasticity, the respective RGP no. is given. Other loci are described by the flanking core genome genes (designations of homologs from reference strain PAO1 are given).
Pseudomonas aeruginosa clone C replacement islands
A specific subgroup among the accessory genome elements are four clusters of functionally well described genes that are present in all P. aeruginosa strains. Genes for LPS biosynthesis (defining the serotype), pyoverdine biosynthesis, flagella glycosylation and the major pilin PilA are found in all genomes, but, unlike core genome parts, are variable elements within the species. The respective counterparts in different strains cannot only be discriminated by high nucleotide substitution rates, the clusters can also differ in gene composition and size. In contrast to accessory elements each genome carries one version of each of these four gene clusters, which are, independently of the respective type or subtype, always located at the same position within the conserved core genome. These gene clusters have been termed replacement islands, which have developed under diversifying selection early in the evolution of P. aeruginosa (Kung, Ozer and Hauser 2010). Within clonal lineages, the subtypes of the four replacement islands are conserved. As listed in Table 3, the clone C reference strain NN2 harbours a LPS biosynthesis serotype 01 gene cluster (Raymond et al. 2002) and a type a1 flagella glycosylation cluster (or ‘a-type long’) (Arora e al. 2004). The pyoverdine biosynthesis gene cluster can be assigned to type II (Smith et al. 2005). The major pilin gene pilA of clone C strains belongs to group II (Spangenberg et al. 1995; Kus et al. 2004).
Table 3.
Replacement island types of P. aeruginosa clone C1.
| Gene (cluster) | RGP2 | Detected type | Reference | Comment |
|---|---|---|---|---|
| LPS biosynthesis (O-antigen) | RGP31 | serotype 01 | (Raymond et al. 2002) | |
| flagella glycosylation | RGP9 | a-type long (a1) | (Arora et al. 2004, Schirm et al. 2004) | |
| pyoverdine biosynthesis | RGP73 | type II | (Smith et al. 2005) | comparably high nucleotide identities (99.1%–99.5%) with all three subtypes (IIa, IIb, IIc) defined in the reference paper |
| major pilin (pilA) | RGP60 | type II | (Voisin et al. 2007) | assignment to type II due to shared gene synteny in this region |
| exotoxin S | RGP68 | exoS | (Kulasekara et al. 2006) | so far no strain was found sharing all clone C markers but harbouring an exoU gene at RGP73 |
Variation of the gene repertoire within Pseudomonas aeruginosa clone C
The strain-specific acquisition of genes generates traits that modulate fitness, virulence, lifestyle or metabolic competence on the level of the individual isolate. An average clone C strain harbors about 100 strain-specific genes in its accessory genome (Fischer et al. 2016) (Table S1, Supporting Information). This gene pool is primarily acquired from phylogenetically related bacteria. For about 80% of these genes the closest orthologues were identified in other P. aeruginosa clones or other Pseudomonas species (Table 4). According to database searches, a further 20% of the genes have their closest homologue among other gamma- (8.7%) or beta-proteobacteria (11.5%) such as Haemophilus somnus, Klebsiella pneumoniae, Salmonella enterica, Achromobacter piechaudii, Achromobacter xylosoxidans or various Burkholderia species (Fischer et al. 2016).
Table 4.
Numbers of strain-specific genes detected in a panel of 58 clone C strains.
| No. | % of Total No. | Median No. per Strain | ||
|---|---|---|---|---|
| In 58 strain panel | 7488 | 100 | 104 | |
| Closest Homolog in | ||||
| other P. aeruginosa | 4349 | 58.08 | 71 | |
| other Pseudomonads | 1620 | 21.63 | 14 | |
| other γ-proteobacteria | 654 | 8.73 | 4 | |
| other origin | 865 | 11.55 | 11 |
In contrast to other common P. aeruginosa clones, clone C strains have enlarged their genetic repertoire for carbohydrate metabolism. P. aeruginosa typically prefers amino acids and fatty acids as carbon source, but this repression of the uptake and catabolism of sugar (‘catabolite repression control’) (Linares et al. 2010) does not apply to the most common clone C strains that may compensate the core genome-predetermined limitations in the utilization of sugars by the horizontal acquisition of genes of carbohydrate metabolism (unpublished results).
Phenotypic variability in Pseudomonas aeruginosa clone C strains
Phenotypes, the timely expression of genetic information, and the regulation of phenotypic traits by environmental and internal signals have been studied mainly in the model strains P. aeruginosa PAO and PA14, both clinical isolates. Thereby, sophisticated regulatory mechanisms, for example, for the secretion of effector proteins of the type III secretion system upon removal of the divalent cation Ca2+, have been unravelled (Lee et al. 2010). However, the generality of regulatory patterns of phenotypic traits within P. aeruginosa has not been established. We exemplarily investigated the expression of two phenotypic traits of P. aeruginosa. Although almost equally virulent as PA14 in the wax moth G. melonella model system, the aquatic isolate SG17M does not secrete type III secretion system effector proteins under promiscuous conditions (Kamal et al. 2019). Variable secretion of type III effector proteins was observed among clinical isolates of clone C and strains of other clonal lineages, however, environmental isolates of clone C, in contrast to non-clone C isolates, did consistently not secrete type III effectors under promiscuous conditions. Likewise, the secretion of the siderophore pyoverdine was not pronounced in environmental isolates of clone C, while two of three clinical isolates showed distinct secretion. However, the ability to produce and secrete pyoverdine is not impaired as pyoverdine secretion is relieved upon deletion of the membrane-bound protease FtsH in the environmental isolate SG17M (Kamal et al. 2019). The underlying molecular regulatory mechanisms of phenotypic variability are to be unravelled, with the reduced variability in clonal isolates to provide a more stringent genetic background, which will facilitate the genetic characterisation.
Microevolution of Pseudomonas aeruginosa clone C during chronic infection of CF airways
The colonization of CF airways with P. aeruginosa is one of the few opportunities to observe the microevolution of a bacterium during chronic infection in real life (Marvig et al. 2015; Winstanley, O'Brien and Brockhurst 2016). When P. aeruginosa conquers the CF lungs, the aquatic bacterium needs to adapt phenotype and genotype to a hostile environment characterized by a plethora of nutrients, but also a large battery of deadly host defenses (Tümmler and Kiewitz 1999; Folkesson et al. 2012; Moradali, Ghods and Rehm 2017). The microevolution of P. aeruginosa clone C strains in CF lungs has been investigated by phenotyping and whole genome sequencing of serial isolates from two patients collected from the onset of chronic colonization over a period of up to 30 years (Cramer et al. 2011; Klockgether et al. 2018). Both patients who became chronically colonized with P. aeruginosa clone C during childhood in the early 1980s had developed rather mild clinical phenotypes. Genome sequencing uncovered from a few hundred to close to a thousand de novo mutations in the serial isolates from the two patients. In parallel, the accessory genome was modified by the loss and acquisition of several DNA blocks. During colonization of the CF lungs isolates became deficient in the secretion of virulence effectors and siderophores. Hence the genotypic and phenotypic conversion of clone C strains is similar to that seen with other P. aeruginosa clones (Marvig et al. 2015; Klockgether et al. 2018).
A transmissible locus for protein quality control in Pseudomonas aeruginosa clone C strains
Genomic islands determine strain-specific traits which are beneficial in terms of virulence, antibiotic resistance, symbiosis, metabolic diversity and adaptation (Juhas et al. 2009). In the aquatic strain SG17M, the P. aeruginosa clone C-specific genomic Island 1 (PACGI-1, a hybrid of two PAGI-like elements; see accessory genome chapter above) is 86 kb in length encoding over 100 genes (Lee et al. 2015, 2016). One border of PACGI-1 constitutes a cluster of protein quality control genes, coding for various holding and disaggregating chaperones, heat-inducible proteases such as FtsH, DegP and HtpX, thioredoxin and other stress resistance genes that seems to be present in all clone C strains (Fig. 8) (Lee et al. 2016). Flanked by mobile elements, this gene cluster named as ‘Transmissible Locus for Protein Quality Control’ (TLPQC-1) (Lee et al. 2016) and alternatively ‘locus of heat resistance’ (LHR) is consistent with protein homeostasis as a major determinant of temperature tolerance (Mercer et al. 2015; Boll et al. 2017).
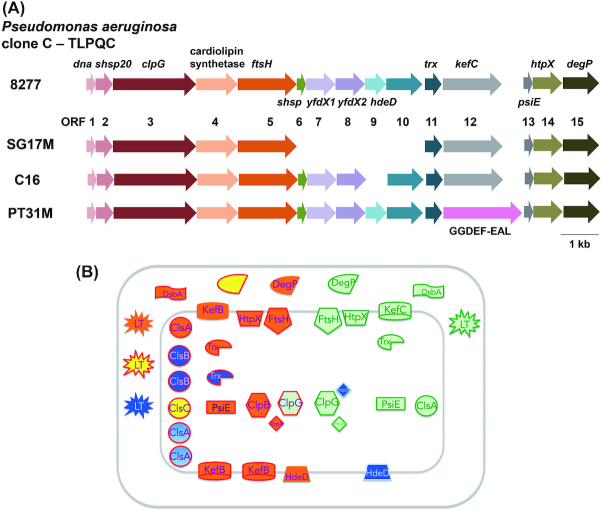
Context of the TLPQC locus in P. aeruginosa clone C strains. (A) Genetic maps of TLPQC loci in selected clone C strains. TLPQC genetic maps from 8277, a clinical isolate from human urine; SG17M, an environmental clone C isolate from river water in Germany; C16, an isolate from a cystic fibrosis patient (unpublished) and PT31M, an isolate from drinking water (unpublished) are described. Open reading frames 1–15 code for the following proteins: Dna, MerR-like transcriptional regulator; sHsp20, small heat shock protein; ClpG, disaggregating chaperone; Cls, cardiolipin synthase; FtsH, metalloprotease; sHsp, small heat shock protein, YfdX1 and YfdX2, antibiotic resistance, anti-virulence protein; HdeD, transmembrane protein involved in acid tolerance; ORF10, hypothetical protein; Ttrx, thioredoxin; KefC, glutathionine-dependent potassium-efflux system and methylglyoxal detoxification; PsiE, putative phosphate starvation-inducible protein; HtpX, inner-membrane associated peptidase; DegP, periplasmic protein with protease and chaperone activity. In PT31M, kefC is replaced by a GGDEF-EAL domain protein encoding ORF. (B) TLPQC locus proteins and homologous core genome gene products. Red line frame, core genome gene products; green line frame, TLPQC gene products of SG17M; blue line frame, TLPQC gene products additionally present in other clone C strains. The P. aeruginosa clone C core genome genes code for one ClpG disaggregase (and the functional homologue ClpB), one FtsH protease, one HtpX protease, one PsiE protein, two DegP-like proteases (AlgW and MucD) and two thioredoxins (Trx1 and Trx2). Furthermore, genes for three KefB-like transporters and six cardiolipin synthase (CLS) and CLS-like proteins are present on the core genome. All these gene products have counterparts on the SG17M TLPQC locus. TLPQC gene products of other P. aeruginosa strains such as the acid resistant protein HdeD, an integral membrane protein, have also counterparts on the SG17M core genome. In addition, genes for a thiol disulfide oxidoreductase DsbA and lytic transglycosylase (LT), also with homologues on the core genome, are encoded elsewhere on PACGI-1.
