Abstract
Free full text

Development of a High-Throughput Homogeneous AlphaLISA Drug Screening Assay for the Detection of SARS-CoV-2 Nucleocapsid
Abstract
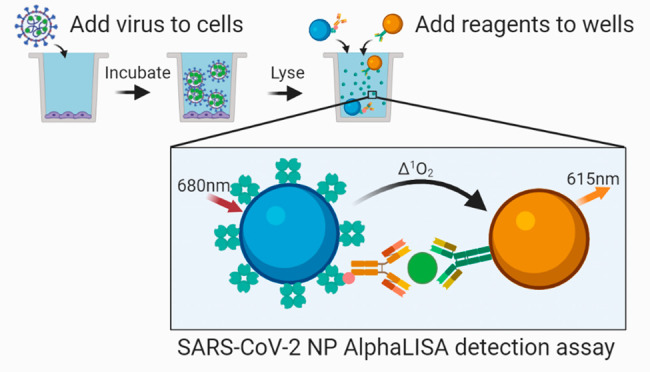
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is in urgent need of therapeutic options. High-throughput screening (HTS) offers an opportunity to rapidly identify such compounds. In this work, we have developed a homogeneous cell-based HTS system using AlphaLISA detection technology for the SARS-CoV-2 nucleocapsid protein (NP). Our assay measures both recombinant and endogenous NP from viral lysates and tissue culture supernatants (TCS) in a sandwich-based format using two monoclonal antibodies against the NP analyte. Viral NP was detected and quantified in both tissue culture supernatants and cell lysates, with large differences observed between 24 and 48 h of infection. We simulated viral infection by spiking recombinant NP into 384-well plates with live Vero-E6 cells and were able to detect the NP with high sensitivity and a large dynamic range. Antiviral agents that inhibit either viral cell entry or replication decrease the AlphaLISA NP signal. Thus, this assay can be used for high-throughput screening of small molecules and biologics in the fight against the COVID-19 pandemic.
The coronavirus disease of 2019, COVID-19, has spread rapidly through the global population since late 2019 due to the newly highly infectious nature of its cause, the deadly severe acute respiratory syndrome coronavirus 2, SARS-CoV-2.1 The unprecedented response from the global research community at every level of the life science enterprise has produced several therapeutic candidates, such as the repurposed investigational Ebola drug remdesivir that targets the SARS-CoV-2 RNA-dependent RNA polymerase,2,3 neutralizing antibodies against the viral spike (S) protein,4 and convalescent plasma from recovered patients containing neutralizing antibodies.5 Additionally, hundreds of vaccines against SARS-CoV-2 are currently under development with over two dozen of them in clinical phase I and II trials.6,7 While these are all viable options, no therapeutic option is yet approved by the United States Food and Drug Administration (US FDA). Therefore, we must continue the search for other alternatives, a task that will benefit from new assays and reagents that facilitate rapid identification of small molecules and biologics that suppress viral infection to treat COVID-19.
High-throughput screening (HTS) is one such method wherein high-density microplates can be used to rapidly screen large libraries of compounds to discover lead compounds for drug development.8 It also can be used for drug-repurposing screens and evaluating therapeutic agents with high confidence, low variability, and generate information regarding an agent’s potency via its half-maximal inhibitory concentration (IC50) and its maximum efficacy. Over the past decade, amplified luminescent proximity homogeneous assay (AlphaLISA) technology has emerged as one of the most reliable screening technologies because of its versatility, sensitivity, and homogeneous format without the need for plate evacuation or washing.9−11
AlphaLISA signal is generated when a donor bead is excited at an appropriate wavelength to general a reactive singlet oxygen radical that when in close proximity to an acceptor bead activates the acceptor bead to produce light at a specific wavelength.9 It can be used to replace ELISA that requires multiple plate-washing steps for the quantitative detection of analytes. The conjugation of antibodies to biotin and streptavidin aids in the creation of a screening system that can be used as a competitive assay or sandwich assay format. In a competitive assay that uses one antibody, unlabeled analyte would compete with a labeled analyte donor or acceptor and decrease signal. In a sandwich assay, two antibodies raised against the same immunogen analyte, albeit at different epitopes, will both bind to the analyte and lead to an increase in signal. For SARS-CoV-2 research, AlphaLISA technology has been utilized to identify inhibitors of the SARS-CoV-2 S protein receptor binding domain (RBD) by attaching the donor and acceptor beads to angiotensin converting enzyme 2 (ACE2) receptor protein and S protein RBD, respectively.12
The SARS-CoV-2 virion is made of four main component structural proteins including S protein, envelope (E) protein, nucleocapsid protein (NP), and membrane (M) protein.13 SARS-CoV-2 S protein is responsible for initial host-cell binding to the angiotensin converting enzyme 2 (ACE2) receptor via the RBD found within the S1 subunit.14 E protein is embedded in the viral membrane and plays a role in viral particle packaging and maturation.15 M protein regulates replication and RNA packaging.13 NP binds M protein and interacts with the viral genome to bind the positive strand RNA viral genome, and NP is the main focus of the work presented here.13
Here, we describe the development of a high-throughput SARS-CoV-2 NP-based immunoassay in the AlphaLISA sandwich assay format that can be used to detect live virus infection and replication in host cells to carry out HTS for drug discovery and development. We show the selection process for identifying the most reliable antibody pair to detect NP, along with the optimization of the AlphaLISA reagents for the highest sensitivity and dynamic range using recombinant NP. We further demonstrate this assay can detect the SARS-CoV-2 NP in cell lysates and tissue culture supernatants (TCS) after SARS-CoV-2 infection. Last, we have simulated viral infection by spiking recombinant NP into Vero-E6 cells grown in in 384-well plates, along with full plate statistics using recombinant NP in media, demonstrating the assay potential for HTS. The results presented here demonstrate a rapid, homogeneous SARS-CoV-2 NP assay that can be used for HTS of compound collections to identify compounds that inhibit SARS-CoV-2 infection and viral replication in live cells.
