Abstract
Free full text

Assessment of Ki67 in Breast Cancer: Updated Recommendations From the International Ki67 in Breast Cancer Working Group
Abstract
Ki67 immunohistochemistry (IHC), commonly used as a proliferation marker in breast cancer, has limited value for treatment decisions due to questionable analytical validity. The International Ki67 in Breast Cancer Working Group (IKWG) consensus meeting, held in October 2019, assessed the current evidence for Ki67 IHC analytical validity and clinical utility in breast cancer, including the series of scoring studies the IKWG conducted on centrally stained tissues. Consensus observations and recommendations are: 1) as for estrogen receptor and HER2 testing, preanalytical handling considerations are critical; 2) a standardized visual scoring method has been established and is recommended for adoption; 3) participation in and evaluation of quality assurance and quality control programs is recommended to maintain analytical validity; and 4) the IKWG accepted that Ki67 IHC as a prognostic marker in breast cancer has clinical validity but concluded that clinical utility is evident only for prognosis estimation in anatomically favorable estrogen receptor–positive and HER2-negative patients to identify those who do not need adjuvant chemotherapy. In this T1-2, N0-1 patient group, the IKWG consensus is that Ki67 5% or less, or 30% or more, can be used to estimate prognosis. In conclusion, analytical validity of Ki67 IHC can be reached with careful attention to preanalytical issues and calibrated standardized visual scoring. Currently, clinical utility of Ki67 IHC in breast cancer care remains limited to prognosis assessment in stage I or II breast cancer. Further development of automated scoring might help to overcome some current limitations.
5% or less, or 30% or more, can be used to estimate prognosis. In conclusion, analytical validity of Ki67 IHC can be reached with careful attention to preanalytical issues and calibrated standardized visual scoring. Currently, clinical utility of Ki67 IHC in breast cancer care remains limited to prognosis assessment in stage I or II breast cancer. Further development of automated scoring might help to overcome some current limitations.
In the era of “precision medicine,” the availability of high-quality tumor biomarker tests with proven analytical validity and clinical utility is critical. For example, estrogen receptor (ER) and HER2 content are strong predictive factors for antiestrogen therapies (1,2) and anti-HER2 therapies, respectively (3,4).
For 30 or more years, measures of cellular proliferation in breast cancer have been proposed as an indication of prognosis and perhaps prediction of benefit from antineoplastic therapies (5). Although several studies have suggested that cancers with high vs low proliferation have a worse prognosis, analytical issues have prevented widespread adoption of these measures to drive patient care (6-9). Cellular proliferation can be measured in several ways, including biological assays, such as analysis of thymidine uptake (10); flow cytometry to determine the percent of cells in S-phase (5), and, most commonly, the use of immunohistochemistry (IHC) assays to measure Ki67 (11), a nuclear marker expressed in all phases of the cell cycle other than the G0 phase (12).
Establishment of the International Ki67 in Breast Cancer Working Group
In 2011, we established the International Ki67 in Breast Cancer Working Group (IKWG) to review methods of determination of Ki67 levels in breast cancer (11). Because of the multiplicity of assays and the apparent poor standardization of them for this marker, we set out to establish internationally acceptable methods for the determination of Ki67. Since 2011, as described in the remainder of this article, we and others have made substantial efforts to address both the technical and scoring aspects of Ki67 assessment. Nonetheless, current guidelines remain skeptical about the technical validity of Ki67 IHC assays. For example, the recent American Joint Committee on Cancer (AJCC) guideline (13) on cancer staging states “As a single factor, Ki-67 was not considered a reliable factor for implementation in clinical practice … because of the known lack of reproducibility (especially between different laboratories).” Consistent or similar statements are present in many national and international guideline documents (8).
The IKWG met in October 2019 to review our own progress in standardizing Ki67 analysis, discuss relevant literature known to the participants, develop recommendations and guidelines for the use of Ki67 IHC to drive patient care, and determine what future research questions remain to be addressed regarding Ki67 assessment. The pivotal question for creating these recommendations is whether there is now sufficient high-level evidence to demonstrate that a robust and analytically validated approach to Ki67 IHC exists, and if so, does this tumor biomarker test have clinical utility for any intended use? Without such evidence, Ki67 analysis should not be considered satisfactory for directing routine clinical care.
Tumor Biomarker Application in the Clinic: Important Semantics
In 2009, Teutsch et al. (14), representing the Evaluation of Genomic Application in Practice and Prevention (EGAPP) initiative, described 3 critical elements to determine if a germ-line genetic test should be used to manage care: 1) analytical validity, 2) clinical validity, and 3) clinical utility. Building on the EGAPP initiative, the United States Food and Drug Administration and the National Institutes of Health convened a group of stakeholders to jointly develop a glossary to better define and harmonize biomarker terminology: the Biomarkers, Endpoints, and Other Tools Resource (15). Broadly, the Biomarkers, Endpoints, and Other Tools Resource defines the acceptability of analytical validity in terms of its sensitivity, specificity, accuracy, precision, and other relevant performance characteristics. The level of acceptability of analytical performance of Ki67 differs among applications and intended uses.
Clinical validity implies that a tumor biomarker test divides 1 population into 2 or more with distinct biological characteristics or clinical outcomes with reasonable accuracy, but it does not imply that the biomarker assay should be used to direct patient care (16). Rather, there are 2 components that determine whether a tumor biomarker test such as Ki67 with established clinical validity should be incorporated into clinical decision making: 1) does it have analytical validity, and if so 2) does it have clinical utility? Clinical utility has been defined by EGAPP as “evidence of improved measurable clinical outcomes and (a test’s) usefulness and added value to patient management decision-making compared with current management without testing.” (14) A similar definition was adopted by the National Academy of Medicine (16).