Protein homeostasis is essential for all living organisms (Hartl, Bracher and Hayer-Hartl 2011; Valastyan and Lindquist 2014) and partially determines cell aging and longevity (Koga, Kaushik and Cuervo 2011). Impairment of protein homeostasis by massive misfolding and aggregating processes results in various proteotoxic human diseases such as Parkinson's, Alzheimer's and Huntington's disease and is connected to cell aging and cytotoxicity (Ross and Poirier 2004). In bacteria, survival of various stresses, antibiotic resistance, adaptation, but also physiological processes such as biofilm formation and virulence, are closely associated with protein homeostasis mechanisms (Marr et al. 2007, Neckers and Tatu 2008, Lee et al. 2016, Pu et al. 2019).
Recent studies have shown that a potent protein quality control system present on an unconventional genomic island contributes to successful survival and adaptation of P. aeruginosa clone C strains with most molecular mechanisms still to be explored in detail (Figs 8 and 9). In this section, we describe the general characteristics of bacterial protein quality control systems as well as report on the initial characterisation of selected gene products of the horizontally acquired clone C specific components for protein quality control.
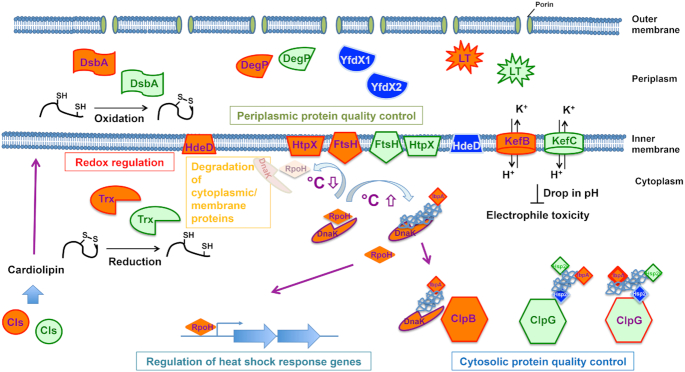
Core functionalities of TLPQC and core genome gene products. Regulation of cytosolic and periplasmic protein quality control, degradation of cytosolic and membrane proteins and redox regulation are major pathways supported by TLPQC gene products. Regulation of the heat shock response by degradation of RpoH is mainly conducted by the core genome membrane-bound protease FtsH. Gene products are described in Fig. 8. For simplicity, one homologous core gene product is displayed.
The chaperone system for protein quality control in bacteria
Environmental stresses such as elevated temperature, detergents, organic solvents and oxidative stress, unfold and denature proteins and eventually lead to reversible and irreversible protein aggregation. Protein (and small molecule) chaperones are key players in protein quality control systems involved not only in folding proteins, but also in preventing protein aggregation and disaggregating protein aggregates for refolding or degradation (Kim et al. 2013; Balchin, Hayer-Hartl and Hartl 2016; Schramm, Schroeder and Jonas 2019). Hydrophobic and electrostatic interactions direct the interactions between chaperones and their client proteins (Kim et al. 2013; Koldewey et al. 2016; Lee, Kim and Bardwell 2018). The central role of chaperones is shown as, for example, Hsp70 (DnaK), Hsp90 (HtpG) and small heat shock proteins are well conserved among all living organisms (Balchin, Hayer-Hartl and Hartl 2016; Mogk, Ruger-Herreros and Bukau 2019).
In the first instance, chaperones are involved in the de novo folding of proteins in the cytosol (Balchin, Hayer-Hartl and Hartl 2016). In bacteria, the nascent peptide chains released from the ribosome are firstly engaged by ribosome-associated trigger factor and the DnaKJE system. Trigger factor is an ATP-independent chaperone. The DnaKJE system consists of Hsp70 (DnaK), its co-chaperone Hsp40 (DnaJ) and nucleotide exchange factor GrpE. The de novo folding of protein is subsequently accelerated by the folding chaperones GroESL and HtpG in a process requiring ATP hydrolysis (Balchin, Hayer-Hartl and Hartl 2016).
Upon stress conditions that eventually lead to protein aggregation, small heat shock proteins (sHsps) act as holding chaperones by forming a complex with client proteins to prevent irreversible protein aggregation (Mogk, Ruger-Herreros and Bukau 2019), but also facilitate the resolublization of aggregated proteins through disaggregating chaperones (Mogk et al. 2003; Mogk, Ruger-Herreros and Bukau 2019). Under stress conditions such as elevated temperature, changes in the secondary and tetrary structure of sHsps lead to a higher binding affinity to client proteins (Mogk, Ruger-Herreros and Bukau 2019).
Once protein aggregates are formed, the ClpB/Hsp100 disaggregating chaperone system unfolds protein aggregates in an ATP-dependent manner cooperatively with sHsps and the DnaKJE system (Mogk, Kummer and Bukau 2015; Mogk, Bukau and Kampinga 2018). ClpB possesses two asymmetric ATPases associated with diverse cellular activities (AAA+) domains and a coiled-coil structured middle domain (M-domain). The M-domain interacts with aggregate-loaded DnaK to elevate the ATPase and disaggregating activity of ClpB (Mogk, Kummer and Bukau 2015). The ATP hydrolysis generates the threading power to unfold the aggregates and pass the peptide chain into the ClpB hexamer (Mogk, Kummer and Bukau 2015).
Although ATP is lacking in the bacterial periplasmic space in contrast to the cytoplasm, diverse ATP-independent chaperones have been identified (Stull, Betton and Bardwell 2018). As porins in the outer membrane allow free diffusion of small molecules less than 600 Da from the extracellular space (Nikaido 2003), periplasmic proteins are more directly exposed to environmental stress than cytoplasmic proteins. While SurA, Skp and DegP can fold outer membrane proteins (McMorran, Brockwell and Radford 2014), Spy and HdeAB are stress-responsive chaperones responding to alcohol and acid stress, respectively (Stull, Betton and Bardwell 2018).
Escherichia coli has been a model organism to investigate the biochemical and physiological role of chaperones. Although P. aeruginosa is a key human pathogen with protein homeostasis as a potential target for antimicrobial treatment, the chaperone system has rarely been adressed in this species. With 5.5–7 Mbp, the genome size of P. aeruginosa is larger than the 4.5–5.5 Mbp of E. coli (Schmidt, Tümmler and Römling 1996; Lee et al. 2006; Lukjancenko, Wassenaar and Ussery 2010; Gordienko, Kazanov and Gelfand 2013). Especially, the successful clonal groups of P. aeruginosa including PA14 and clone C have distinct genome characteristics such as to display instant double crossover homologous recombination and to flexibly acquire genomic islands through horizontal gene transfer (Römling, Schmidt and Tümmler1997b; Fischer et al. 2016; Lee, Kamal and Römling 2019). These observations suggests P. aeruginosa, especially common clonal strains, to possess a large cargo of distinct genes, which may include unique genes involved in protein homeostasis, allowing them to survive successfully under a variety of host and environmental conditions. Of note, those gene products, such as disaggregases, might serve as potent disaggregases also for aggregates causing the above mentioned human diseases such as Alzheimer (Gao et al. 2015).
The TLPQC-1 island of Pseudomonas aeruginosa clone C encodes xenologues of core genome genes
As a remarcable hallmark of TLPQC, many of the encoded gene products are xenologues of conserved core genome genes of P. aeruginosa (discussed below; Fig. 8B; (Lee et al. 2015, 2018)). TLPQCs islands, classified in three classes with minimal diversification, but variable gene content, are found not only in common clones of P. aerguinosa such as clone C and clone J, but also in clinical and food-derived strains of species from diverse genera, such as E. coli, K. pneumoniae and Cronobacter sakazakii (Bojer et al. 2010; Lee et al. 2016; Nguyen et al. 2017). However, the origin of TLPQC is most likely an environmental bacterium such as Cupriavidus necator (Ralstonia eutropha) thriving under extreme conditions. TLPQC encoded on either the genome or a plasmid can be efficiently transferred by horizontal gene transfer (Lee et al. 2016; Nguyen et al. 2017) and integrated into the proximal region of a tRNA gene, a common insertion site of a genomic island. Consistently, in clone C strains, PACGI-1/TLPQC is inserted at the 3′ end of a tRNAGly gene. However, in SG17M, a strain isolated from river water in Germany, TLPQC is uniquely inserted into the 7th spacer region of the CRISPR locus that encodes an adaptive bacterial immune system against plasmid and phages (Lee et al. 2015).
Despite of efficient horizontal gene transfer, only 2% of sequenced E. coli strains contain TLPQC, suggesting a detrimental effect in strain backgrounds beyond phylogroup A (Mercer et al. 2015). Of note, after preheating of milk to 55–60°C, 36% E. coli contained TLPQC (Boll et al. 2017). Equally, only 5% of the E. coli strains have been found TLPQC positive in untreated wastewater, but 59% E. coli strains were TLPQC positive in chlorine-treated wastewater (Zhi et al. 2016). These results strongly suggest that TLPQC is selected to aid survival upon protein stress. Consistently, P. aeruginosa clone C strains exhibit higher tolerance against lethal heat stress than other epidemic and non-epidemic P. aeruginosa strains which do not harbour TLPQC (Lee et al. 2015). The core unit of TLPQC is composed of the three genes dna, shsp20GI and clpGGI (Lee et al. 2016; Nguyen et al. 2017). Overexpression of the dna-shsp20GI-clpGGI operon can potently enhance heat tolerance even in unrelated P. aeruginosa strains, suggesting that these genes are crucial elements for heat tolerance (Lee et al. 2015).
The small heat shock protein sHsp20GI
The sHsp20GI belongs to class B of small heat shock proteins in conjunction with other bacterial and fungal small heat shock proteins. Within the B subclass, sHsp20GI is the founding member of a subfamily of TLPQC encoded sHsp20s, horizontally transferred small heat shock proteins of bacterial isolates of various species. Structural modeling showed that a core of two anti-parallel β-sheets consisting of seven anti-parallel β-strands characteristic for the α-crystallin core region of sHsp family proteins is also present in sHsp20GI. sHsp20GI uniquely has an extended N-terminal region compared to the well-characterized class A sHsps EcIbpA, EcIbpB and PaIbpA (Lee et al. 2015). Biochemical analyses and electron microscopy indicated that the sphere-like sHsp20GI oligomer, probably constituting an inactive state, is composed of 24 monomers consistent with the amino acid sequence core regions II and I required for dimerization and oligomerization, respectively (Lee et al. 2015). sHsps typically alter their oligomeric state upon stress such as elevated temperature (Mogk, Ruger-Herreros and Bukau 2019). Although the secondary structure of sHsp20GI shows thermal stability up to 60°C (Lee et al. 2015), alternations in the tertiary structure of sHsp20GI upon temperature or other stress challenges has not been characterized yet.