Results
Best NP–Antibody Pair Selection
NP is one of the four structural proteins present in the SARS-CoV-2 particle, and its levels increase with viral replication as more viral particles are produced. Thus, the level of NP can serve as an indicator of virus infection and replication in host cells. The cell-based AlphaLISA SARS-CoV-2 NP detection assay relies on the proximity of two labeled antibodies that bind to NPs in cell lysate and supernatant in high-density microplates to generate a luminescent signal. This assay detects viral proteins inside the cell and the virus that is released into the medium.
The biotinylated antibody is bound to the streptavidin-coated donor bead that when excited with 680 nm light induces a singlet oxygen radical to activate an acceptor bead conjugated to the second antibody, resulting in a 615 nm luminescent signal (Figure Figure11A). A successful reaction depends on the close proximity of the donor and acceptor. Multiple antibodies were matrixed together to determine which antibody pair produced the highest AlphaLISA signal (counts) from recombinant NP at 10 000 pg/mL (Figure Figure11B,C). The three best antibody pairs were 1 + 4, 11 + 4, and 11 + 10. Each pair was then tested against a titration of recombinant NP at 10 000, 1000, 100, and 0 pg/mL, and produced a concentration dependent signal increase, with signal to background ratios (S/B, 10 000–0 pg/mL) of 66.2, 72.8, and 51.5, respectively (Figure S-1). On the basis of these results, pairs 1 + 4, and 11 + 4 were selected for further optimization. Interestingly, the pairs consisting of antibody 11 did not detect the histidine (His)-tagged NP (Figure Figure11B) but generated the best signal with untagged NP (Figure Figure11C), suggesting the His-tag interferes with binding of antibody 11 to NP. The other antibody pairs produced relatively similar counts regardless of the NP protein used in the optimization.

Best-pair antibody selection for SARS-CoV-2 NP AlphaLISA sandwich assay. (A) Schematic diagram of homogeneous cell-based AlphaLISA sandwich assay. (B) AlphaLISA signal (counts) for matrixed antibody pairs using His-tagged NP. High values indicate efficient AlphaLISA reaction. (C) AlphaLISA signal (counts) for matrixed antibody pairs using untagged NP. High values indicate efficient AlphaLISA reaction. N = duplicate wells. Error bars indicate SD.
Optimization of Acceptor to Donor Ratio and Biotinylated Anitbody Concentration
We next determined the S/B at a greater range of untagged recombinant NP (20 000–81.9 pg/mL) using donor/SA acceptor ratios of 10–40 μg/mL and 20–20 μg/mL and different concentrations of biotinylated antibody from 0.5 to 5.0 nM (Figure S-2). We selected a donor/SA ratio of 10–40 μg/mL for both pairs and 1.0 and 5.0 nM biotinylated antibody for Pairs 1 + 4 and 11 + 4, respectively, based on the S/B calculations (Figure S-2). The standard curves were generated using recombinant NP for these conditions, and no plateau was observed at the higher concentrations, indicating that the upper limit of detection was more than 20 000 pg/mL (20 ng/mL) (Figure S-3).
Lysis Buffer Selection and Viral Lysate Testing
To determine the limits of detection and dynamic range of the assay, we next generated standard curves using concentrations of NP from 200 000 to 0.02 ng/mL for both pairs (Figure Figure22A,C; Table S-1). We observed a significant hook effect at concentrations of recombinant NP above 320 ng/mL for pair 1 + 4 and 1600 ng/mL for pair 11 + 4 (Figure Figure22; Table S-1). The hook effect indicates the concentration of analyte at which an overabundance of analyte titrates the donor and acceptor away from each other, and this hooking dictates the upper limit of detection before the signal will start to decrease. Two different lysis buffers were used to optimize any lysis buffer effects: The NCATS lysis buffer consisted of 0.5% Triton-X 100 with protease inhibitor; the other was the AlphaLISA lysis buffer from the manufacturer. For pair 1 + 4, both lysis buffers performed equally, while the AlphaLISA lysis buffer performed better for pair 11 + 4 in generating the standard curve. The overall S/B was higher for pair 11 + 4 at higher concentrations of NP, but pair 1 + 4 had a higher sensitivity as seen by the larger S/B values at lower concentrations (Figure Figure22B,D; Table S-1).
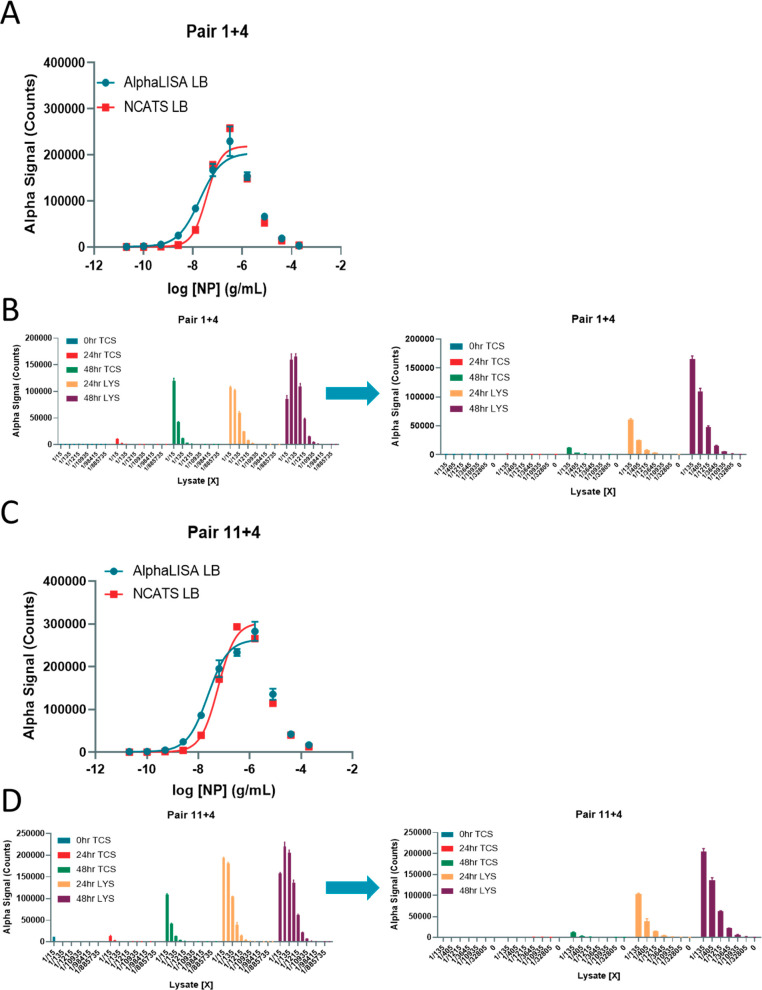
SARS-CoV-2 NP detection in virus-infected cell lysates and tissue culture supernatants. (A) Standard curve for Pair 1 + 4 demonstrating hook effect using AlphaLISA or NCATS lysis buffer. (B) (Left) AlphaLISA signal (counts) for 24 and 48 h virus-infected cell lysates and 0, 24, and 48 h tissue culture supernatants at various dilutions. (Right) Same as left panel with a narrower dilution range. (C) Standard curve for Pair 11 + 4 demonstrating hook effect using AlphaLisA or NCATS lysis buffer. (D) (Left) AlphaLISA signal (counts) for 24 and 48 h virus-infected cell lysates and 0, 24, and 48 h tissue culture supernatants at various dilutions. (Right) Same as left panel with a narrower dilution range. Data points beyond the hook effect are shown but not included in the range for the nonlinear regression curve fit. N = triplicate wells. Error bars indicate SD.