There are several potential intended uses for Ki67, and clinical utility must be determined for each. We have largely focused on the most prominent potential application, estimation of prognosis in the absence of a subsequent treatment, which may be better designated “residual risk.” There are a limited number of studies that indicate that Ki67 may also be predictive of whether further therapy, if needed, is likely to work, and serial levels may provide a real-time indication of whether it is working. For example, low expression of Ki67 may be associated with prediction of poor or no benefit to cytotoxic chemotherapy (17). Furthermore, serial assessment is used by some to monitor the effects of presurgical systemic treatment. Analysts should consider how the acceptable limits may differ for different intended uses.
The efforts of the IKWG over the last decade aimed first to determine analytical validity for Ki67 IHC and to promote standardization. The IKWG has not yet formally addressed clinical utility. At the October 2019 meeting, the IKWG addressed several questions that were considered essential in order for Ki67 to be used to manage patient care.
Does Ki67 IHC Have Analytical Validity? Methodological Issues in Ki67
As outlined in our original article (11), the key element preventing implementation of Ki67 as a diagnostic assay has been a lack of analytical validity. For any tumor biomarker test, including Ki67, there are many factors that may affect the result, including collection, processing, and archiving of the specimen (preanalytical) to staining, analysis, and reporting (analytical), and finally to ensuring ongoing quality of the analytical assessment.
Preanalytical Considerations
As with all diagnostic procedures performed on formalin-fixed paraffin-embedded pathology blocks, the handling and processing of tissues before analysis is critical. For example, the criteria for appropriate collection, fixation, and processing of breast cancer specimens for HER2 and hormone receptor testing are clearly described in guidelines developed jointly by the American Society of Clinical Oncology and the College of American Pathologists (ASCO and CAP) (18,19). These recommendations include minimization of prefixation delays, division of surgical specimens to 5- to 10-mm slices for fixation, and fixation in neutral buffered formalin for 6-72 hours. No specific additional requirements for Ki67 testing have been identified at this time. Ki67 is a robust antigen originally selected in part for its favorable properties for IHC analyses from among the many available proliferation-associated proteins expressed in human cells. Ki67 IHC appears to be more sensitive than ER or HER2 to variabilities in fixation. Ki67 index values decrease with use of fixatives other than neutral buffered formalin, with delays of 16
hours. No specific additional requirements for Ki67 testing have been identified at this time. Ki67 is a robust antigen originally selected in part for its favorable properties for IHC analyses from among the many available proliferation-associated proteins expressed in human cells. Ki67 IHC appears to be more sensitive than ER or HER2 to variabilities in fixation. Ki67 index values decrease with use of fixatives other than neutral buffered formalin, with delays of 16 hours or more before fixation, or with overly short (3
hours or more before fixation, or with overly short (3 hours) or long (14
hours) or long (14 days) fixation times (20). The IKWG recommends that breast cancer samples for Ki67 testing be processed in line with ASCO and CAP guidelines for HER2 and hormone receptors, and ideally tested on core needle biopsies in preference to excision specimens, because doing so will preclude many fixation problems. Ki67 is also more sensitive to antigen decay with long-term storage in paraffin (21), and for this reason the IKWG recommends that Ki67 IHC be performed within 5 years from the time of tumor placement into paraffin blocks. Because the exact mechanism and timing of the epitope degradation are unknown, we are unable to make a more specific recommendation but express concern about the accuracy of Ki67 assessment in tissue collected many years ago.
days) fixation times (20). The IKWG recommends that breast cancer samples for Ki67 testing be processed in line with ASCO and CAP guidelines for HER2 and hormone receptors, and ideally tested on core needle biopsies in preference to excision specimens, because doing so will preclude many fixation problems. Ki67 is also more sensitive to antigen decay with long-term storage in paraffin (21), and for this reason the IKWG recommends that Ki67 IHC be performed within 5 years from the time of tumor placement into paraffin blocks. Because the exact mechanism and timing of the epitope degradation are unknown, we are unable to make a more specific recommendation but express concern about the accuracy of Ki67 assessment in tissue collected many years ago.
Analytical Considerations: Visual Interpretation of Ki67 Index
The major focus of the IKWG has been interpretation of already processed and stained tissue. For immunohistochemical assays (both fluorometric and colorimetric), analytical validity requires both robust assay performance with universal standards (staining) and reporting (scoring). During the first IKWG workshop in 2011, it was clear that there were several different methods of scoring IHC-stained slides to determine Ki67 values (11). Therefore, the IKWG undertook a series of carefully planned, incremental, multi-institutional studies, and the results are summarized below (Figure 1):
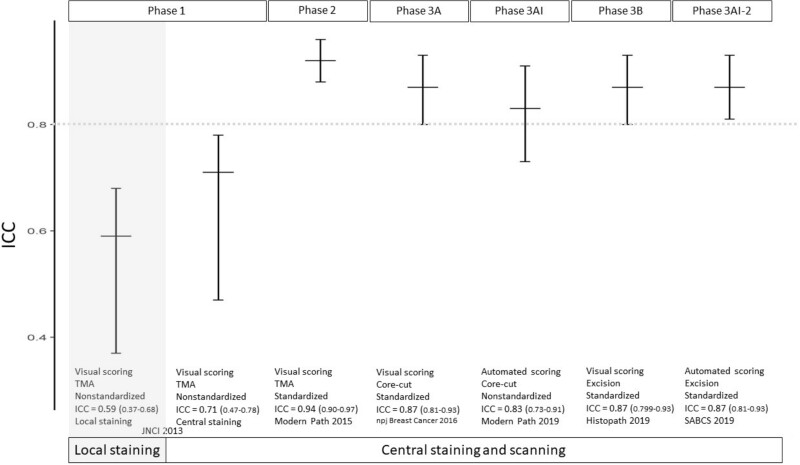
The series of International Ki67 Working Group (IKWG) studies to standardize methods for visual scoring of Ki67 index in breast cancer. Intraclass correlation coefficient (ICC) through the 3 study phases (1, 2, 3 A visual and automated [3AI], 3B visual and automated [3AI-2]) are shown with error bars representing the lower and upper 95% credible intervals. The numeric values of the various ICCs are shown at the x-axis labels with the 95% credible intervals in parentheses. The horizontal bar lines represent observed ICCs. The extent of the vertical lines indicates 95% credible interval. The dotted grey color line indicates ICC = 0.8. TMA = tissue microarray.