Conventionally, heat shock proteins including sHsps are specifically transcriptionally induced at elevated temperature under the transcriptional control of the heat shock sigma factor 32 (RpoH) (Yura and Nakahigashi 1999). This tight control extends to the posttranscriptional level as the ROSE element of repression of heat shock gene expression in the 5′-untranslated region of sHsps inhibits expression at temperatures below 30 °C (Kortmann and Narberhaus 2012; Krajewski, Nagel and Narberhaus 2013). Of note, however, sHsp20GI, as other TLPQC gene products (see below, (Lee et al. 2018)), shows an unconventional expression pattern. sHsp20GI is produced between 20°C and 42°C from mid-logarithmic phase with maximum expression in stationary phase in both minimal and rich medium (Lee et al. 2015). Oxidative stress can further enhance production of sHsp20GI (Lee et al. 2015). Of note, sHsp20GI is one of the most highly expressed proteins of P. aeruginosa clone C strains under standard growth conditions (Sriramulu, Nimtz and Römling 2005).
Conventionally, sHsp20s are holding chaperones, which can prevent the thermal aggregation of client proteins (Fig. 10). Such an activity was also demonstrated for sHsp20GI using the model substrate citrate synthase (Lee et al. 2015). Furthermore, as for other sHsps, deletion of shsp20GI slightly, but significantly reduces heat tolerance. Redundancy in thermotolerance functionality is demonstrated upon co-deletion of the P. aeruginosa gene ibpA coding for the core genome sHsp20. In E. coli, the sHsps IbpA and IbpB work cooperatively with disaggregating and refolding chaperones such as ClpB, DnaK and GroELS (Zolkiewski 1999; Mogk et al. 2003; Mogk, Ruger-Herreros and Bukau 2019). Upon thermo and other stress conditions, sHsps create a reservoir of client proteins to prevent irreversible aggregation, which provides an amenable protein state for disaggregation and subsequent refolding (Mogk et al. 2003; Mogk, Ruger-Herreros and Bukau 2019). Although the operon context of shsp20GI in combination with clpGGI encoding the disaggregating chaperone suggests cooperativity, such a functionality has not yet been examined.
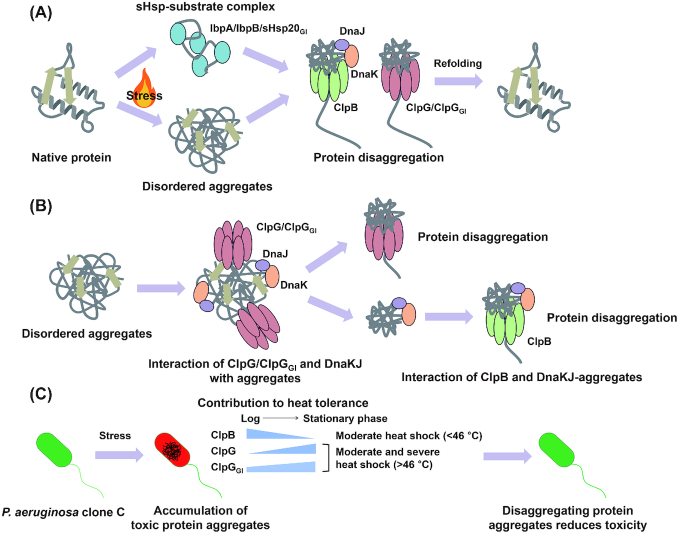
Processing of stress-induced protein aggregates by ClpG and ClpB chaperones. (A) Various stresses, such as elevated temperature, can lead to disordered aggregation of native proteins. sHsps form a complex with denatured proteins to prevent irreversible protein aggregation as well as to facilitate the disaggregating and refolding by ClpB-DnaKJ/ClpG chaperones. (B) ClpG, ClpGGI and the DnaK chaperone directly bind disordered aggregates. ClpG and ClpGGI bind to the disordered aggregates with their N-terminal domain to subsequently perform disaggregation. Although the DnaK/DnaJ/GrpE system can dissolve aggregates to some extent, aggregate-loaded DnaK/DnaJ/GrpE associates with the M-domain of the disaggregase ClpB to activate ATP-ase activity and disaggregation. (C) Aggregation of proteins, which have a vital role in the cell, has a detrimental effect on cell physiology. The ClpB-DnaKJ disaggregating chaperone complex functions efficiently at moderate heat shock condition, which is up to 46°C, in the logarithmic phase. ClpG and ClpGGI mainly work in the stationary phase of growth and their potent threading power allows them to disaggregate proteins formed under severe heat shock conditions (Lee et al. 2018). Unlike ClpG, ClpGGI contributes to heat tolerance in the logarithmic growth phase and backs up the ClpB-DnaKJ system.
The stand-alone disaggregase ClpG/ClpGGI
Together with the Mer-like transcriptional regulator dna and shsp20GI, clpGGI composes the core unit of any TLPQC locus. As other Hsp100 family members, ClpGGI contains two distinct AAA + domains with a M-domain integrated into the first AAA + domain to form a hexameric structure (Lee et al. 2016, 2018). ClpGGI shows the highest amino acid homology to the well characterized class I AAA + chaperones ClpC and the disaggregase ClpB, but has distinctively longer N- and C-terminal domains and a unique M-domain sequence. Although a species specific protease interaction motif cannot be excluded (Miller, Chaudhary and Marsee 2018), VGF protease interaction motif present in Bacillus subtilis ClpC, which couples protein disaggregation with proteolytic digest (Trentini et al. 2016), is missing. Accordingly, ClpGGI has been characterized as a disaggregating chaperone with distinct features compared to the ClpB-DnaK/DnaJ/GrpE bichaperone system. Most characteristic, in contrast to ClpB, purified ClpGGI is able to potently disaggregate client proteins without requiring the assistance of the accessory DnaK/DnaJ/GrpE helper chaperone system (Fig. 9; (Lee et al. 2018)). Consistent with in vitro results, clpGGI expression confers a heat tolerant phenotype and solubilizes the heat-induced protein aggregates in a clpB or dnaK deletion background.
The molecular basis of these unique features of ClpGGI is based on the high basal ATPase activity and the extended N-terminal region that directly binds the substrate (Lee et al. 2018). These unique biochemical features of ClpGGI have two consequences. First, the high ATPase activity of ClpGGI calls for a repressive type of tight physiological regulation of ClpG disaggregases besides activation by aggregate loaded DnaK of the disaggregation activity of ClpBin vivo. Indeed, we have observed high molecular weight structures of ClpGGI (Lee, Curth, Carroni and Römling, unpublished data) that probably represent an inactive state as it has been observed in other systems (Carroni et al. 2017).
Second, the feed-forward loop of the heat shock response regulon is disrupted as transcription of clpGGI is independent of sigma 32 and, at the same time, direct substrate binding uncouples protein disaggregation by ClpGGI from activation by the aggregate-loaded anti-sigma factor DnaK/DnaJ/GrpE. The lack of this regulatory circuit suggests that ClpGGI mainly disassembles protein aggregates beyond saturation of the capacity of the DnaK/DnaJ/GrpE system (as DnaK and ClpGGI compete for a certain type of aggregates (Katikaridiset al.2019)), beyond recognition by DnaK/DnaJ/GrpE or created by severe stress conditions alternative to elevated temperature that do not induce the heat shock regulon.
Furthermore, we have found that P. aeruginosa harbours a clpGGI homologue in the core genome. ClpG shows similar features as ClpGGI such as the high ATPase activity and substrate binding by the extended N-terminal domain suggesting that those two features are hallmarks of the ClpG family within the Hsp100 superfamily (Lee et al. 2016, 2018). Nevertheless, dissection of the function of the extended N-terminal domain of ClpG/ClpGGI showed that the extended part of the N-terminal domain (N2) is required for the disaggregation activity. Despite high homology, upon deletion, distinct functionality of the N2 domain between ClpG and ClpGGI is observed. While the N2 domain of ClpG was required for substrate binding, the N2 domain of ClpGGI dramatically repressed the ATPase activity (Lee et al.2018).
Pseudomonas aeruginosa is the predominant human pathogen in the Pseudomonas genus, and the only species harbouring a monocistronic clpG gene in the core genome. Together with an altered GC content of the ORF, clpG has been acquired by horizontal gene transfer upon speciation of P. aeruginosa. Why does P. aeruginosa redundantly possess both a ClpB and a ClpG disaggregating chaperone? Indeed, while clpB is active in the logarithmic phase of growth, clpG regulated by the oxygen-sensing transcriptional regulator Anr and under direct or indirect control of the PhoP/PhoQ two-component system mainly shows activity in the stationary phase of growth ((Gooderham et al. 2009; Trunk et al. 2010; Babin et al. 2016); (Fig. 9C)). Of note, core genome clpG seems to encode a multifunctional gene product deeply involved in P. aeruginosa physiology as it is also required for dispersion-responsive biofilm formation in P. aeruginosa (Petrova and Sauer 2012), is produced in elevated amounts in human urinary catheter biofilms (Lassek et al. 2015) and has been found to be required for virulence in a rat model of chronic infection in a transposon screen (Potvin et al. 2003). Of note, as clpG is consistently upregulated under low oxygen and anaerobiosis with expression induced by nitrate (Filiatrault et al. 2005; Alvarez-Ortega and Harwood 2007) cumulatively those data suggest that clpG provides an advantage to P. aeruginosa upon oxygen limitation. And why do P. aeruginosa clone C strains redundantly possess even two ClpG-like disaggregating chaperones? Heat shock sensitivity experiments with mutants have shown that clpGGIhas a major role in the logarithmic growth phase where it backs up mainly clpB, while it backs up mainly clpG in the stationary phase of growth (Fig. 10C; (Lee, Kim and Bardwell 2018)). Whether clpGGI also has a backup function during oxygen limiting conditions, needs to be further demonstrated.
How do ClpB and ClpGs differently contribute to bacterial protein homeostasis? The production of ClpB and ClpGs is distinct suggesting that the timing and regulation of expression contributes to the differential role of these diaggregases. ClpB expression is neglectable at 37°C, but induced upon heat shock by RpoH (Kitagawa et al. 1991; Lee et al.2018). On the other hand, both ClpGs are highly and constitutively expressed in the stationary phase of growth from 20°C to 42°C. Furthermore, ClpG confers superior heat tolerance at higher temperature compared to ClpB (Katikaridis et al. 2019). Consistently, in vitro ClpG exhibits robust activity towards diverse protein aggregates formed at elevated temperature whereby the ClpB system shows poor activity. These results suggest that ClpG exerts potent disaggregation to tight aggregates formed during severe thermal stress.
ClpB is widespread in bacteria, but the occurrence of ClpG class proteins is restricted mainly to single strains within a species or to extremely heat-tolerant strains (Mercer et al. 2015). Why is ClpG not as relevant as ClpB despite superior biochemical characteristics? As discussed above, the ClpB/DnaK system works as a very efficient dissaggreage at moderate heat shock condition up to 46°C (Katikaridis et al. 2019) and is tightly integrated into the heat shock regulon with respect to production, functionality and regulation. Thus, it can be argued that sudden elevation of temperature to sublethal and lethal temperatures seem to be rare. However, microorganisms are exposed to elevated temperature stress (57–68°C) during food such as cheese and milk production and sterilization of medical devices. These environmental settings lead to selection of ClpGGI-harboring E. coli strains and 2/3 of Klebsiella pneumoniae strains from clinical environment harbour ClpG (ClpK) (Bojer et al. 2010; Jorgensen et al. 2016). Furthermore, in E. coli, the TLPQC island promotes also tolerance to other type of stresses such as pressure and oxidative stress (Li et al. 2020; Wang et al. 2020).