To determine whether the detection system could detect native NP in SARS-CoV-2-infected lysates, TCS collected at 0, 24, and 48 h along with cell lysates collected at 24 and 48 h postinfection were tested at multiple dilutions. Pair 11 + 4 produced absolute AlphaLISA signal (counts) than greater that of pair 1 + 4 at the highest concentrations, but both pairs exhibited a hook effect at 1/15 lysate dilutions (Figure Figure22D). NP was better detected in TCS by pair 11 + 4 than by pair 1 + 4. The concentrations of NP in ng/mL were interpolated from the standard curves. For both pairs, the concentration of NP was approximately 9-fold greater in 48 h TCS than in 24 h TCS. For pair 11 + 4, the 24 and 48 h TCS concentrations were 80.3 and 708 ng/mL, respectively. The 24 and 48 h lysate NP concentrations were significantly higher at 7380 and 30 200 ng/mL, respectively. These concentrations were comparable, although not exactly the same, when comparing pairs 1 + 4 and 11 + 4.
Sequential Two- versus Three-Step Assay Protocol Optimization
We next optimized the sequential addition of assay reagents including lysis buffer, biotinylated antibody, acceptor beads, and donor beads in the two- versus three-step assay protocol (Figure S-4). For pairs 1 + 4 and 11 + 4, the two-step assay produced a greater dynamic range for recombinant NP detection than did the three-step assay without exhibiting a hook effect (Figure S-4) as determined by the S/B ratios. NCATS lysis buffer performed slightly better than AlphaLISA lysis buffer. We further optimized the two- and three-step assays for recombinant NP spiked into Vero-E6 cells grown in 384-well plates (Figure S-5). In most cases, the two-step assays performed better with 20 000 cells/well as measured by S/B ratios. The effect of media and cell density was also tested for assay interference. Media affected pair 1 + 4 to a greater extent than pair 11 + 4, while cell density did not significantly affect S/B ratios. The lower limit of detection for both pairs was 0.22 ng/mL with an upper limit at or beyond 400 ng/mL (Figure S-6).
Assay Volume Optimization
Next, we optimized the total assay volume in a 384-well plate format using recombinant NP in AlphaLISA LB and found that higher total assay volumes produced lower absolute AlphaLISA signal (counts), but 100 uL total volume performed better than 50 uL total volume (Figure Figure33A,B; Table S-2). Again, pair 11 + 4 exhibited a higher limit of detection than pair 1 + 4 (Figure Figure33B,D). In this assay, the 20 μL condition utilized the 384-well Proxiplate, while the 50 and 100 uL conditions utilized the 384-well CulturPlate. The main difference between the two is the shallower circle wells in the Proxiplate compared to the deeper square wells in the CulturPlate, suggesting the plate type and well dimensions change the S/B ratios.
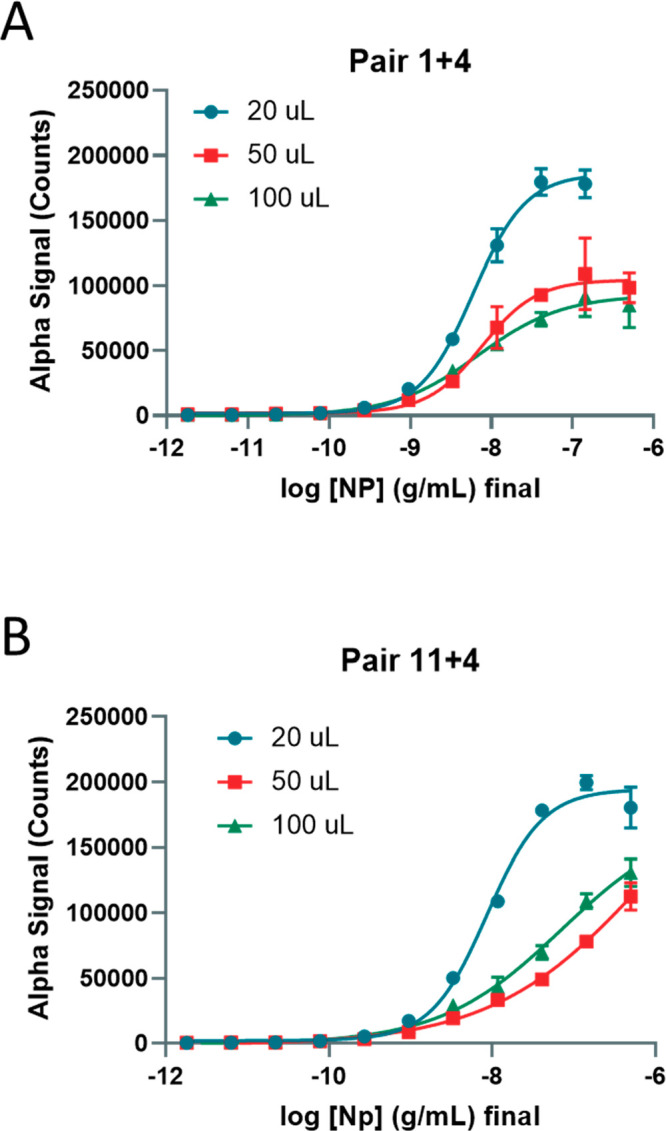
Optimization of assay volumes for 384-well plate using recombinant NP. (A) Detection of titrated NP in wells with different final volumes of 20, 50, and 100 μL using Pair 1 + 4. (B) Detection of titrated NP in wells with different starting volumes 20, 50, and 100 μL using Pair 11 + 4. N = triplicate wells. Error bars indicate SD.