A visual and automated [3AI], 3B visual and automated [3AI-2]) are shown with error bars representing the lower and upper 95% credible intervals. The numeric values of the various ICCs are shown at the x-axis labels with the 95% credible intervals in parentheses. The horizontal bar lines represent observed ICCs. The extent of the vertical lines indicates 95% credible interval. The dotted grey color line indicates ICC = 0.8. TMA = tissue microarray.
Phase 1: Intra- and Inter-Laboratory Scoring of Ki67 IHC Stained Tissue Microarrays in the Absence of Standardized Methodology (22). In this study, a series of staining and scoring experiments was conducted among several expert laboratories that were provided a tissue microarray of 100 breast cancer cases. Ki67 scoring was internally consistent, with an intraclass correlation (ICC) of 0.94, but inter-observer variability in scoring of centrally stained slides (ICC = 0.71) and inter-laboratory variability that reflected both local staining and scoring (ICC = 0.59) were not satisfactory. Indeed, the IKWG data suggested that among the many sources of variability in Ki67, scoring differences were considerably more important than preanalytical or even staining issues.
Phase 2: Intra- and Inter-Laboratory Scoring of Centrally Stained Tissue Microarrays Using a Uniform Method of Scoring (23). In this study, an online calibration exercise (for color thresholds and tumor cell selection) and a standardized nuclei counting method were introduced to improve reproducibility. Inter-observer variability (ICC = 0.92, 95% credible interval [CI] = 0.88 to 0.96) then met the prespecified criteria for success.
Phase 3: Intra- and Inter-Laboratory Scoring of Centrally Stained Core Cut Biopsies and Full Face Excision Sections Using the Uniform Method of Scoring (24). We next extended the uniform scoring method from tissue microarrays to clinical diagnostic formats: core needle biopsies and excision specimens (25), and considered the variability from field selection and the impact of “hot spot” vs “global” counting (26). The prespecified criterion for success was met with the global method in core biopsies (ICC = 0.87, 95% CI = 0.81 to 0.93); similar values were obtained on excisions (Figure 1). In contrast, the hot spot method was associated with higher variability and lower ICC (core biopsy: ICC = 0.84, 95% CI = 0.77 to 0.92; excision: ICC = 0.83, 95% CI = 0.74 to 0.90). The scoring software application for these methods is publicly available (https://www.ki67inbreastcancerwg.org/).
Support for IKWG Scoring System for Ki67. Taken together, this logical progression of studies demonstrates that the IKWG scoring system (Box 1) is reproducible across observers. The IKWG proposes that the following criteria should be applied to achieve analytical validity when developing a method to read Ki67 IHC (or for that matter, any IHC assay):
Studies should include a sufficiently large number of participating scorers to represent variability inherent in a broad cross-section of pathology interpretations;
Observers doing the scoring in test validation studies need to follow prespecified training methods and score independently and in a fashion blinded to others’ scores;
A sufficient number of specimens should be included to have adequate statistical power, and the specimens should represent the entire dynamic range of the assay (in the case of Ki67 IHC, 0%-100%);
Although the expected implementation of tests is often categorical, based on 1 or more cutpoint(s), most tumor biomarkers (including Ki67) are continuous variables, and data for assessing analytical validity should be captured as such. Doing so will allow for parametric tests that maximize information, and for results to be transposed to alternative cut-points of clinical relevance. The data distribution for Ki67 is log-normal, meaning that log transformation is required to satisfy the normal distribution and constant variance assumptions underlying common parametric statistical tests.;
Studies using biospecimens or linking to prognosis should adhere to Biospecimen Reporting for Improved Study Quality (BRISQ) (27) and Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) guidelines (28,29), including such important features as transparent and detailed reporting of scoring methods so others can apply the system exactly and prespecified metrics of success, ideally with independent statistical analysis.
We do not imply that the IKWG system is the only scoring system that has analytical validity. However, it fulfils these criteria. Many other scoring systems have been published (30-38). Some of these provide faster and more convenient methods by counting fewer cells, making global estimations without specific cell counting, or using hot spot scoring methods. However, these published scoring systems have not met all the above criteria.
Multiple caveats remain even for the IKWG method. The final scoring method (Box 1) is arguably tedious and requires training on the calibration set, installation of software, and a median of 9 minutes scoring time per case. This level of attention to training, and time required to perform each assay, may be challenging to achieve in the routine pathology laboratory setting. The absolute magnitude of this residual variability, particularly in the clinically relevant Ki67 index range of 10%-20%, could still cause misassignment of some cases.
minutes scoring time per case. This level of attention to training, and time required to perform each assay, may be challenging to achieve in the routine pathology laboratory setting. The absolute magnitude of this residual variability, particularly in the clinically relevant Ki67 index range of 10%-20%, could still cause misassignment of some cases.
Importantly, in our studies, kappa values in this cutoff range were near 0.6. Indeed, all participating laboratories agreed on categorizing a case as “low” or “high” Ki67 only for cases with a median Ki67 index of 5% or less or 30% or greater, respectively (25), consistent with results from other groups (39,40), and this observation has strongly influenced our recommendations regarding the clinical utility of Ki67 IHC, discussed below.
5% or less or 30% or greater, respectively (25), consistent with results from other groups (39,40), and this observation has strongly influenced our recommendations regarding the clinical utility of Ki67 IHC, discussed below.
Other Methodological Considerations for Ki67 IHC That May Affect Analytical Validity
Staining Protocols
The efforts of the IKWG have been focused on standardizing the scoring of prestained tissue for Ki67. However, it should be recognized that there are multiple sources of inter-laboratory variation due to differences in staining methodologies, including staining platform, antigen retrieval, primary antibody, detection system, and counterstain (Table 1). Published results from external quality assessment schemes generally focus on “quality” of staining as opposed to the impact on assay results (39,41,44-47). Kappa values for dichotomized comparisons of IHC for different antibodies were “good” despite evidence of statistically significant differences between labelling indices for these same comparisons (45). Further, some multi-parameter prognostic signatures that incorporate Ki67 may be more tolerant of laboratory-to-laboratory variation because Ki67 constitutes only part of the respective prognostic score (34). These factors should be considered when setting criteria for assessment of the impact of variations in staining on Ki67 scores and in particular to ensure that external quality assurance (EQA) schemes, as discussed below, set standards reflecting these observations.