Within the Pseudomonas genus, the human pathogen P. aeruginosa is the only species with a growth temperature up to 42°C, which implies a potential role of clpG in bacterial virulence, antibiotic resistance and host–microbe interaction. Additionally, revealing physiological substrates of ClpG will give us insight to understand more precisely the role of ClpG in the species P. aeruginosa. Regulatory mechanism to control the constitutive ATPase activity and cooperativity with the functionally associated sHsp20GI still remains to be investigated.
The membrane-bound protease FtsH
The multifunctional inner membrane anchored protease FtsH (Ito and Akiyama 2005) is ubiquituously present in Gram-negative and Gram-positive bacteria, archeae, chloroplasts and mitochondria. FtsH is a multidomain protein composed of an N-terminal periplasmic region flanked by transmembrane helices, a distinct AAA domain with a characteristic second region of homology and a C-terminal M41 proteinase domain. Aided by the transmembrane helices and the AAA domains FtsH assembles into a hexamer which unfolds and translocates substrates into the proteolytic chamber at the C-terminus for hydrolysis into 5–28 amino acid long peptides (Ito and Akiyama 2005). The catalytic site of the C-terminal M41 proteinase contains a conserved HEXXH motif coordinated Zn2+−ion (Ito and Akiyama 2005).
Functionality and regulation of the bacterial FtsH protease has been mainly explored in the model bacterium Escherichia coli K-12. The hexameric FtsH is part of a multi-protein complex and associates with modulator proteins such as HflC and HflK, with prohibitins as eukaryotic homologues, to regulate proteolytic activity towards membrane proteins (Kihara, Akiyama and Ito 1996). Major substrates of FtsH are (i) out-of-context membrane proteins, (ii) truncated and misfolded ssrA tagged proteins and (iii) proteins with partially unfolded loops. As one of the physiologically most relevant substrates FtsH mediates the efficient degradation of the heat shock responsive sigma factor RpoH at non-stress temperature (Fig. 10; (Tomoyasu et al. 1995)). Another key physiological feature of FtsH is regulation of the balance between LPS and phospholipids with the UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase LpxC, a key enzyme involved in biosynthesis of the lipid A anchor of lipopolysaccharide (Ogura et al. 1999), as a major substrate. In addition, FtsH is involved e.g. in the decision between lysis and lysogeny upon bacteriophage infection by degrading CII and CIII and the regulation of basic energy and secretion processes by degradation of the subunit alpha of the F1F0 ATP synthase complex and the type 2 secretion system translocon protein SecY (Ito and Akiyama 2005).
Despite intensive investigations in E. coli, functionality of ftsH in P. aeruginosa is less well characterized. Of note, ftsH has a broad effect on physiology and metabolism as ftsH encoded on the core genome in P. aeruginosa clone C strains is required for the optimal growth of the organism in rich and defined medium and intrinsic resistance as well as tolerance of biofilms against clinically relevant aminoglycosides (Hinz et al. 2011; Kamal et al. 2019). Furthermore, ftsH is involved in a multitude of phenotypes from promotion of motility and biofilm formation to tolerance against hypochlorous acid and the production of secondary metabolites such as pyoverdine, phenazines and the P. aeruginosa quinolone signal (PQS) molecules (Kamal et al. 2019). Upregulated production in the Australien epidemic strain AES-1R compared with PAO and PA14 indicates a role of FtsH in facilitating early infection and transmission (Hare et al. 2012). Recent analysis also showed that FtsH is one of the few gene products that aids the survival of P. aeruginosa under both prolonged carbon and oxygen starvation in interplay with other proteases (Basta, Bergkessel and Newman 2017; Basta et al. 2020) Whether the multitude of phenotypes affected by FtsH requires the disaggregation and proteolytic activity and/or one of the alternative functions of FtsH, such as its reported chaperone or translocation activity (Schumann 1999; Chauleau et al. 2011), has not yet been fully sorted out.
In clone C strains, ftsH2, a xenologue of core genome ftsH1, is encoded on TLPQC-1 downstream of clpGGI. Of note, ftsH2 backs up above described phenotypes in the absence of the ftsH1 core genome copy. Nevertheless, FtsH2 on TLPQC-1 is constitutively produced throughout the growth phase with maximum level in the late stationary growth phase (Kamal et al. 2019) suggesting distinct mechanisms of regulation for ftsH2. The neglectible production of FtsH1 in late stationary phase suggests a unique role of FtsH2 in stationary phase with specific substrates (Kamal et al. 2019). In addition, experimental indications for hetero oligomer suggest broadened substrate specificity or proteolytic activity upon co-expression of FtsH1 and FtsH2 (Kamal et al. 2019). In conclusion, FtsH is involved in various phenotypes, important for bacterial fitness and adaptation in both environmental and clinical niches.
The heat shock sigma factor Sigma 32 is a major substrate of core genome FtsH1 indicating that in P. aeruginosa FtsH1 is a major regulator of the heat shock response. Similar as with the phenotypes, at most, FtsH2 backs up degradation of Sigma 32 in the absence of FtsH1. Pull-down of cross-linked tagged FtsH1 and FtsH2 protein complexes identified a number of interacting proteins several of them consistently substrates of FtsH in E. coli. Phenotypes and degradation studies confirmed PhzC, which channels precurser compounds into the synthesis of the phenazine pyocyanin, to be a substrate of FtsH1 (Kamal et al. 2019).
Description of additional TLPQC-1 core gene products
Besides the core operon dna-shsp20GI-clpGGIand ftsH, additional horizontally transferred genes are encoded on TLPQC-1. The gene products show consistency in physiological function,which indicates involvement in stress response, primarily towards high temperature, and antimicrobial treatment. Many of those genes are xenologues of conserved core genome genes, and consequently possess a distant homologue on the core genome. For example, three proteases, with identified core genome homologues required for high temperature physiology, are encoded on the island; FtsH (described above), DegOGI (a homologue of E. coli DegQ/DegP/DegS (HtrA (high temperature requirement A)); Fig. 11) and HtpX. Besides TLPQC-1 located DegOGI, core genome DegS (AlgW) and DegT (MucD) are present in P. aeruginosa (Fig. 11A and B). The three proteins have a similar domain structure with a N-terminal trypsin_2 peptidase domain and one or two C-terminal PDZ substrate binding domains (Fig. 11C). Both, AlgW and MucD, are involved in the repression of alginate production, the characteristic exopolysaccharide of biofilm-forming P. aeruginosa CF lung isolates (Boucher et al. 1996). Of note, close similarity to the functionally well-characterized protease and chaperone DegP (which includes closely related DegQ and DegS serine peptidase homologues) in E. coli is restricted to the 42.6% identitical DegS/AlgW which seems to be a member of the DegS subfamily. In contrast, DegOGI and DegT belong to distinct subfamilies (Fig. 11A and B). HtpX, another membrane-bound Zn2+ dependent peptidase, shows overlapping functionality with FtsH in E. coli (Yoshitani, Hizukuri and Akiyama 2019).
Phylogenetic analysis showing the position of the three HtrA homologues of P. aeruginosa clone C. (A) Phylogenetic analysis of HtrA/Deg proteases of Pseudomonas aeruginosa SG17M with closest homologues from different taxonomic groups of bacteria for each of the proteins. P. aeruginosa possesses the two HtrA/Deg homologues AlgW (DegS) and MucD (DegT), while P. aeruginosa clone C possesses DegOGI as an additional HtrA/Deg homologue. Proteins DegS, DegP and DegQ of E. coli K-12 were included as reference. DegP of Oryza sativa is included as outgroup. (B) A corresponding 16S rRNA phylogenetic tree was constructed for each of the HtsA/Deg subgroups. Branch lengths correspond to substitutions per site, bootstrap values are indicated in %. The sequence accession numbers are shown in parenthesis. Maximum likelihood method from MEGA 7 program was used for phylogenetic analysis. (C) Domain structure of the three HtrA/Deg homologues of P. aeruginosa clone C. Trypsin_2 is the trypsin-like peptidase domain with conserved residues H/D/S involved in catalysis indicated; PDZ indicates the PDZ_serine_protease domain associated domain involved in substrate binding. Predicted protein binding sites (https://blast-ncbi-nlm-nih-gov.proxy.kib.ki.se/Blast.cgi) are indicated.
Another example of a gene product, present on TLPQC-1, and conserved throughout the phylogenetic tree is the ubiquitous redox protein thioredoxin. The thioredoxin copy on TLPQC-1 is a distant homologue of thioredoxin 2 of E. coli and coexists with the core genome E. coli homologues of thioredoxin 1 and thioredoxin 2. Introduction of xenologues by horizontally transferred elements have been observed previously in P. aeruginosa (Liang et al. 2001). A prominent example of functionality diversification in bacteria is the occurrence and task distribution of four FtsH proteases in the cyanobacterium Synechocystis sp. PCC 6803 (Mann et al. 2000; Zhang et al. 2007). Furthermore, many, including the above mentioned TLPQC-1 encoded gene products are conserved not only in bacteria, but throughout the phylogenetic tree in archaea, mitochondria and chloroplasts suggesting that those distinct functionalities of protein quality control have a global impact on persistence and survival of organisms.
Concluding remarks
The species P. aeruginosa thrives in various habitats facing highly diverse environmental conditions. While early highly discriminatory molecular typing approaches defined the epidemic population structure of P. aeruginosa characterized by frequent recombination with few highly abundant clones thriving in various habitats, whole genome sequencing opened up to trace the evolution of individual isolates within a clonal complex. As highly successful clones are characterized by matching population-wide variability of accessory genetic elements, potentially a combination of core genome features and strain variability contributes to their success. The horizontal acquisition of a common gene cluster encoding protein homeostasis elements is believed to be a factor that contributes to clone C persistence in environmental and clinical niches. As not all successful P. aeruginosa clones harbor this genomic island, the question whether there are common genetic and phenotypic traits that characterize successful clones still remains open. Whether, and, if so, how invididual strains of successful clones coexist in a microniche is another open question.
ACKNOWLEDGEMENTS
Burkhard Tmmler and Ute Römling shoudl be considered as corresponding authors. Work in the authors’ laboratories was supported by the Bundesministerium für Bildung und Forschung (programme ‘Medical Infection Genomics’, 0315827A), the Deutsche Forschungsgemeinschaft (SFB 900, projects A2 and Z1) and the Swedish Research Council (K2012–56X-22034–01-3).
Contributor Information
Changhan Lee, Department of Microbiology, Tumor and Cell Biology, Biomedicum C8, Karolinska Institutet, SE-171 77 Stockholm, Sweden.
Jens Klockgether, Clinic for Paediatric Pneumology, Allergology and Neonatology, Clinical Research Group ‘Pseudomonas Genomics’, Hannover Medical School, D-30625 Hannover, Germany.
Sebastian Fischer, Clinic for Paediatric Pneumology, Allergology and Neonatology, Clinical Research Group ‘Pseudomonas Genomics’, Hannover Medical School, D-30625 Hannover, Germany.
Janja Trcek, Faculty of Natural Sciences and Mathematics, Department of Biology, University of Maribor, Maribor, 2000, Slovenia.
Burkhard Tümmler, Clinic for Paediatric Pneumology, Allergology and Neonatology, Clinical Research Group ‘Pseudomonas Genomics’, Hannover Medical School, D-30625 Hannover, Germany.
Ute Römling, Department of Microbiology, Tumor and Cell Biology, Biomedicum C8, Karolinska Institutet, SE-171 77 Stockholm, Sweden.