We further optimized media conditions and found that cell culture media itself had a profound effect on the S/B ratios, while the concentration of FBS had little to no impact on the assay performance (Figure S-7).
Batch to Batch Reagent Reproducibility
A second batch of reagents was produced, and the concentration of biotinylated antibody was matched to generate the same performance as the first batch for pair 1 + 4 and pair 11 + 4 (Figure S-8). We tested the second batch of reagents with the recombinant NP spiked into Vero-E6 cell culture in 384-well plates and found better performance with pair 11 + 4 when cells were present in the wells (Figure Figure44A–D, Table S-3). The lower limit of detection was 0.2 ng/mL, and the upper limit was greater than 500 ng/mL NP. Altogether, pair 11 + 4 performed better than did pair 1 + 4 and was selected as the final pair for future assays. To confirm that the second batch of reagents performed well, we tested the same conditions for optimization as before with cell densities ranging from 20 000 to 0 cells/well, along with a no media control in PBS.
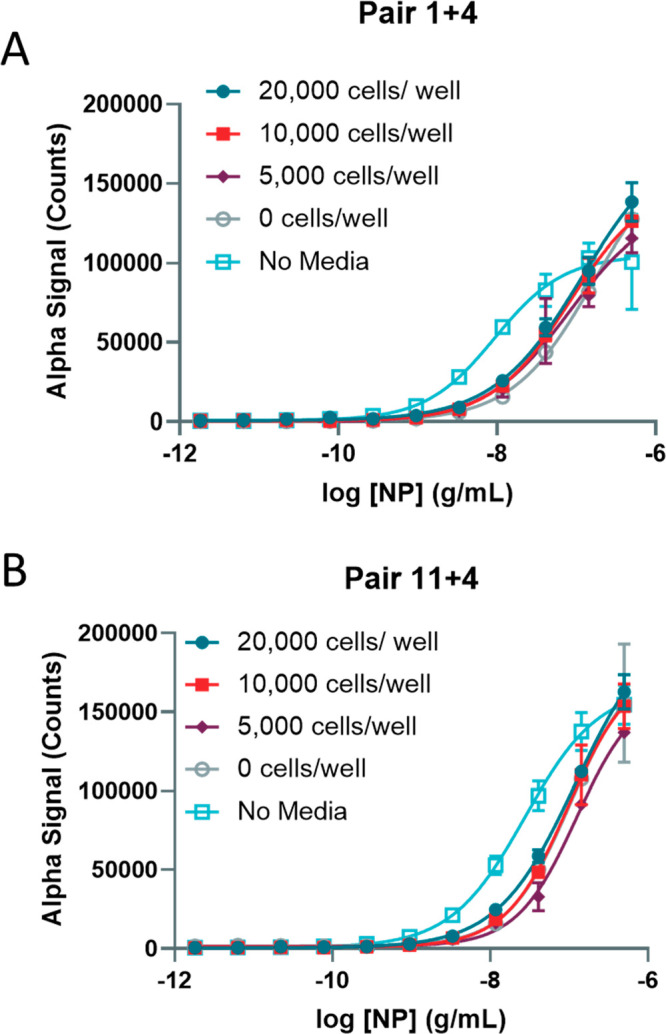
Optimization of 384-well plate cell number in simulated infection using second preparation of reagents. (A) Detection of titrated NP in wells culturing 20 000, 10 000, 5000, or 0 cells/well along with a no-media control in Vero-E6 cells using Pair 1 + 4 (second batch). (B) Detection of titrated NP in wells culturing 20 000, 10 000, 5000, or 0 cells/well along with a no-media control in Vero-E6 cells using Pair 11 + 4 (second batch). N = triplicate wells. Error bars indicate SD.
Lysis Buffer and Incubation Time Optimization, And Determination of Cell Lysate Interference
The AlphaLISA buffer and NCATS lysis buffers (LB) were compared in plates incubated for 30 min versus 1 h and for the AlphaLISA buffer there was a slight increase in signal after 1 h incubation (Figure S-9). The incubation time had little effect on the S/B for the NCATS LB condition. NP detection in the presence of lysed 10 000 and 5000 cells/well performed similarly to NP detection in media and better than in 20 000 cells/well. A concentration of 20 000 cells/well would be considered over-confluent, beyond 100%. The no-media PBS control had a lower S/B ratio overall. The main difference between the LBs was that a signal plateau was observed for the NCATS LB indicating an upper limit of detection of approximately 1500 ng/mL, whereas the AlphaLISA buffer could potentially detect higher concentrations. Importantly, the sensitivity of the assay with NCATS LB reveals a similar lower limit of detection of 0.23 ng/mL as in previous experiments, whereas the AlphaLISA buffer was observed be less sensitive, for example, with a lower limit of detection of 2.06 ng/mL for 10 000 cells/well.