Table 1.
Factors that may affect Ki67 IHCa
| Setting | Factor | Variables | Comments |
|---|---|---|---|
| Preanalytical | Type of specimen | Core vs excision | Both are suitable, but core biopsies are preferred. Use case must be specimen type specific, eg, cutpoint for core cut may differ from excision; changes in Ki67 at multiple time points must be based on measurement on the same specimen type. |
| Fixation | Prefixation delays (warm and cold ischemia time); tissue thickness; fixative type; time spent in fixative) | Affects morphologic nuclear integrity and intensity of nuclear IHC stain. Inadequate fixation decreases Ki67 (20). Ethanol-fixed or decalcified preparations should not be used. ASCO/CAP guidelines for breast tissue handling for ER/HER apply (19). | |
| Means of storage | Tissue in paraffin block vs unstained slides | Prolonged storage of formalin-fixed paraffin-embedded tissue block at room temperature has little effect on Ki67 (21). Avoid prolonged exposure to air of cut sections on glass slides. | |
| Analytical | Antigen retrieval | Yes vs no | Required. High-temperature antigen retrieval mandatory. |
| Specific antibody | MIB1 vs other antibodies against Ki67 antigen | MIB1 is the most widely validated antibody; 30-9, K2, MM1, and SP6 are also commonly used. Particular automated immunostainers have recommended antibodies (eg, MIB1 for Dako, 30-9 for Ventana, K2 for Leica). Some evidence indicates poor performance of MM1 (41), although this might be confined to its use on non-Leica platforms (42). | |
| Colorimetric detection system | Avidin-biotin immunoperoxidase vs polymer detection vs amplified systems | Avidin-biotin systems have substantially lower sensitivity and have largely been replaced by polymer detection (43) on automated platforms. Amplified systems such as OptiView+Amp (Ventana) produce powerful, open-ended amplification that is difficult to standardize (UK NEQAS internal observations). | |
| Counterstain | Completeness and intensity of stain | Important that all negative nuclei are counterstained (otherwise apparent Ki67 index can be falsely high). | |
| Quality assurance/quality control |
— | Should be established in each laboratory and systematically maintained. Quantitative external quality assessment should be established and participation should be mandatory. | |
| Interpretation and scoring | Method of scoring | Cellular component, staining intensity |
1) Count all positive invasive carcinoma cells within region in which all nuclei have been stained. 2) Scoring requires determination of percentage cells positive among total number of invasive cancer cells. 3) No interpretation of intensity. |
| Area of slide read | Average value across slide vs value in hot spot | Controversial: global (average) scores across the section had higher reproducibility than hot spot methods in IKWG studies, although differences were not statistically significant. | |
| Digital imaging | Visual vs automated analysis | IKWG-standardized visual counting (Box 1) under light microscopy or from a digital image is validated. Automated scoring is still investigational, but evidence to date suggests that automated score is not worse than standardized visual scoring for core-cuts. | |
| Data format and cutpoints | Categorical or continuous | Capture Ki67 data as a continuous percentage variable rather than in bins relative to specific cutpoint(s). Log transformation is required for parametric statistical testing. |
In summary, the IKWG maintains that the different but commonly used Ki67 staining protocols need to be evaluated for consistent, inter-laboratory reproducibility. The results of such protocols then need to be coupled with existing EQA schemes, as described below, if one is to implement these assays in a clinical setting.
Non-IHC Methods for Determination of Proliferative Index
The IKWG considered other tools for the assessment of proliferation in malignancies, such as measurement by gene expression rather than IHC, or of IHC by automated technology.
Multigene Expression Signatures.
As assessed by quantitative mRNA measurement techniques, these methods are already stringently validated at a technical level (48,49). Many provide broader assessment of proliferation than a single marker, such as is provided by Ki67 IHC, and may provide a more robust, precise, and ultimately informative solution. However, beyond cost and accessibility issues, the relationship of the specific proliferative component of these signatures with clinical outcome varies between them (50).
Automated Scoring by Digital Image Analysis.
The variability in scoring of Ki67 IHC suggests that its advancement to the clinic as a useful biomarker may be aided by automated scoring (51). There are many commercial or open-source platform approaches to image analysis and quantification (52-54).
Similar to our studies of visual scoring, the IKWG undertook studies of the use of a range of platforms and software to assess the feasibility of the introduction of automation to scoring of Ki67 (Figure 1). We investigated 10 different software platforms using 7 different scanners and observed an ICC = 0.83 (95% CI = 0.73 to 0.91). Different scanners and analysis systems provided different scores from one another. Nonetheless, for 8 sites using the same scanner, the ICC for average automated scores was 0.89 (95% CI = 0.81 to 0.96), which exceeded our prestudy criterion for success and was similar to the ICC of 0.87 (95% CI = 0.81 to 0.93) achieved in our optimized pathologist-based scoring analysis (Figure 1).
In a pilot study of a low-cost, broadly inclusive, single open-source platform (QuPath), 10 sites had a preliminary ICC in the 0.95 range (55). In a follow-up study involving 17 sites, only 6 were able to perform the entire analysis without central laboratory assistance. However, when successfully incorporated, this software again resulted in ICCs of approximately 0.9 (53). None of the automated analysis studies included plans for inter-platform standardization.