REFERENCES
- Adesemoye AO, Obini M, Ugoji EO. Comparison of plant growth-promotion with Pseudomonas aeruginosa and Bacillus subtilis in three vegetables. Braz J Microbiol. 2008;39:423–6. [Europe PMC free article] [Abstract] [Google Scholar]
- Ahlen C, Mandal LH, Iversen OJ. The impact of environmental Pseudomonas aeruginosa genotpes on skin infections in occupational saturation diving systems. Scand J Infect Dis. 2001;33:413–9. [Abstract] [Google Scholar]
- Alvarez-Ortega C, Harwood CS. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol Microbiol. 2007;65:153–65. [Europe PMC free article] [Abstract] [Google Scholar]
- Arora SK, Wolfgang MC, Lory S et al. Sequence polymorphism in the glycosylation island and flagellins of Pseudomonas aeruginosa. J Bacteriol. 2004;186:2115–22. [Europe PMC free article] [Abstract] [Google Scholar]
- Babin BM, Bergkessel M, Sweredoski MJ et al. SutA is a bacterial transcription factor expressed during slow growth in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2016;113:E597–605. [Europe PMC free article] [Abstract] [Google Scholar]
- Balchin D, Hayer-Hartl M, Hartl FU. In vivo aspects of protein folding and quality control. Science. 2016;353:aac4354. [Abstract] [Google Scholar]
- Basta DW, Angeles-Albores D, Spero MA et al. Heat-shock proteases promote survival of Pseudomonas aeruginosa during growth arrest. Proc Natl Acad Sci USA. 2020;117:4358–67. [Europe PMC free article] [Abstract] [Google Scholar]
- Basta DW, Bergkessel M, Newman DK. Identification of fitness determinants during energy-limited growth arrest in Pseudomonas aeruginosa. mBio. 2017;8 e01170–17. [Europe PMC free article] [Abstract] [Google Scholar]
- Bjerklund Johansen TE, Cek M et al. Prevalence of hospital-acquired urinary tract infections in urology departments. Eur Urol. 2007;51:1100–11.; discussion 1112. [Abstract] [Google Scholar]
- Bojer MS, Struve C, Ingmer H et al. Heat resistance mediated by a new plasmid encoded Clp ATPase, ClpK, as a possible novel mechanism for nosocomial persistence of Klebsiella pneumoniae. PLoS One. 2010;5:e15467. [Europe PMC free article] [Abstract] [Google Scholar]
- Boll EJ, Marti R, Hasman H et al. Turn up the heat-food and clinical Escherichia coli isolates feature two transferrable loci of heat resistance. Front Microbiol. 2017;8:579. [Europe PMC free article] [Abstract] [Google Scholar]
- Bosshammer J, Fiedler B, Gudowius P et al. Comparative hygienic surveillance of contamination with pseudomonads in a cystic fibrosis ward over a 4-year period. J Hosp Infect. 1995;31:261–74. [Abstract] [Google Scholar]
- Boucher JC, Martinez-Salazar J, Schurr MJ et al. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J Bacteriol. 1996;178:511–23. [Europe PMC free article] [Abstract] [Google Scholar]
- Bouza E, San Juan R, Munoz P et al. Co-operative Group of the European Study Group on Nosocomial Infections. A European perspective on nosocomial urinary tract infections II. Report on incidence, clinical characteristics and outcome (ESGNI-004 study). European Study Group on Nosocomial Infection. Clin Microbiol Infect. 2001;7:532–42. [Abstract] [Google Scholar]
- Broder UN, Jaeger T, Jenal U. LadS is a calcium-responsive kinase that induces acute-to-chronic virulence switch in Pseudomonas aeruginosa. Nat Microbiol. 2016;2:16184. [Abstract] [Google Scholar]
- Brouwer S, Pustelny C, Ritter C et al. The PqsR and RhlR transcriptional regulators determine the level of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa by producing two different pqsABCDE mRNA isoforms. J Bacteriol. 2014;196:4163–71. [Europe PMC free article] [Abstract] [Google Scholar]
- Brumlik MJ, Storey DG. Post-transcriptional control of Pseudomonas aeruginosa lasB expression involves the 5' untranslated region of the mRNA. FEMS Microbiol Lett. 1998;159:233–9. [Abstract] [Google Scholar]
- Burrus V, Pavlovic G, Decaris B et al. Conjugative transposons: the tip of the iceberg. Mol Microbiol. 2002;46:601–10. [Abstract] [Google Scholar]
- Carroni M, Franke KB, Maurer M et al. Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control. Elife. 2017;6:e30120. [Europe PMC free article] [Abstract] [Google Scholar]
- Chastre J, Fagon JY. Ventilator-associated pneumonia. Am J Respir Crit Care Med. 2002;165:867–903. [Abstract] [Google Scholar]
- Chauleau M, Mora L, Serba J et al. FtsH-dependent processing of RNase colicins D and E3 means that only the cytotoxic domains are imported into the cytoplasm. J Biol Chem. 2011;286:29397–407. [Europe PMC free article] [Abstract] [Google Scholar]
- Cooke EM, Shooter RA, O'Farrell SM et al. Faecal carriage of Pseudomonas aeruginosa by newborn babies. Lancet. 1970;2:1045–6. [Abstract] [Google Scholar]
- Cramer N, Klockgether J, Wrasman K et al. Microevolution of the major common Pseudomonas aeruginosa clones C and PA14 in cystic fibrosis lungs. Environ Microbiol. 2011;13:1690–704. [Abstract] [Google Scholar]
- Cramer N, Wiehlmann L, Ciofu O et al. Molecular epidemiology of chronic Pseudomonas aeruginosa airway infections in cystic fibrosis. PLoS One. 2012;7:e50731. [Europe PMC free article] [Abstract] [Google Scholar]
- Curran B, Jonas D, Grundmann H et al. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol. 2004;42:5644–9. [Europe PMC free article] [Abstract] [Google Scholar]
- D'Arrigo I, Bojanovic K, Yang X et al. Genome-wide mapping of transcription start sites yields novel insights into the primary transcriptome of Pseudomonas putida. Environ Microbiol. 2016;18:3466–81. [Abstract] [Google Scholar]
- Das K, Mukherjee AK. Crude petroleum-oil biodegradation efficiency of Bacillus subtilis and Pseudomonas aeruginosa strains isolated from a petroleum-oil contaminated soil from North-East India. Bioresour Technol. 2007;98:1339–45. [Abstract] [Google Scholar]
- De Soyza A, Hall AJ, Mahenthiralingam E et al. Developing an international Pseudomonas aeruginosa reference panel. Microbiologyopen. 2013;2:1010–23. [Europe PMC free article] [Abstract] [Google Scholar]
- Dettman JR, Rodrigue N, Kassen R. Genome-wide patterns of recombination in the opportunistic human pathogen Pseudomonas aeruginosa. Genome Biol Evol. 2014;7:18–34. [Europe PMC free article] [Abstract] [Google Scholar]
- Dinesh SD, Grundmann H, Pitt TL et al. European-wide distribution of Pseudomonas aeruginosa clone C. Clin Microbiol Infect. 2003;9:1228–33. [Abstract] [Google Scholar]
- Dossel J, Meyer-Hoffert U, Schröder JM et al. Pseudomonas aeruginosa-derived rhamnolipids subvert the host innate immune response through manipulation of the human beta-defensin-2 expression. Cell Microbiol. 2012;14:1364–75. [Abstract] [Google Scholar]
- Dötsch A, Eckweiler D, Schniederjans M et al. The Pseudomonas aeruginosa transcriptome in planktonic cultures and static biofilms using RNA sequencing. PLoS One. 2012;7:e31092. [Europe PMC free article] [Abstract] [Google Scholar]
- Fenibo EO, Ijoma GN, Selvarajan R et al. Microbial Surfactants: The next generation multifunctional biomolecules for applications in the petroleum industry and its associated environmental remediation. Microorganisms. 2019;7:581. [Europe PMC free article] [Abstract] [Google Scholar]
- Filiatrault MJ, Wagner VE, Bushnell D et al. Effect of anaerobiosis and nitrate on gene expression in Pseudomonas aeruginosa. Infect Immun. 2005;73:3764–72. [Europe PMC free article] [Abstract] [Google Scholar]
- Fischer S, Klockgether J, Moran Losada P et al. Intraclonal genome diversity of the major Pseudomonas aeruginosa clones C and PA14. Environ Microbiol Rep. 2016;8:227–34. [Europe PMC free article] [Abstract] [Google Scholar]
- Folkesson A, Jelsbak L, Yang L et al. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nature Rev Microbiol. 2012;10:841–51. [Abstract] [Google Scholar]
- Fothergill JL, Walshaw MJ, Winstanley C. Transmissible strains of Pseudomonas aeruginosa in cystic fibrosis lung infections. Eur Respir J. 2012;40:227–38. [Abstract] [Google Scholar]
- Freschi L, Vincent AT, Jeukens J et al. The Pseudomonas aeruginosa pan-genome provides new insights on its population structure, horizontal gene transfer, and pathogenicity. Genome Biol Evol. 2019;11:109–20. [Europe PMC free article] [Abstract] [Google Scholar]
- Fukushima J, Ishiwata T, You Z et al. Dissection of the promoter/operator region and evaluation of N-acylhomoserine lactone mediated transcriptional regulation of elastase expression in Pseudomonas aeruginosa. FEMS Microbiol Lett. 1997;146:311–8. [Abstract] [Google Scholar]
- Gaillard M, Pernet N, Vogne C et al. Host and invader impact of transfer of the clc genomic island into Pseudomonas aeruginosa PAO1. Proc Natl Acad Sci USA. 2008;105:7058–63. [Europe PMC free article] [Abstract] [Google Scholar]
- Gao X, Carroni M, Nussbaum-Krammer C et al. Human Hsp70 disaggregase reverses Parkinson's-linked alpha-Synuclein amyloid fibrils. Mol Cell. 2015;59:781–93. [Europe PMC free article] [Abstract] [Google Scholar]
- Garcia M, Morello E, Garnier J et al. Pseudomonas aeruginosa flagellum is critical for invasion, cutaneous persistence and induction of inflammatory response of skin epidermis. Virulence. 2018;9:1163–75. [Europe PMC free article] [Abstract] [Google Scholar]
- Gooderham WJ, Gellatly SL, Sanschagrin F et al. The sensor kinase PhoQ mediates virulence in Pseudomonas aeruginosa. Microbiology. 2009;155:699–711. [Abstract] [Google Scholar]
- Gordienko EN, Kazanov MD, Gelfand MS. Evolution of pan-genomes of Escherichiacoli, Shigella spp., and Salmonella enterica. J Bacteriol. 2013;195:2786–92. [Europe PMC free article] [Abstract] [Google Scholar]
- Gould IM, Wise R. Pseudomonas aeruginosa: clinical manifestations and management. Lancet. 1985;2:1224–7. [Abstract] [Google Scholar]
- Grobe S, Wingender J, Truper HG. Characterization of mucoid Pseudomonas aeruginosa strains isolated from technical water systems. J Appl Bacteriol. 1995;79:94–102. [Abstract] [Google Scholar]
- Grosso-Becerra MV, Croda-Garcia G, Merino E et al. Regulation of Pseudomonas aeruginosa virulence factors by two novel RNA thermometers. Proc Natl Acad Sci USA. 2014;111:15562–7. [Europe PMC free article] [Abstract] [Google Scholar]