Estimation of NP Concentration in Cell Lysates Using Standard Curve Interpolation
Finally, we estimated the concentration of NP in TCS and lysate from cells infected with live SARS-CoV-2 for 24 and 48 h compared to the noninfected (0 h) controls using interpolation from the 10 000 cells/well standard curve (Figure S-10). Dilutions of TCS and lysates from 15-fold to over 1900-fold were created, and NCATS LB was added to the samples. The linear portion of the curves was used for interpolation. The concentrations of NP in lysates was determined to be 7300 and 34 092 ng/mL for 24 and 48 h lysates, respectively (Figure S-10). In correspondence with the NP concentration, the viral titers were calculated to be 8.2 × 1006 and 1.1 × 1009 focus forming units (FFU)/mL for 24 and 48 h TCS, respectively, indicating increasing amounts of viral particles with longer infection times. This data indicated that the NP in viral lysates could be quantified using the AlphaLISA NP detection reagents and standard curve interpolation.
384-Well Plate Assay Statistics for HTS
We further calculated the assay plate statistics using a small-volume 384-well microtiter plate to demonstrate the assay performance for HTS (Figure Figure55). Using media only as a positive control to mimic a drug that reduces viral titer in cells and media with recombinant NP added to mimic a virus only condition, we utilized the BioRAPTR FRD liquid-dispensing workstation to automatically dispense the media and reagents for AlphaLISA detection of NP. After reading the plate on the BMG Labtech Pherastar using the AlphaLISA optical module, we determined the Z-factor to be 0.86, calculated using the standard deviation and average values of the positive control (columns 1 and 2) containing just media and the negative control containing 500 ng/mL of recombinant NP (columns 3–24). The coefficient of variation (CV) for the positive control was 20.2%, and the negative control CV was 4.0%. Between the negative control and the positive control, the S/B was 86.9. On the basis of these plate statistics, the assay performance has an “excellent” rating using recombinant NP and should be well-suited for live-virus optimization and HTS.
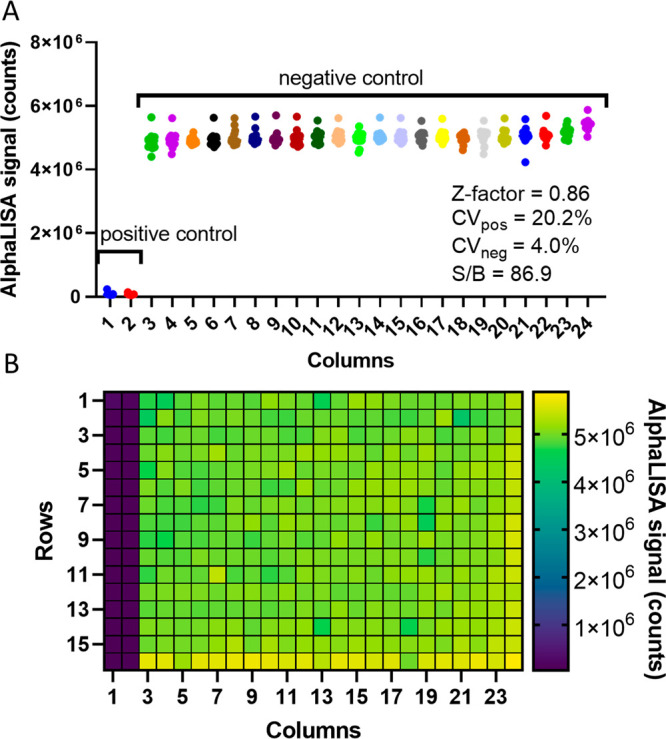
Robust assay plate statistics in 384-well plate format. (A) Graph illustrating the AlphaLISA signal counts per well in each column and the calculated plate statistics for a 384-well plate using a total volume of 16 μL. The positive control is media only, and the negative control is media with 500 ng/mL recombinant NP. (B) Heat map demonstrating the AlphaLISA signal counts of each well.
We further tested the stability of the assay after a 24 h incubation at 4 °C (Figure S-11). After letting the plate reach room temperature, we scanned the plate once more at the same settings used for the 30 min time point. The plate statistics were virtually identical, with a Z-factor of 0.86, CVs for the positive and negative controls of 20.2 and 4.0%, respectively, and a slight reduction of S/B at 71.7. Therefore, the assay signal is stable using the aforementioned conditions up to 24 h, and perhaps longer, following the protocol execution and reagent addition. This will enable a biosafety level 3/4 (BSL 3/4) facility to execute the assay and then read the plates over a longer period of time, without worrying about assay performance degradation due to long incubations before reading.
Discussion
The ability to rapidly detect viral proteins in a homogeneous cell culture system provides researchers with an easy to use method for screening thousands of potential antiviral compounds, both newly synthesized analogs and existing FDA-approved drugs alike. In this work, we have designed and developed through meticulous optimization an AlphaLISA sandwich-based detection system to identify the SARS-CoV-2 NP. Here, two antibodies were identified to detect recombinant and virally generated NP both in solution and in cell culture. Importantly, we demonstrated a significant increase in NP with longer incubation of virus-infected Vero-E6 cells and were able to quantitatively assess the NP concentration using standard curve interpolation. This assay system should be able to identify compounds that decrease the presence of SARS-CoV-2 NP as a measure of antiviral activity that prevents either viral entry or replication leading to the translation of NP in cells. Our add-and-read system requires no wash steps or removal of cell culture media, providing an excellent methodology for the use in BSL 3/4 facilities that require simple and easy to use assay technologies for drug screening and validation.
The selection of SARS-CoV-2 NP was a strategic one in that mutations of the SARS-CoV-2 viral genome will likely occur in coding regions that are selected for during viral–host interactions. The SARS-CoV-2 S protein that protrudes from the envelope binds ACE2 with high affinity as the first step in viral infection,16 whereas the viral NP is located deeper within the virion core. Indeed, mutations in S protein have already been identified, with one of those being the more infectious D614G mutation.17−19 By targeting NP, this detection system will maintain its reliability for drug screening of multiple viral strains, barring significant changes to the epitopes to which the antibodies used in the AlphaLISA sandwich assay bind.