Internal Standards
In other areas of laboratory medicine, absolute standards with known quantities of the analyte are used to produce internal standard curves. In contrast, IHC has been historically dependent on the number of cells that are positive at any intensity without internal standards. In this regard, the ASCO and CAP guidelines call for inclusion of proper controls, if not standards, in the IHC evaluation of ER, PgR, and HER2 (18,56). Ki67 represents a further challenge because small quantitative errors in assay results may markedly affect patient treatment and outcome. Members of the IKWG are cooperating with commercial vendors to achieve such a standard that is low cost, inexhaustible, and accurate. The IKWG likewise recommends use of internal on-slide and batch-to-batch controls (positive and negative), regardless of whether the reading is performed manually or by automated systems. Although such a resource is not currently available, the IKWG is currently working to develop a cell-line–based standard Ki67 index tissue microarray for such a purpose.
Role of EQA
EQA is the process of centrally assessing the results of a procedure, such as IHC staining for Ki67, from multiple laboratories so that each can benchmark their results against those of their peers either locally or beyond. EQA is a critical prerequisite to allowing Ki67 to be widely used for clinical management; regular participation in such schemes is known to markedly enhance between-laboratory concordance (57). Unfortunately, although a number of schemes exist at the moment, none provides a comprehensive assessment that includes comparison of the values of Ki67 reported in different laboratories. CAP and UK National External Quality Assessment Scheme are in the planning and implementation stages for such schemes, but they will not be available until late 2021 or 2022.
The UK National External Quality Assessment Scheme for Immunocytochemistry and In-Situ-Hybridisation (58) has assessed the quality of staining for Ki67 to determine differences between the quality of staining with different antibody clones in general and when used on particular analytical platforms (42). Other countries have also established EQA systems, including the Nordic immunohistochemical Quality Control (59), which has reported statistically significant differences in the mean Ki67 score obtained using different antibody clones, different formats of those clones, and different platforms (41,60). Likewise, the Qualitätssicherungs-Initiative Pathologie, a joint venture between the German Society of Pathology and the German Association of Pathologists, has conducted Ki67 assessment in breast cancer specimens (61) on an annual basis since 2002. On average, 95% of participants reached the benchmark of over 80% concordance rates, with the Ki67 category preestablished by the panel (39). Despite the potential inter-laboratory and inter-observer variance of Ki67, its prognostic effect remained demonstrable in a large dataset from Bavaria (62). The results of these studies support the major impact that EQA could have on achieving the necessary precision for KI67 to be used as a clinical tool with confidence.
Ki67 IHC Clinical Utility
Although other indications (eg, predicting benefit from radiation therapy) are the subject of active investigation, at present there are fundamentally 3 intended uses for Ki67 IHC: 1) to estimate prognosis in early-stage disease regarding whether further adjuvant chemotherapy is warranted, 2) to predict whether chemotherapy may or may not be active, and 3) to monitor patients during or after neoadjuvant endocrine or chemotherapy to determine if the regimen chosen is working or an alternative should be considered.
Ki67 to Estimate Prognosis in Early-Stage Disease to Inform Whether Further Adjuvant Chemotherapy Is Warranted
Adjuvant systemic therapy clearly improves outcomes for patients with early-stage invasive breast cancer (2,63,64). Selection of adjuvant endocrine therapy and adjuvant anti-HER2 therapy is based on the predictive factors ER and HER2, respectively. The choice of adjuvant chemotherapy is principally based on estimation of prognosis, also designated residual risk, given that the absolute (as opposed to proportional) benefit from chemotherapy is determined by baseline prognosis.
Several multi-parameter, gene expression molecular profiling assays (eg, OncotypeDX, Prosigna, EndoPredict, Mammaprint, Breast Cancer Index, Genomic Grade Index) have been developed to estimate the residual risk for distant recurrence if patients with ER-positive early-stage breast cancer are treated with endocrine therapy alone. With this information, the clinician can estimate if a patient has such a favorable prognosis that she can safely forego adjuvant chemotherapy (8,65). However, rather than performing highly sophisticated molecular assays, Ki67 by IHC has been considered to play a similar role, because higher levels relate to poorer prognosis of ER-positive breast cancer. Investigators have proposed that analysis of 4 proteins by IHC, namely Ki67, ER, PgR, and HER2, might be able to reflect prognosis with sufficient accuracy. Such an assay, designated IHC4, might be more generally accessible and provide similar results at a lower cost (66) if these assays were sufficiently controlled and standardized.
Many studies confirm that Ki67 is prognostic in early-stage breast cancer (11,67). However, these studies display substantial variability in both analytical validity and choice of cutoffs, and most have been studies of convenience in which selection of patients, the intrinsic subtypes of the cancers, and application of therapies have been mixed or often not even reported (67). Even when applied to clinical trials in more carefully designed studies (68-70), analytical validity of the specific Ki67 methodologies used has not been formally proven across distributed laboratories, limiting the clinical utility of Ki67 determined by IHC (8,9,71).
Overall, the IKWG does conclude that Ki67 IHC using a highly analytically validated assay and scoring system, as described above, might be used for this specific intended use, but in a very limited fashion. As noted, concordance among the IKWG investigators was extremely high for specimens that were 5% or less or 30% or more (nearly unanimous agreement). Therefore, we agreed that KI67 IHC could be used to withhold or proceed with chemotherapy if the results are below or above these thresholds, respectively, without the need for more expensive commercial multi-parameter gene expression assays. We do not recommend making this decision for patients with Ki67 IHC between 5% and 30%, because concordance was less than acceptable.
To Predict Whether Chemotherapy May or May Not Be Active
If a patient is considered to have a sufficiently high residual risk of subsequent recurrence, the clinician must then decide which adjuvant systemic therapies to recommend. As noted, ER and HER2 are potent predictive factors for activity of endocrine treatment (ET) and anti-HER2, and their use is widely recommended independent of any measure of proliferation, including Ki67 (72,73). In contrast, identification of a reliable predictive factor for chemotherapy in general, or for specific chemotherapy agents, has been problematic.