- Hare NJ, Solis N, Harmer C et al. Proteomic profiling of Pseudomonas aeruginosa AES-1R, PAO1 and PA14 reveals potential virulence determinants associated with a transmissible cystic fibrosis-associated strain. BMC Microbiol. 2012;12:16. [Europe PMC free article] [Abstract] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–32. [Abstract] [Google Scholar]
- Hatipoglu M, Mutluoglu M, Uzun G et al. The microbiologic profile of diabetic foot infections in Turkey: a 20-year systematic review: diabetic foot infections in Turkey. Eur J Clin Microbiol Infect Dis. 2014;33:871–8. [Abstract] [Google Scholar]
- He J, Baldini RL, Deziel E et al. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc Natl Acad Sci USA. 2004;101:2530–5. [Europe PMC free article] [Abstract] [Google Scholar]
- Hilker R, Munder A, Klockgether J et al. Interclonal gradient of virulence in the Pseudomonas aeruginosa pangenome from disease and environment. Environ Microbiol. 2015;17:29–46. [Abstract] [Google Scholar]
- Hilliam Y, Moore MP, Lamont IL et al. Pseudomonas aeruginosa adaptation and diversification in the non-cystic fibrosis bronchiectasis lung. Eur Respir J. 2017;49:1602108. [Europe PMC free article] [Abstract] [Google Scholar]
- Hinz A, Lee S, Jacoby K et al. Membrane proteases and aminoglycoside antibiotic resistance. J Bacteriol. 2011;193:4790–7. [Europe PMC free article] [Abstract] [Google Scholar]
- Huangyutitham V, Guvener ZT, Harwood CS. Subcellular clustering of the phosphorylated WspR response regulator protein stimulates its diguanylate cyclase activity. mBio. 2013;4:e00242–00213. [Europe PMC free article] [Abstract] [Google Scholar]
- Ito K, Akiyama Y. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu Rev Microbiol. 2005;59:211–31. [Abstract] [Google Scholar]
- Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. [Europe PMC free article] [Abstract] [Google Scholar]
- Jorgensen SB, Bojer MS, Boll EJ et al. Heat-resistant, extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in endoscope-mediated outbreak. J Hosp Infect. 2016;93:57–62. [Abstract] [Google Scholar]
- Juhas M, van der Meer JR, Gaillard M et al. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev. 2009;33:376–93. [Europe PMC free article] [Abstract] [Google Scholar]
- Kamal SM, Rybtke ML, Nimtz M et al. Two FtsH proteases contribute to fitness and adaptation of Pseudomonas aeruginosa clone C strains. Front Microbiol. 2019;10:1372. [Europe PMC free article] [Abstract] [Google Scholar]
- Kamarainen J, Huokko T, Kreula S et al. Pyridine nucleotide transhydrogenase PntAB is essential for optimal growth and photosynthetic integrity under low-light mixotrophic conditions in Synechocystis sp. PCC 6803. New Phytol. 2017;214:194–204. [Abstract] [Google Scholar]
- Katikaridis P, Meins L, Kamal SM et al. ClpG provides increased heat resistance by acting as superior sisaggregase. Biomolecules. 2019;9:815. [Europe PMC free article] [Abstract] [Google Scholar]
- Kerckhoffs AP, Ben-Amor K, Samsom M et al. Molecular analysis of faecal and duodenal samples reveals significantly higher prevalence and numbers of Pseudomonas aeruginosa in irritable bowel syndrome. J Med Microbiol. 2011;60:236–45. [Abstract] [Google Scholar]
- Khan NH, Ishii Y, Kimata-Kino N et al. Isolation of Pseudomonas aeruginosa from open ocean and comparison with freshwater, clinical, and animal isolates. Microb Ecol. 2007;53:173–86. [Abstract] [Google Scholar]
- Kidd TJ, Grimwood K, Ramsay KA et al. Comparison of three molecular techniques for typing Pseudomonas aeruginosa isolates in sputum samples from patients with cystic fibrosis. J Clin Microbiol. 2011;49:263–8. [Europe PMC free article] [Abstract] [Google Scholar]
- Kidd TJ, Ritchie SR, Ramsay KA et al. Pseudomonas aeruginosa exhibits frequent recombination, but only a limited association between genotype and ecological setting. PLoS One. 2012;7:e44199. [Europe PMC free article] [Abstract] [Google Scholar]
- Kiewitz C, Larbig K, Klockgether J et al. Monitoring genome evolution ex vivo: reversible chromosomal integration of a 106 kb plasmid at two tRNA(Lys) gene loci in sequential Pseudomonas aeruginosa airway isolates. Microbiology. 2000;146:((Pt 10):):2365–73. [Abstract] [Google Scholar]
- Kiewitz C, Tümmler B. Sequence diversity of Pseudomonas aeruginosa: impact on population structure and genome evolution. J Bacteriol. 2000;182:3125–35. [Europe PMC free article] [Abstract] [Google Scholar]
- Kihara A, Akiyama Y, Ito K. A protease complex in the Escherichia coli plasma membrane: HflKC (HflA) forms a complex with FtsH (HflB), regulating its proteolytic activity against SecY. EMBO J. 1996;15:6122–31. [Europe PMC free article] [Abstract] [Google Scholar]
- Kim YE, Hipp MS, Bracher A et al. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–55. [Abstract] [Google Scholar]
- Kitagawa M, Wada C, Yoshioka S et al. Expression of ClpB, an analog of the ATP-dependent protease regulatory subunit in Escherichia coli, is controlled by a heat shock sigma factor (sigma 32). J Bacteriol. 1991;173:4247–53. [Europe PMC free article] [Abstract] [Google Scholar]
- Klockgether J, Cramer N, Fischer S et al. Long-term microevolution of Pseudomonas aeruginosa differs between mildly and severely affected Cystic Fibrosis lungs. Am J Respir Cell Mol Biol. 2018;59:246–56. [Abstract] [Google Scholar]
- Klockgether J, Cramer N, Wiehlmann L et al. Pseudomonas aeruginosa genomic structure and diversity. Front Microbiol. 2011;2:150. [Europe PMC free article] [Abstract] [Google Scholar]
- Klockgether J, Reva O, Larbig K et al. Sequence analysis of the mobile genome island pKLC102 of Pseudomonas aeruginosa C. J Bacteriol. 2004;186:518–34. [Europe PMC free article] [Abstract] [Google Scholar]
- Klockgether J, Wurdemann D, Reva O et al. Diversity of the abundant pKLC102/PAGI-2 family of genomic islands in Pseudomonas aeruginosa. J Bacteriol. 2007;189:2443–59. [Europe PMC free article] [Abstract] [Google Scholar]
- Klockgether J, Wurdemann D, Wiehlmann L et al. Transcript profiling of the Pseudomonas aeruginosa genomic islands PAGI-2 and pKLC102. Microbiology. 2008;154:1599–604. [Abstract] [Google Scholar]
- Koga H, Kaushik S, Cuervo AM. Protein homeostasis and aging: The importance of exquisite quality control. Ageing Res Rev. 2011;10:205–15. [Europe PMC free article] [Abstract] [Google Scholar]
- Koldewey P, Stull F, Horowitz S et al. Forces driving chaperone action. Cell. 2016;166:369–79. [Europe PMC free article] [Abstract] [Google Scholar]
- Kortmann J, Narberhaus F. Bacterial RNA thermometers: molecular zippers and switches. Nat Rev Microbiol. 2012;10:255–65. [Abstract] [Google Scholar]
- Krajewski SS, Nagel M, Narberhaus F. Short ROSE-like RNA thermometers control IbpA synthesis in Pseudomonas species. PLoS One. 2013;8:e65168. [Europe PMC free article] [Abstract] [Google Scholar]
- Kresse AU, Blöcker H, Römling U. ISPa20 advances the individual evolution of Pseudomonas aeruginosa clone C subclone C13 strains isolated from cystic fibrosis patients by insertional mutagenesis and genomic rearrangements. Arch Microbiol. 2006;185:245–54. [Abstract] [Google Scholar]
- Kresse AU, Dinesh SD, Larbig K et al. Impact of large chromosomal inversions on the adaptation and evolution of Pseudomonas aeruginosa chronically colonizing cystic fibrosis lungs. Mol Microbiol. 2003;47:145–58. [Abstract] [Google Scholar]
- Kulasekara BR, Kulasekara HD, Wolfgang MC et al. Acquisition and evolution of the exoU locus in Pseudomonas aeruginosa. J Bacteriol. 2006;188:4037–50. [Europe PMC free article] [Abstract] [Google Scholar]
- Kung VL, Ozer EA, Hauser AR. The accessory genome of Pseudomonas aeruginosa. Microbiol Mol Biol Rev. 2010;74:621–41. [Europe PMC free article] [Abstract] [Google Scholar]
- Kus JV, Tullis E, Cvitkovitch DG et al. Significant differences in type IV pilin allele distribution among Pseudomonas aeruginosa isolates from cystic fibrosis (CF) versus non-CF patients. Microbiology. 2004;150:1315–26. [Abstract] [Google Scholar]
- Lanini S, D'Arezzo S, Puro V et al. Molecular epidemiology of a Pseudomonas aeruginosa hospital outbreak driven by a contaminated disinfectant-soap dispenser. PLoS One. 2011;6:e17064. [Europe PMC free article] [Abstract] [Google Scholar]
- Larbig K, Kiewitz C, Tümmler B. Pathogenicity islands and PAI-like structures in Pseudomonas species. Curr Top Microbiol Immunol. 2002a;264:201–11. [Abstract] [Google Scholar]
- Larbig KD, Christmann A, Johann A et al. Gene islands integrated into tRNA(Gly) genes confer genome diversity on a Pseudomonas aeruginosa clone. J Bacteriol. 2002b;184:6665–80. [Europe PMC free article] [Abstract] [Google Scholar]
- Lassek C, Burghartz M, Chaves-Moreno D et al. A metaproteomics approach to elucidate host and pathogen protein expression during catheter-associated urinary tract infections (CAUTIs). Mol Cell Proteomics. 2015;14:989–1008. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee C, Franke KB, Kamal SM et al. Stand-alone ClpG disaggregase confers superior heat tolerance to bacteria. Proc Natl Acad Sci USA. 2018;115:E273–82. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee C, Kamal SM, Römling U. High frequency of double crossover recombination facilitates genome engineering in Pseudomonas aeruginosa PA14 and clone C strains. Microbiology. 2019;165:757–60. [Abstract] [Google Scholar]
- Lee C, Kim H, Bardwell JCA. Electrostatic interactions are important for chaperone-client interaction in vivo. Microbiology. 2018;164:992–7. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee C, Peters V, Ö M et al. Draft genome sequence of Pseudomonas aeruginosa SG17M, an environmental isolate belonging to Clone C, prevalent in patients and aquatic habitats. Genome Announc. 2014;2:e00186–14. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee C, Wigren E, Lünsdorf H et al. Protein homeostasis-more than resisting a hot bath. Curr Opin Microbiol. 2016;30:147–54. [Abstract] [Google Scholar]