We anticipate rapid developments in HTS screening following the adoption and application of the AlphaLISA NP detection system in BSL 3/4 laboratories. Future work will optimize assay conditions for live virus experiments to establish the appropriate multiplicity of infection and incubation time of the virus in order to match the wide dynamic range and high sensitivity of the assay.
Methods
Reagents and Materials
Vero-E6 cells (CRL-1586, RRID:CVCL_0574) were purchased from ATCC. The following items were purchased from Corning TM: EMEM (10–009-CV), HI FBS (35-016-VC, and 0.25% Trypsin (25053CI). The untagged NP (Z03501) was purchased from Genscript. His-tagged NP (40588-V08B) was purchased from SinoBiological.
Pen/Strep (15140–122)was purchased from Gibco. PBS (SH30256FS) was purchased from HyClone. The following items were purchased from PerkinElmer: ProxiPlate-384 Plus (Cat# 6008280), CulturPlate-384 (Cat#: 6007680), Alpha Streptavidin Donor beads (6760002), and AlphaLISA lysis buffer (AL003C). The following custom labeling was performed by PerkinElmer: Donor antibodies were biotinylated using NHS-activated biotinylating reagent (ChromaLink #B-1001–105), and acceptor antibodies were conjugated to Alpha Acceptor Beads (PerkinElmer #6760137M).
Preparation of Antibody Pair Matrixing
A matrix with all antibodies provided by PerkinElmer was tested for the ability to detect NP.
The untagged NP and His-tagged NP were diluted in AlphaLISA lysis buffer at 10 000, 1000, and 100 pg/mL. For initial testing, 20 μg/mL AlphaLISA Acceptor (final), 1 nM (final) biotinylated antibody, and 20 μg/mL donor (final) were used. A two-step assay was performed (data points in duplicate) using the following protocol:
- 1.
Dispense 5 μL of recombinant protein.
- 2.
Dispense 10 μL of mix of acceptor beads and biotinylated antibody.
- 3.
Incubate 60 min at room temperature (RT).
- 4.
Dispense 5 μL of SA-Donor Beads.
- 5.
Incubate 30 min at room temperature.
- 6.
Read plates on EnVision (PerkinElmer).
Antibody Concentration Optimizations
Best pairs 1 + 4 and 11 + 4 were optimized using the following parameters: Biotinylated antibody concentrations tested were 0.5, 1, 2, and 5 nM. Acceptor bead–SA donor bead concentrations tested were at 20–20 μg/mL and 10–40 μg/mL. The concentration of untagged NP recombinant protein started at 20 000 pg/mL followed by 2.5-fold dilutions. The dispensing protocol was the same as for the antibody pair matrixing.
Vero-E6 Cell Culture
Vero-E6 (grown in EMEM, 10% FBS, and 1% penicillin/streptomycin), were cultured in T175 flasks and passaged at 95% confluency. Briefly, cells were washed once with PBS and dissociated from the flask using 0.25% Trypsin. Cells were counted prior to seeding.
Preparation of Viral Lysate and Tissue Culture Supernatant Production
Vero-E6 cells were plated in 12-well plates at 450 000 cells/well in 1.25 mL of growth media. Cells were incubated for 24 h at 37 °C. A 250 μL aliquot of SARS-CoV-2 [USA-WA1/2020 strain (Gen Bank: MN985325.1)] was used at an MOI of 0.05. Cells were inoculated for 45 min at 37 °C. Supernatant was collected and pooled for 0 h TCS. A final concentration of 0.5% Triton-X 100 and 1× protease inhibitor was used for all samples. TCS samples were stored at −20 °C until needed. Next, 1.5 mL of fresh media was added to each well, and cells were incubated for 24 and 48 h at 37 °C. Six wells were harvested at each time point. Supernatant was collected from 6 wells each at 24 and 48 h. Samples were pooled for 24 or 48 h TCS. For viral titer calculations, 200 μL of TCS was removed and diluted with equal volume of lysis buffer. Samples were stored at −20 °C until needed. Lysate was collected from 6 wells each at 24 and 48 h. Cells were rinsed once with ice-cold PBS, and 250 μL/well of cell lysis buffer + PI was added to lyse cells. Protease inhibitor was added to the lysis buffer before lysing cells. Cells were scraped to the bottom of the well, and pooled lysates were collected into 1.5 mL tubes on ice. Tubes were vortexed for 10 s and placed on ice for 10 min. This was repeated three times. Lysates were spun down for 30 min at 13.2 000 rpm at 4 °C and stored at −20 °C until further use.
Optimization of Assay Using Viral Lysates and Tissue Culture Supernatant
The best conditions of pairs 1 + 4 and 11 + 4 were used to expand the standard curve and test TCS and lysate samples: 1 + 4: 10–40 μg/mL with 1 nM biotinylated antibody, 11 + 4: 10–40 μg/mL with 5 nM biotinylated antibody. An 11-point curve of recombinant untagged NP started at 200 000 ng/mL, followed by 5-fold dilutions. Lysates and TCS were diluted in PBS + Triton-X 100 + protease inhibitors 1:15 (1×) followed by 3-fold dilutions. The dispensing protocol was the same as above.
Simulation of Viral Infection Using Recombinant NP
The best conditions of pairs 1 + 4 and 11 + 4 were used to test the assay in a cell-based format: 1 + 4: 10–40 μg/mL with 1 nM biotinylated antibody and 11 + 4: 10–40 μg/mL with 5 nM biotinylated antibody. Vero E6 cells were plated at 20 000 and 50 000 cells/well in 384-well plates (TC) and incubated overnight. An 11-point curve of recombinant NP started at 4000 ng/mL initial (400 ng/mL final) followed by 3.5-fold dilutions in AlphaLISA lysis buffer.