It has been hypothesized that tumors with low proliferative thrust might be resistant to chemotherapy, because most cytotoxic agents require cells to be in the cell cycle (74). In general, ER-negative breast cancers, which are generally more highly proliferative, are more likely to respond to chemotherapy than those that are ER positive in both the metastatic and neoadjuvant settings (75-78). Furthermore, retrospective studies of additional or more aggressive vs standard adjuvant chemotherapy have suggested that either low ER or high HER2 is associated with better apparent responses to adjuvant chemotherapy than are seen in patients with ER-positive or HER2-negative disease (79).
However, this hypothesis has not been sufficiently validated to change clinical practice (74). Of particular note, results from the worldwide Early Breast Cancer Trialists’ Collaborative Group suggest that the relative reduction in distant recurrences is the same regardless of hormone receptor status or grade (64,80).
Once again, though, poor analytical validity and inconsistent study designs have confounded studies of this issue. It is not clear that patients with anatomically poor prognosis (positive nodes, large tumor size, etc) but favorable biology (ER rich, HER2 low, and Ki67 low) cancers can safely avoid adjuvant chemotherapy based on the theory that it will not work. This question is currently the focus of an ongoing prospective randomized trial (RxPonder) (74). The IKWG does not recommend use of Ki67 IHC to direct care, such as withholding chemotherapy based on low Ki67 from patients with poor anatomic prognosis.
Ki67 in Neoadjuvant Therapy
Neoadjuvant Endocrine Therapy.
Downstaging of breast tumors using presurgical (also designated “neoadjuvant”) ET for ER-positive disease is most frequently practiced in elderly women who may not be sufficiently robust to tolerate chemotherapy. Measurement of Ki67 before neoadjuvant treatment has not been considered to play a role in selection of neoadjuvant ET for other patients in standard care. However, in clinical trials in patients of all ages, reductions in Ki67 in serial biopsies during short-term (2-4 weeks) or long-term (>3
weeks) or long-term (>3 months) ET, compared with lack of reduction, are associated with improved outcomes. In addition, Ki67 expression after 2
months) ET, compared with lack of reduction, are associated with improved outcomes. In addition, Ki67 expression after 2 weeks of ET has been found to be more strongly prognostic than baseline values, probably because it is a derivative of both the prognostic value of baseline Ki67 and the suppressive effect that ET has on Ki67 in responsive patients (81). It is important to keep these 2 uses separate (early change vs residual value of Ki67), but they can overlap in interpretation. The first is used to determine if a patient is likely to benefit from current (and hence ongoing) therapy, whereas the second relates to whether that patient has a sufficiently high residual risk after several months to justify additional treatment, such as chemotherapy.
weeks of ET has been found to be more strongly prognostic than baseline values, probably because it is a derivative of both the prognostic value of baseline Ki67 and the suppressive effect that ET has on Ki67 in responsive patients (81). It is important to keep these 2 uses separate (early change vs residual value of Ki67), but they can overlap in interpretation. The first is used to determine if a patient is likely to benefit from current (and hence ongoing) therapy, whereas the second relates to whether that patient has a sufficiently high residual risk after several months to justify additional treatment, such as chemotherapy.
In the PeriOperative Endocrine Therapy-Individualised Care trial, short-term changes in Ki67 were determined using a method closely similar to the IKWG-endorsed global method within a single central laboratory at baseline and after 2 weeks of aromatase inhibitor therapy (82). In the HER2-negative subpopulation, women with Ki67 greater than 10% at baseline that converted to less than 10% at 2 weeks had lower 5-year absolute recurrence risk (8.4%) than patients with Ki67 IHC that remained greater than 10% (21.5%). Assessment of Ki67 after 2 weeks of aromatase inhibitor provided substantially more prognostic information for those who had high baseline Ki67. In the Z1031 trial, patients with ER-positive primary breast cancers received 2-4
weeks had lower 5-year absolute recurrence risk (8.4%) than patients with Ki67 IHC that remained greater than 10% (21.5%). Assessment of Ki67 after 2 weeks of aromatase inhibitor provided substantially more prognostic information for those who had high baseline Ki67. In the Z1031 trial, patients with ER-positive primary breast cancers received 2-4 weeks of preoperative aromatase inhibition. If Ki67 was greater than 10% at the end of this period, patients were switched to chemotherapy, based on the assumption that the therapy had not achieved sufficient benefit (83). However, despite the relatively high residual Ki67, the rate of pathological complete response after chemotherapy was only 5.7%, arguing against a “predictive” effect of Ki67 for chemotherapy response. Other studies addressing this issue are ongoing (84).
weeks of preoperative aromatase inhibition. If Ki67 was greater than 10% at the end of this period, patients were switched to chemotherapy, based on the assumption that the therapy had not achieved sufficient benefit (83). However, despite the relatively high residual Ki67, the rate of pathological complete response after chemotherapy was only 5.7%, arguing against a “predictive” effect of Ki67 for chemotherapy response. Other studies addressing this issue are ongoing (84).
Besides indicating a decision after short-term exposure to ET regarding chemotherapy or not, changes in Ki67 during or after neoadjuvant systemic therapy have been used as a pharmacodynamic approach to drive selection and/or assess potential benefit of other treatments, such as CDK4/6 inhibitors (85-87), chloroquine (88), vitamin D (89), the AKT inhibitor capivasertib (90), the CXCR1/2 inhibitor reparixin (91), the tyrosine kinase inhibitor lapatinib (92), and the cyclooxygenase-2 inhibitor celecoxib (93). However, the mode of action for several of these agents is not antiproliferative. In such cases, although changes in Ki67 may be of interest, they should not be considered to be valid endpoints of pharmacologic or therapeutic activity.
Ki67 changes after long-term preoperative ET (several months) have been investigated as an indicator of sufficiently high residual risk to justify additional treatments after surgery. In the Z1031 trial, the preoperative prognostic index (PEPI) is a weighted multi-factoral algorithm consisting of residual tumor size, node status, ER status, and Ki67 (94). One-quarter of patients had a PEPI score of 0 (highly favorable), and these patients had exceptionally good prognosis. These Ki67 triage and PEPI approaches are being studied further in the ALTERNATE trial (NCT01953588).