- Lee C, Wigren E, Trcek J et al. A novel protein quality control mechanism contributes to heat shock resistance of worldwide-distributed Pseudomonas aeruginosa clone C strains. Environ Microbiol. 2015;17:4511–26. [Abstract] [Google Scholar]
- Lee DG, Urbach JM, Wu G et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006;7:R90. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee PC, Stopford CM, Svenson AG et al. Control of effector export by the Pseudomonas aeruginosa type III secretion proteins PcrG and PcrV. Mol Microbiol. 2010;75:924–41. [Europe PMC free article] [Abstract] [Google Scholar]
- Liang X, Pham XQ, Olson MV et al. Identification of a genomic island present in the majority of pathogenic isolates of Pseudomonas aeruginosa. J Bacteriol. 2001;183:843–53. [Europe PMC free article] [Abstract] [Google Scholar]
- Li H, Mercer R, Behr J et al. Heat and pressure resistance in Escherichia coli relates to protein folding and aggregation. Front Microbiol. 2020;11:111. [Europe PMC free article] [Abstract] [Google Scholar]
- Linares JF, Moreno R, Fajardo A et al. The global regulator Crc modulates metabolism, susceptibility to antibiotics and virulence in Pseudomonas aeruginosa. Environ Microbiol. 2010;12:3196–212. [Abstract] [Google Scholar]
- Liu S, Skory C, Liang X et al. Increased ethanol tolerance associated with the pntAB locus of Oenococcus oeni and Lactobacillus buchneri. J Ind Microbiol Biotechnol. 2019;46:1547–56. [Abstract] [Google Scholar]
- Li Y, Du X, Lu ZJ et al. Regulatory feedback loop of two phz gene clusters through 5'-untranslated regions in Pseudomonas sp. M18. PLoS One. 2011;6:e19413. [Europe PMC free article] [Abstract] [Google Scholar]
- Long CP, Gonzalez JE, Feist AM et al. Dissecting the genetic and metabolic mechanisms of adaptation to the knockout of a major metabolic enzyme in Escherichia coli. Proc Natl Acad Sci USA. 2018;115:222–7. [Europe PMC free article] [Abstract] [Google Scholar]
- Lukjancenko O, Wassenaar TM, Ussery DW. Comparison of 61 sequenced Escherichia coli genomes. Microb Ecol. 2010;60:708–20. [Europe PMC free article] [Abstract] [Google Scholar]
- Mann NH, Novac N, Mullineaux CW et al. Involvement of an FtsH homologue in the assembly of functional photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 2000;479:72–77. [Abstract] [Google Scholar]
- Ma Q, Zhai Y, Schneider JC et al. Protein secretion systems of Pseudomonas aeruginosa and P fluorescens. Biochim Biophys Acta. 2003;1611:223–33. [Abstract] [Google Scholar]
- Marr AK, Overhage J, Bains M et al. The Lon protease of Pseudomonas aeruginosa is induced by aminoglycosides and is involved in biofilm formation and motility. Microbiology. 2007;153:474–82. [Abstract] [Google Scholar]
- Marvig RL, Dolce D, Sommer LM et al. Within-host microevolution of Pseudomonas aeruginosa in Italian cystic fibrosis patients. BMC Microbiol. 2015;15:218. [Europe PMC free article] [Abstract] [Google Scholar]
- Mathee K, Narasimhan G, Valdes C et al. Dynamics of Pseudomonas aeruginosa genome evolution. Proc Natl Acad Sci USA. 2008;105:3100–5. [Europe PMC free article] [Abstract] [Google Scholar]
- McMorran LM, Brockwell DJ, Radford SE. Mechanistic studies of the biogenesis and folding of outer membrane proteins in vitro and in vivo: what have we learned to date? Arch Biochem Biophys. 2014;564:265–80. [Europe PMC free article] [Abstract] [Google Scholar]
- Mena A, Smith EE, Burns JL et al. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J Bacteriol. 2008;190:7910–7. [Europe PMC free article] [Abstract] [Google Scholar]
- Mercer RG, Zheng J, Garcia-Hernandez R et al. Genetic determinants of heat resistance in Escherichia coli. Front Microbiol. 2015;6:932. [Europe PMC free article] [Abstract] [Google Scholar]
- Mergeay M, Monchy S, Vallaeys T et al. Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: towards a catalogue of metal-responsive genes. FEMS Microbiol Rev. 2003;27:385–410. [Abstract] [Google Scholar]
- Middleton MA, Layeghifard M, Klingel M et al. Epidemiology of clonal Pseudomonas aeruginosa infection in a Canadian Cystic Fibrosis population. Ann Am Thorac Soc. 2018;15:827–36. [Abstract] [Google Scholar]
- Migula W. Über ein neues System der Bakterien. Arb Bakteriol Inst Karlsruhe. 1894;1:235–328. [Google Scholar]
- Miller JM, Chaudhary H, Marsee JD. Phylogenetic analysis predicts structural divergence for proteobacterial ClpC proteins. J Struct Biol. 2018;201:52–62. [Abstract] [Google Scholar]
- Mogk A, Bukau B, Kampinga HH. Cellular handling of protein aggregates by disaggregation machines. Mol Cell. 2018;69:214–26. [Abstract] [Google Scholar]
- Mogk A, Deuerling E, Vorderwulbecke S et al. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol Microbiol. 2003;50:585–95. [Abstract] [Google Scholar]
- Mogk A, Kummer E, Bukau B. Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front Mol Biosci. 2015;2:22. [Europe PMC free article] [Abstract] [Google Scholar]
- Mogk A, Ruger-Herreros C, Bukau B. Cellular functions and mechanisms of action of small heat shock proteins. Annu Rev Microbiol. 2019;73:89–110. [Abstract] [Google Scholar]
- Moradali MF, Ghods S, Rehm BH. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol. 2017;7:39. [Europe PMC free article] [Abstract] [Google Scholar]
- Munder A, Tümmler B. Assessing Pseudomonas virulence using mammalian models: acute infection model. Methods Mol Biol. 2014;1149:773–91. [Abstract] [Google Scholar]
- Neckers L, Tatu U. Molecular chaperones in pathogen virulence: emerging new targets for therapy. Cell Host Microbe. 2008;4:519–27. [Europe PMC free article] [Abstract] [Google Scholar]
- Nguyen SV, Harhay GP, Bono JL et al. Genome sequence of the thermotolerant foodborne pathogen Salmonella enterica Serovar Senftenberg ATCC 43845 and phylogenetic analysis of loci encoding increased protein quality control mechanisms. mSystems. 2017;2:e00190–16. [Europe PMC free article] [Abstract] [Google Scholar]
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. [Europe PMC free article] [Abstract] [Google Scholar]
- Ogura T, Inoue K, Tatsuta T et al. Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol Microbiol. 1999;31:833–44. [Abstract] [Google Scholar]
- Ojima M, Toshima Y, Koya E et al. Hygiene measures considering actual distributions of microorganisms in Japanese households. J Appl Microbiol. 2002;93:800–9. [Abstract] [Google Scholar]
- Oliver A, Canton R, Campo P et al. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 2000;288:1251–4. [Abstract] [Google Scholar]
- Parkins MD, Somayaji R, Waters VJ. Epidemiology, biology, and impact of clonal Pseudomonas aeruginosa infections in cystic fibrosis. Clin Microbiol Rev. 2018;31;e00019–18. [Europe PMC free article] [Abstract] [Google Scholar]
- Petrova OE, Sauer K. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc Natl Acad Sci USA. 2012;109:16690–5. [Europe PMC free article] [Abstract] [Google Scholar]
- Pirnay JP, Bilocq F, Pot B et al. Pseudomonas aeruginosa population structure revisited. PLoS One. 2009;4:e7740. [Europe PMC free article] [Abstract] [Google Scholar]
- Pirnay JP, Matthijs S, Colak H et al. Global Pseudomonas aeruginosa biodiversity as reflected in a Belgian river. Environ Microbiol. 2005;7:969–80. [Abstract] [Google Scholar]
- Poigbo P, Wolf YI, Koonin EV. Genome-wide comparative analysis of trees: the prokaryotic forest of life. Evolutionary Genomics: Statistical and Computational Methods, Vol. 2, (Anisimova M, editor. ed). Springer Science+Business Media, Humana Press, New York: 2012, pp. 53–79. [Google Scholar]
- Potvin E, Lehoux DE, Kukavica-Ibrulj I et al. In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ Microbiol. 2003;5:1294–308. [Abstract] [Google Scholar]
- Pustelny C, Brouwer S, Musken M et al. The peptide chain release factor methyltransferase PrmC is essential for pathogenicity and environmental adaptation of Pseudomonas aeruginosa PA14. Environ Microbiol. 2013;15:597–609. [Abstract] [Google Scholar]
- Pu Y, Li Y, Jin X et al. ATP-dependent dynamic protein aggregation regulates bacterial dormancy depth critical for antibiotic tolerance. Mol Cell. 2019;73:143–156 e144. [Abstract] [Google Scholar]
- Rahme LG, Stevens EJ, Wolfort SF et al. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–902. [Abstract] [Google Scholar]
- Ratnam S, Hogan K, March SB et al. Whirlpool-associated folliculitis caused by Pseudomonas aeruginosa: report of an outbreak and review. J Clin Microbiol. 1986;23:655–9. [Europe PMC free article] [Abstract] [Google Scholar]
- Ravatn R, Zehnder AJ, van der Meer JR. Low-frequency horizontal transfer of an element containing the chlorocatechol degradation genes from Pseudomonas sp. strain B13 to Pseudomonas putida F1 and to indigenous bacteria in laboratory-scale activated-sludge microcosms. Appl Environ Microbiol. 1998;64:2126–32. [Europe PMC free article] [Abstract] [Google Scholar]
- Raymond CK, Sims EH, Kas A et al. Genetic variation at the O-antigen biosynthetic locus in Pseudomonas aeruginosa. J Bacteriol. 2002;184:3614–22. [Europe PMC free article] [Abstract] [Google Scholar]
- Reetz MT, Jaeger KE. Overexpression, immobilization and biotechnological application of Pseudomonas lipases. Chem Phys Lipids. 1998;93:3–14. [Abstract] [Google Scholar]
- Rello J, Allegri C, Rodriguez A et al. Risk factors for ventilator-associated pneumonia by Pseudomonas aeruginosa in presence of recent antibiotic exposure. Anesthesiology. 2006;105:709–14. [Abstract] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nature Med. 2004;10:Suppl: S10–17. [Abstract] [Google Scholar]
- Rutherford V, Yom K, Ozer EA et al. Environmental reservoirs for exoS+ and exoU+ strains of Pseudomonas aeruginosa. Environ Microbiol Rep. 2018;10:485–92. [Europe PMC free article] [Abstract] [Google Scholar]
- Römling U, Fiedler B, Bosshammer J et al. Epidemiology of chronic Pseudomonas aeruginosa infections in cystic fibrosis. J Infect Dis. 1994. b;170:1616–21. [Abstract] [Google Scholar]
- Römling U, Kader A, Sriramulu DD et al. Worldwide distribution of Pseudomonas aeruginosa clone C strains in the aquatic environment and cystic fibrosis patients. Environ Microbiol. 2005;7:1029–38. [Abstract] [Google Scholar]