For second batch preparation, the best conditions of pairs 1 + 4 and 11 + 4 were used to test the assay in a cell-based format (newly conjugated and biotinylated Ab were used and optimized to reproduce previous antibodies): 1 + 4: 10–40 μg/mL with 0.5 nM Biotinylated antibody, 11 + 4: 10–40 μg/mL with 1 nM Biotinylated antibody. Vero E6 cells were plated at 20 000, 10 000, and 5000 cells/well in 384-well plate (TC) and incubated overnight. The assay was also tested without cells and without media as a control. An 11-point curve of recombinant NP started at 2000 ng/mL initial (500 ng/mL final) followed by 3.5-fold dilutions in media.
384-Well Plate Assay Statistics
Whole-plate statistics were calculated using recombinant NP at a concentration of 500 ng/mL in media using a scaled down protocol to conserve reagents in a white small-volume 384-well plate (Greiner 784075). Reagents were added using a fully automated BioRAPTR FRD Workstation (Beckman Coulter). Briefly, 8 uL of cell culture media with or without NP was added to columns 3–24 and columns 1–2, respectively. Next, 2 uL of 5X AlphaLISA Lysis buffer was then dispensed to wells and agitated for 30 min. Then, 4 uL of mixed biotinylated anti-NP antibody and AlphaLISA acceptor beads were dispensed into each well and incubated for 60 min. Last, 2 uL of streptavidin donor beads were dispensed and incubated for 30 min at room temperature. The plate was then read on the Pherastar with an AlphaLISA module from BMG Labtech. Z-factor, CV, and S/B was calculated for the entire plate.
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Center for Advancing Translational Sciences, NIH. We thank Dr. Arturo Gonzalez-Moya and his team at PerkinElmer for assay development support.
Glossary
Abbreviations
| HTS | high-throughput screening |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus 2 |
| NP | nucleocapsid protein |
| TCS | tissue culture supernatants |
| COVID-19 | coronavirus disease of 2019 |
| AlphaLISA | amplified luminescent proximity homogeneous assay |
| IC50 | half-maximal inhibitory concentration |
| His | histidine tag |
Notes
This article is made available via the ACS COVID-19 subset for unrestricted RESEARCH re-use and analyses in any form or by any means with acknowledgement of the original source. These permissions are granted for the duration of the World Health Organization (WHO) declaration of COVID-19 as a global pandemic.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.0c00122.
Detection of titrated NP using best antibody pairs. Optimization of best antibody pair and reagent concentrations. Standard curves using best conditions for best antibody pairs. SARS-CoV-2 NP detection in virus-infected cell lysates and tissue culture supernatants. Assay optimization for best antibody pairs and conditions. Simulation of viral infection in 384-well plates using recombinant NP spiked into Vero-E6 wells. Optimization of lysis buffer and cell density for 384-well plate viral infection simulation using recombinant NP. Optimization of assay volumes for 384-well plate using recombinant NP. Optimization of AlphaLISA assay using different FBS percentages in cell culture media. Optimization of best antibody pair and reagent concentrations of second preparation to match first preparation. Optimization of 384-well plate cell number in simulated infection using second preparation of reagents. Optimization of lysis buffer, cell density, and incubation time for 384-well plate viral infection simulation using recombinant NP and second batch of reagents. Estimation of viral NP concentration in viral lysates. Stability of the assay after 24 h. (PDF)
Author Contributions
# K.G. and C.Z.C. contributed equally to this work. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. Experimental contributions: K.G., C.Z.C., M.X., J.C.T., L.M.-S., and T.M. Project management: K.G. and C.Z.C. Initial conception and design: K.G., C.Z.C., and W.Z. Manuscript writing and editing: K.G., C.Z.C., and W.Z.
References
- Dong E.; Du H.; Gardner L. (2020) An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 20, 533–534. 10.1016/S1473-3099(20)30120-1. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wang M.; Cao R.; Zhang L.; Yang X.; Liu J.; Xu M.; Shi Z.; Hu Z.; Zhong W.; Xiao G. (2020) Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 30, 269–271. 10.1038/s41422-020-0282-0. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Eastman R. T.; Roth J. S.; Brimacombe K. R.; Simeonov A.; Shen M.; Patnaik S.; Hall M. D. (2020) Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. ACS Cent. Sci. 6, 672–683. 10.1021/acscentsci.0c00489. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Jiang S.; Hillyer C.; Du L. (2020) Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 41, 355–359. 10.1016/j.it.2020.03.007. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Duan K.; Liu B.; Li C.; Zhang H.; Yu T.; Qu J.; Zhou M.; Chen L.; Meng S.; Hu Y.; Peng C.; Yuan M.; Huang J.; Wang Z.; Yu J.; Gao X.; Wang D.; Yu X.; Li L.; Zhang J.; Wu X.; Li B.; Xu Y.; Chen W.; Peng Y.; Hu Y.; Lin L.; Liu X.; Huang S.; Zhou Z.; Zhang L.; Wang Y.; Zhang Z.; Deng K.; Xia Z.; Gong Q.; Zhang W.; Zheng X.; Liu Y.; Yang H.; Zhou D.; Yu D.; Hou J.; Shi Z.; Chen S.; Chen Z.; Zhang X.; Yang X. (2020) Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. U. S. A. 117, 9490–9496. 10.1073/pnas.2004168117. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Amanat F.; Krammer F. (2020) SARS-CoV-2 vaccines: status report. Immunity 52, 583–589. 10.1016/j.immuni.2020.03.007. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lurie N.; Saville M.; Hatchett R.; Halton J. (2020) Developing Covid-19 vaccines at pandemic speed. N. Engl. J. Med. 382, 1969–1973. 10.1056/NEJMp2005630. [Abstract] [CrossRef] [Google Scholar]