It should be emphasized that these uses of Ki67 in neoadjuvant ET are currently investigational, and the IKWG do not recommend their use to optimize treatment for individuals in standard care. In each of these trials, Ki67 was assayed and scored in a single, central laboratory for each trial in which high concordance among the readers was determined.
Neoadjuvant Chemotherapy.
Like other measures of tumor cell proliferation, Ki67 before treatment is associated with a greater likelihood of pathological complete response to chemotherapy (95). However, as noted, baseline Ki67 has not been used as a major criterion for selecting patients for chemotherapy because it is not clear that the lower pathologic complete response rate for tumors with low Ki67 is sufficient to withhold it. Furthermore, unlike the situation with neoadjuvant ET, early reductions in Ki67 are modest in the early phases of neoadjuvant chemotherapy and have not been found to have a sufficiently close relationship with outcome to merit their use for subsequent modification of treatment (96). Nevertheless, at the end of neoadjuvant therapy, Ki67 in residual disease has a strong correlation with long-term outcome (97,98). For example, the residual proliferative cancer burden index integrates postneoadjuvant Ki67 with the residual cancer burden after neoadjuvant chemotherapy, and it provides a more robust estimate of the risk of subsequent recurrence than either the residual cancer burden or Ki67 measurement alone (99,100). Therefore, neoadjuvant studies are now being conducted in which patients who have substantial clinically or radiographically manifested residual disease after chemotherapy are treated with innovative therapies for a short period of time and then undergo surgical excision. Reductions in Ki67 are considered evidence of potential activity of these agents (PHOENIX, NCT03740893).
Special Issues for Assessment of Ki67 in Neoadjuvant Treatment.
There are several important issues that need to be considered before Ki67 IHC is incorporated into standard clinical practice for any of the described presurgical uses. First, as noted, for all the investigational neoadjuvant studies described, Ki67 measurements were performed by a selected group of highly trained observers using strict preanalytical and analytical protocols. One study has compared digital image analysis with exact counting in a large clinical trial cohort and found a strong prognostic impact of both methods (101). Sequential Ki67 analyses during treatment frequently involve comparison of biopsies at surgery with those taken at baseline or during early treatment. Whereas the excised biopsy is a convenient source for such evaluations, the longer time for full fixation of excision compared with core-cut needle biopsies may lead to lower values of Ki67 (82). It is therefore strongly recommended that all samples for Ki67 IHC be taken by core-cut, which may involve taking cores close to, or at the time of, surgery for the end of treatment measurement. If the excision biopsy must be used, time to fixation should be minimized and studies undertaken to identify if meaningful differences in Ki67 need to be acknowledged.
Using Ki67 IHC to compare likelihood of activity between 2 antiproliferative agents with confidence may require larger numbers of cells to be counted than for prognostic estimates. Like most other biomarkers, there is substantial variation in Ki67 measurement across a single hot spot (25). The potential for this heterogeneity to confound differences between sequential measures made in the same tumor should be borne in mind.
Cut-Point Selection for Clinical Application of Ki67
The issue of cutoff selection for IHC determination of Ki67 is particularly relevant. In the IKWG phase 1 and 2 exercises reported above, substantial inter-observer/laboratory variability is observed in the range of >5 to <30%, which is where most investigators have selected cutoffs. As discussed above, based on the results of all 3 IKWG study phases, particularly the residual assay variability in this critical range for treatment decisions, the consensus at the 2019 workshop was that, without improvements in standardization, only very low (≤5%) and very high (≥30%) values can be reliably categorized as low or high by visual scoring of Ki67 IHC in routine, nontrial settings (as opposed to clinical trial settings, where Ki67 IHC is usually performed in a single, central testing site) (22). Therefore, the IKWG recommends that Ki67 analysis only be used to drive patient care for cases with 5% or less or 30% or more unless centers have carefully calibrated their assay performance against clinical outcome for a specific intended use with high levels of evidence to support doing so.
Our overall consensus findings regarding Ki67 IHC are provided in Figure 2. In summary, when considering Ki67 IHC in breast cancer, the IKWG strongly recommends careful attention to preanalytical and staining protocols as well as use of a highly standardized and now validated scoring system. Because staining protocols are not as standardized and validated as scoring, we strongly recommend use of internal standards and participation in well-designed and conducted quality assurance (QA) and quality control (QC) programs. We also recommend careful evaluation and if necessary, enhancement of existing QA and QC programs for their adequacy in dealing with variabilities specific to Ki67 IHC. Educational resources such as training module and scoring protocols as well as literature updates from IKWG are available at https://www.ki67inbreastcancerwg.org/.
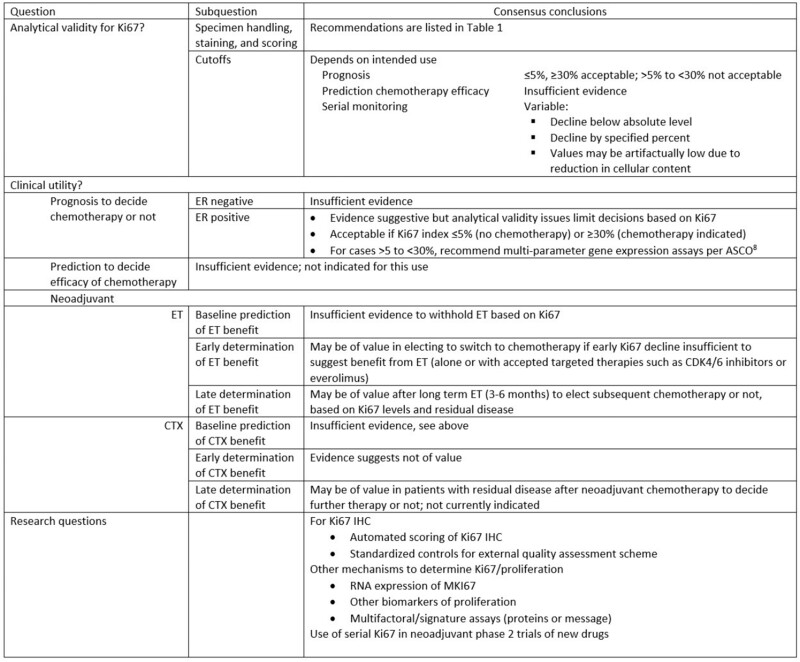
Conclusions regarding the questions for Ki67 in breast cancer. ET = endocrine therapy; CTX = chemotherapy; ASCO = American Society of Clinical Oncology.