- Römling U, Schmidt KD, Tümmler B. Large chromosomal inversions occur in Pseudomonas aeruginosa clone C strains isolated from cystic fibrosis patients. FEMS Microbiol Lett. 1997a;150:149–56. [Abstract] [Google Scholar]
- Römling U, Schmidt KD, Tümmler B. Large genome rearrangements discovered by the detailed analysis of 21 Pseudomonas aeruginosa clone C isolates found in environment and disease habitats. J Mol Biol. 1997b;271:386–404. [Abstract] [Google Scholar]
- Römling U, Wingender J, Müller H et al. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl Environ Microbiol. 1994a;60:1734–8. [Europe PMC free article] [Abstract] [Google Scholar]
- Schirm M, Arora SK, Verma A et al. Structural and genetic characterization of glycosylation of type a flagellin in Pseudomonas aeruginosa. J Bacteriol. 2004;186:2523–31. [Europe PMC free article] [Abstract] [Google Scholar]
- Schmidt KD, Tümmler B, Römling U. Comparative genome mapping of Pseudomonas aeruginosa PAO with P. aeruginosa C, which belongs to a major clone in cystic fibrosis patients and aquatic habitats. J Bacteriol. 1996;178:85–93. [Europe PMC free article] [Abstract] [Google Scholar]
- Schramm FD, Schroeder K, Jonas K. Protein aggregation in bacteria. FEMS Microbiol Rev. 2019;44:54-72. [Europe PMC free article] [Abstract] [Google Scholar]
- Schroth MN, Cho JJ, Green SK et al. Epidemiology of Pseudomonas aeruginosa in agricultural areas. J Med Microbiol. 2018;67:1191–201. [Abstract] [Google Scholar]
- Schumann W. FtsH–a single-chain charonin? FEMS Microbiol Rev. 1999;23:1–11. [Abstract] [Google Scholar]
- Schwartz DC, Cantor CR. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984;37:67–75. [Abstract] [Google Scholar]
- Scott FW, Pitt TL. Identification and characterization of transmissible Pseudomonas aeruginosa strains in cystic fibrosis patients in England and Wales. J Med Microbiol. 2004;53:609–15. [Abstract] [Google Scholar]
- Sentchilo V, Zehnder AJ, van der Meer JR. Characterization of two alternative promoters for integrase expression in the clc genomic island of Pseudomonas sp. strain B13. Mol Microbiol. 2003;49:93–104. [Abstract] [Google Scholar]
- Sheng S, Xin L, Yam JKH et al. The MapZ-mediated methylation of chemoreceptors contributes to pathogenicity of Pseudomonas aeruginosa. Front Microbiol. 2019;10:67. [Europe PMC free article] [Abstract] [Google Scholar]
- Silvestre JF, Betlloch MI. Cutaneous manifestations due to Pseudomonas infection. Int J Dermatol. 1999;38:419–31. [Abstract] [Google Scholar]
- Smith EE, Sims EH, Spencer DH et al. Evidence for diversifying selection at the pyoverdine locus of Pseudomonas aeruginosa. J Bacteriol. 2005;187:2138–47. [Europe PMC free article] [Abstract] [Google Scholar]
- Spangenberg C, Fislage R, Sierralta W et al. Comparison of type IV-pilin genes of Pseudomonas aeruginosa of various habitats has uncovered a novel unusual sequence. FEMS Microbiol Lett. 1995;125:265–73. [Abstract] [Google Scholar]
- Sriramulu DD, Nimtz M, Römling U. Proteome analysis reveals adaptation of Pseudomonas aeruginosa to the cystic fibrosis lung environment. Proteomics. 2005;5:3712–21. [Abstract] [Google Scholar]
- Starkey M, Rahme LG. Modeling Pseudomonas aeruginosa pathogenesis in plant hosts. Nature Prot. 2009;4:117–24. [Europe PMC free article] [Abstract] [Google Scholar]
- Stewart L, Ford A, Sangal V et al. Draft genomes of 12 host-adapted and environmental isolates of Pseudomonas aeruginosa and their positions in the core genome phylogeny. Pathog Dis. 2014;71:20–25. [Abstract] [Google Scholar]
- Stull F, Betton JM, Bardwell JCA. Periplasmic chaperones and prolyl isomerases. EcoSal Plus. 2018;8:ESP–0005-2018. [Abstract] [Google Scholar]
- Tielen P, Narten M, Rosin N et al. Genotypic and phenotypic characterization of Pseudomonas aeruginosa isolates from urinary tract infections. Int J Med Microbiol. 2011;301:282–92. [Abstract] [Google Scholar]
- Tomoyasu T, Gamer J, Bukau B et al. Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor sigma 32. EMBO J. 1995;14:2551–60. [Europe PMC free article] [Abstract] [Google Scholar]
- Treepong P, Kos VN, Guyeux C et al. Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin Microbiol Infect. 2018;24:258–66. [Abstract] [Google Scholar]
- Trentini DB, Suskiewicz MJ, Heuck A et al. Arginine phosphorylation marks proteins for degradation by a Clp protease. Nature. 2016;539:48–53. [Europe PMC free article] [Abstract] [Google Scholar]
- Trunk K, Benkert B, Quack N et al. Anaerobic adaptation in Pseudomonas aeruginosa: definition of the Anr and Dnr regulons. Environ Microbiol. 2010;12:1719–33. [Abstract] [Google Scholar]
- Tümmler B, Kiewitz C. Cystic fibrosis: an inherited susceptibility to bacterial respiratory infections. Mol Med Today. 1999;5:351–8. [Abstract] [Google Scholar]
- Tümmler B, Koopmann U, Grothues D et al. Nosocomial acquisition of Pseudomonas aeruginosa by cystic fibrosis patients. J Clin Microbiol. 1991;29:1265–7. [Europe PMC free article] [Abstract] [Google Scholar]
- Valastyan JS, Lindquist S. Mechanisms of protein-folding diseases at a glance. Dis Model Mech. 2014;7:9–14. [Europe PMC free article] [Abstract] [Google Scholar]
- Voisin S, Kus JV, Houliston S et al. Glycosylation of Pseudomonas aeruginosa strain Pa5196 type IV pilins with mycobacterium-like alpha-1,5-linked d-Araf oligosaccharides. J Bacteriol. 2007;189:151–9. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang Z, Fang Y, Zhi S et al. The locus of heat resistance confers resistance to chlorine and other oxidizing chemicals in Escherichia coli. Appl Environ Microbiol. 2020;86:e02123–19. [Europe PMC free article] [Abstract] [Google Scholar]
- Wiehlmann L, Cramer N, Tümmler B. Habitat-associated skew of clone abundance in the Pseudomonas aeruginosa population. Environ Microbiol Rep. 2015;7:955–60. [Abstract] [Google Scholar]
- Wiehlmann L, Wagner G, Cramer N et al. Population structure of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2007;104:8101–6. [Europe PMC free article] [Abstract] [Google Scholar]
- Willcox MD. Management and treatment of contact lens-related Pseudomonas keratitis. Clin Ophthalmol. 2012;6:919–24. [Europe PMC free article] [Abstract] [Google Scholar]
- Winstanley C, O'Brien S, Brockhurst MA. Pseudomonas aeruginosa evolutionary adaptation and diversification in Cystic Fibrosis chronic lung infections. Trends Microbiol. 2016;24:327–37. [Europe PMC free article] [Abstract] [Google Scholar]
- Wurtzel O, Yoder-Himes DR, Han K et al. The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature. PLoS Pathog. 2012;8:e1002945. [Europe PMC free article] [Abstract] [Google Scholar]
- Xu H, Lin W, Xia H et al. Influence of ptsP gene on pyocyanin production in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2005;253:103–9. [Abstract] [Google Scholar]
- Yoshitani K, Hizukuri Y, Akiyama Y. An in vivo protease activity assay for investigating the functions of the Escherichia coli membrane protease HtpX. FEBS Lett. 2019;593:842–51. [Abstract] [Google Scholar]
- Yura T, Nakahigashi K. Regulation of the heat-shock response. Curr Opin Microbiol. 1999;2:153–8. [Abstract] [Google Scholar]
- Zhang P, Sicora CI, Vorontsova N et al. FtsH protease is required for induction of inorganic carbon acquisition complexes in Synechocystis sp. PCC 6803. Mol Microbiol. 2007;65:728–40. [Abstract] [Google Scholar]
- Zhi S, Banting G, Li Q et al. Evidence of naturalized stress-tolerant strains of Escherichia coli in municipal wastewater treatment plants. Appl Environ Microbiol. 2016;82:5505–18. [Europe PMC free article] [Abstract] [Google Scholar]
- Zolkiewski M. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli. J Biol Chem. 1999;274:28083–6. [Abstract] [Google Scholar]
Articles from FEMS Microbiology Reviews are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/femsre/fuaa029
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/femsre/article-pdf/44/6/740/34489493/fuaa029.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
Adaptive Evolution of Pseudomonas aeruginosa in Human Airways Shows Phenotypic Convergence Despite Diverse Patterns of Genomic Changes.
Mol Biol Evol, 41(2):msae022, 01 Feb 2024
Cited by: 0 articles | PMID: 38366124 | PMCID: PMC10883414
The membrane-cytoplasmic linker defines activity of FtsH proteases in Pseudomonas aeruginosa clone C.
J Biol Chem, 300(2):105622, 03 Jan 2024
Cited by: 0 articles | PMID: 38176647 | PMCID: PMC10850787
Insights into the Synergistic Antibacterial Activity of Silver Nitrate with Potassium Tellurite against Pseudomonas aeruginosa.
Microbiol Spectr, 11(4):e0062823, 06 Jul 2023
Cited by: 4 articles | PMID: 37409940 | PMCID: PMC10433965
Structural genome variants of Pseudomonas aeruginosa clone C and PA14 strains.
Front Microbiol, 14:1095928, 13 Mar 2023
Cited by: 1 article | PMID: 36992927 | PMCID: PMC10040652
Optimization of Transposon Mutagenesis Methods in Pseudomonas antarctica.
Microorganisms, 11(1):118, 01 Jan 2023
Cited by: 0 articles | PMID: 36677410 | PMCID: PMC9864612
Go to all (14) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A recently isolated human commensal Escherichia coli ST10 clone member mediates enhanced thermotolerance and tetrathionate respiration on a P1 phage-derived IncY plasmid.
Mol Microbiol, 115(2):255-271, 12 Oct 2020
Cited by: 18 articles | PMID: 32985020 | PMCID: PMC7984374
Intraclonal genome diversity of the major Pseudomonas aeruginosa clones C and PA14.
Environ Microbiol Rep, 8(2):227-234, 28 Jan 2016
Cited by: 29 articles | PMID: 26711897 | PMCID: PMC4819714
Diversity of the abundant pKLC102/PAGI-2 family of genomic islands in Pseudomonas aeruginosa.
J Bacteriol, 189(6):2443-2459, 28 Dec 2006
Cited by: 82 articles | PMID: 17194795 | PMCID: PMC1899365
The accessory genome of Pseudomonas aeruginosa.
Microbiol Mol Biol Rev, 74(4):621-641, 01 Dec 2010
Cited by: 172 articles | PMID: 21119020 | PMCID: PMC3008168
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Bundesministerium für Bildung und Forschung
Deutsche Forschungsgemeinschaft (2)
Grant ID: SFB 900
Grant ID: A2 and Z1
Swedish Research Council (1)
Grant ID: K2012–56X-22034–01-3