- Inglese J.; Auld D. S.; Jadhav A.; Johnson R. L.; Simeonov A.; Yasgar A.; Zheng W.; Austin C. P. (2006) Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc. Natl. Acad. Sci. U. S. A. 103, 11473–11478. 10.1073/pnas.0604348103. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Bielefeld-Sevigny M. (2009) AlphaLISA immunoassay platform—the “no-wash” high-throughput alternative to ELISA. Assay Drug Dev. Technol. 7, 90–92. 10.1089/adt.2009.9996. [Abstract] [CrossRef] [Google Scholar]
- Yasgar A.; Jadhav A.; Simeonov A.; Coussens N. P. (2016) AlphaScreen-Based Assays: Ultra-High-Throughput Screening for Small-Molecule Inhibitors of Challenging Enzymes and Protein-Protein Interactions. Methods Mol. Biol. 1439, 77–98. 10.1007/978-1-4939-3673-1_5. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Ott C. A.; Baljinnyam B.; Zakharov A. V.; Jadhav A.; Simeonov A.; Zhuang Z. (2017) Cell Lysate-Based AlphaLISA Deubiquitinase Assay Platform for Identification of Small Molecule Inhibitors. ACS Chem. Biol. 12, 2399–2407. 10.1021/acschembio.7b00543. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Hanson Q. M., Wilson K. M., Shen M., Itkin Z., Eastman R. T., Shinn P., and Hall M. D. (June 16, 2020) Targeting ACE2-RBD interaction as a platform for COVID19 therapeutics: Development and drug repurposing screen of an AlphaLISA proximity assay, bioRxiv (Biochemistry) 2020.06.16.154708, DOI: 10.1101/2020.06.16.154708. [Europe PMC free article] [Abstract]
- Yoshimoto F. K. (2020) The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the Cause of COVID-19. Protein J. 39, 198–216. 10.1007/s10930-020-09901-4. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Hoffmann M.; Kleine-Weber H.; Schroeder S.; Krüger N.; Herrler T.; Erichsen S.; Schiergens T. S.; Herrler G.; Wu N.-H.; Nitsche A.; et al. (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280. 10.1016/j.cell.2020.02.052. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Tilocca B.; Soggiu A.; Sanguinetti M.; Babini G.; De Maio F.; Britti D.; Zecconi A.; Bonizzi L.; Urbani A.; Roncada P. (2020) Immunoinformatic analysis of the SARS-CoV-2 envelope protein as a strategy to assess cross-protection against COVID-19. Microbes Infect. 22, 182–187. 10.1016/j.micinf.2020.05.013. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Li W.; Moore M. J.; Vasilieva N.; Sui J.; Wong S. K.; Berne M. A.; Somasundaran M.; Sullivan J. L.; Luzuriaga K.; Greenough T. C.; Choe H.; Farzan M. (2003) Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454. 10.1038/nature02145. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K. E., Tompkins-Tinch C., Nyalile T., Wang Y., Baum A., Diehl W. E., Dauphin A., Carbone C., Veinotte K., Egri S. B., Schaffner S. F., Lemieux J. E., Munro J., Rafique A., Barve A., Sabeti P. C., Kyratsous C., Dudkina N., Shen K., and Luban J. (July 4, 2020) Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant bioRxiv (Microbiology) 2020.2007.2004.187757, version 2,10.1101/2020.07.04.187757. [Europe PMC free article] [Abstract] [CrossRef]
- Bhattacharyya C., Das C., Ghosh A., Singh A. K., Mukherjee S., Majumder P. P., Basu A., and Biswas N. K. (May 5, 2020) Global Spread of SARS-CoV-2 Subtype with Spike Protein Mutation D614G is Shaped by Human Genomic Variations that Regulate Expression of TMPRSS2 and MX1 Genes. bioRxiv (Genomics) 2020.05.04.075911, DOI: 10.1101/2020.05.04.075911.
- Korber B.; Fischer W.; Gnanakaran S.; Yoon H.; Theiler J.; Abfalterer W.; Hengartner N.; Giorgi E.; Bhattacharya T.; Foley B.; et al. (2020) Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182, 812–827. 10.1016/j.cell.2020.06.043. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
Articles from ACS Pharmacology & Translational Science are provided here courtesy of American Chemical Society
Full text links
Read article at publisher's site: https://doi.org/10.1021/acsptsci.0c00122
Read article for free, from open access legal sources, via Unpaywall:
https://pubs.acs.org/doi/pdf/10.1021/acsptsci.0c00122
Citations & impact
Impact metrics
Article citations
Cell-Based Covalent-Capture Deubiquitinase Assay for Inhibitor Discovery.
ACS Pharmacol Transl Sci, 7(9):2827-2839, 16 Aug 2024
Cited by: 0 articles | PMID: 39296272
Development of an AlphaLISA assay for sensitive and accurate detection of influenza B virus.
Front Med (Lausanne), 10:1155551, 05 May 2023
Cited by: 2 articles | PMID: 37215702 | PMCID: PMC10196263
Computational and Experimental Approaches Identify Beta-Blockers as Potential SARS-CoV-2 Spike Inhibitors.
ACS Omega, 7(32):27950-27958, 08 Aug 2022
Cited by: 5 articles | PMID: 35983371 | PMCID: PMC9380819
Development of a rapid homogeneous immunoassay for detection of rotavirus in stool samples.
Front Public Health, 10:975720, 04 Aug 2022
Cited by: 1 article | PMID: 35991049 | PMCID: PMC9386352
Organoid and microfluidics-based platforms for drug screening in COVID-19.
Drug Discov Today, 27(4):1062-1076, 23 Dec 2021
Cited by: 19 articles | PMID: 34954328 | PMCID: PMC8695520
Review Free full text in Europe PMC
Go to all (8) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences
- (1 citation) ENA - MN985325
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
SARS-CoV-2 Nucleocapsid Protein TR-FRET Assay Amenable to High Throughput Screening.
ACS Pharmacol Transl Sci, 5(1):8-19, 03 Jan 2022
Cited by: 3 articles | PMID: 35036857 | PMCID: PMC8751018
SARS-CoV-2 cytopathic effect (CPE)
National Center for Advancing Translational Sciences (NCATS), Bethesda (MD), 06 May 2022
Cited by: 0 articles | PMID: 35512046
ReviewBooks & documents Free full text in Europe PMC
Funding
Funders who supported this work.
National Center for Advancing Translational Sciences (1)
Grant ID: Intramural Research Program


 and
and 