The general consensus of the IKWG is that Ki67 IHC does have clinical validity for the determination of prognosis in patients with early stage breast cancer. However, we determined that its clinical utility has been demonstrated only for a very small, intended use: to eliminate the need for multi-parameter gene expression assays in women with ER-positive and favorable anatomic prognoses, if the Ki67 levels are 5% or less or 30% or more. We would like to point out, however, that use of Ki67 as suggested here would lead to reduced costs in the health-care system as well as faster clinical decision making. For other intended uses, insufficient high levels of evidence are available to support its routine use. The IKWG does recognize the value of using Ki67 IHC in clinical trial settings for both prediction and monitoring, but this use is investigational. Positive studies using Ki67 as a companion diagnostic can only be brought into generalized use if the analytical methods used have been validated in a distributed setting. The following research issues remain open: 1) better determination of cut points related to particular clinical outcomes; 2) improvement in precision achieved by application of automated scoring systems; 3) determination of the value of measures of proliferation as determined by gene expression for Ki67 or other proliferation proteins that might replace Ki67 IHC; and 4) use of serial Ki67 analyses as an attractive early endpoint to determine if novel therapeutic agents have evidence of activity.
Finally, we believe that the work of IKWG over the past decade, the lessons learned, and the processes gone through can be used as a template for the adoption of additional molecular biomarkers in the care of breast cancer patients.
Funding
This work was supported by a generous grant from the Breast Cancer Research Foundation (DFH).
Notes
Role of the funder: Dr. Flowers, representing BCRF, is a co-author of this commentary.
Disclosures: Torsten O. Nielsen (T.O.N.) received royalty from NanoString Technologies. T.O.N. has intellectual property rights and holds patent with Bioclassifier LLC. David L. Rimm (D.L.R.) had consulting or advisory role with: Amgen, Astra Zeneca, Cell Signaling Technology, Cepheid, Daiichi Sankyo, Danaher, GSK, Konica/Minolta, Merck, Nanostring, NextCure, Odonate, Perkin Elmer, PAIGE.AI, Roche, Sanofi, Ventana and Ultivue. D.L.R. received honoraria from BMS. D.L.R. received research/instrument support from: Akoya/Perkin Elmer, Amgen, Cepheid, Nanostring, Navigate BioPharma, NextCure, PixelGear, Konica/Minolta, Lilly, Ultivue and Ventana. D.L.R. received royalty from Rarecyte. Carsten Denkert (C.D.) has ownership of Sividon Diagnostics (now Myriad). C.D. received honoraria from Novartis and Roche. C.D. had consulting or advisory role with MSD Oncology, Daiichi Sankyo and Molecular Health. C.D. received research funding from Myriad Genetics. C.D. holds the following intellectual property/patents: VMScope digital pathology software, patent application EP18209672 (cancer immunotherapy), patent application EP20150702464 (therapy response) and patent application EP20150702464 (therapy response). Matthew J. Ellis (M.J.E.) has ownership of Bioclassifier LLC that licenses intellectual property (IP) to Veracyte for the breast cancer prognostic test Prosigna. M.J.E. received honorarium from Lilly for consulting on Ki67-based assays. M.J.E. is a McNair Scholar supported by the McNair Medical Institute and receives support from the Susan G Komen Foundation. Susan Fineberg served on expert advisory panel for Genomic Health in 2017. Frédérique M. Penault-Llorca received honoraria and travel grants from the following: AstraZeneca, Roche, Lilly Novartis, Pfizer, Sanofi, Nanostring, Myriad and Genomic Health. John M.S. Bartlett (J.M.S.B) participated in consulting or advisory role with: Breast Cancer Society of Canada, MedcomXchange Communications, Inc, Insight Genetics, Inc, BioNTech AG, Biotheranostics, Inc, Pfizer, Rna Diagnostics, Inc, OncoXchange/MedcomXchange Communications, Inc, Herbert Smith French Solicitors and OncoCyte Corporation. J.M.S.B received honoraria from: NanoString Technologies, Inc, Oncology Education, Biotheranostics, Inc and MedcomXchange Communications, Inc. J.M.S.B received research funding from: Thermo Fisher Scientific, Genoptix, Agendia, NanoString Technologies, Inc, Stratifyer GmbH and Biotheranostics, Inc. J.M.S.B. holds the following patents: (Jan 2017) Methods and Devices for Predicting Anthracycline Treatment Efficacy, US utility—15/325,472 (EPO—15822898.1; Canada—not yet assigned); (Jan 2017) Systems, Devices and Methods for Constructing and Using a Biomarker, US utility –15/328,108 (EPO—15824751.0; Canada—not yet assigned); (Oct 2016) Histone gene module predicts anthracycline benefit, PCT/CA2016/000247; (Dec 2016) 95‐Gene Signature of Residual Risk Following Endocrine Treatment, PCT/CA2016/000304; (Dec 2016) Immune Gene Signature Predicts Anthracycline Benefit, PCT/CA2016/000305; (June 2020) Use of Molecular Classifiers to Diagnose, Treat and Prognose Prostate Cancer, US Provisional 63/040.692. Mitch Dowsett received lecture fees from Nanostring and Myriad; participated in advisory/consultancy role with Radius, Lilly, AbbVie, H3 Biomedicine and Zentalis. His institution received grants from Pfizer and Lilly on studies that includes Ki67 analysis. Daniel F. Hayes (D.F.H.) reports research support from Menarini Silicon BioSystems (MSB). The University of Michigan (UM) holds patent US 8,790,878 B2 for which D.F.H. is designated as inventor, and that is licensed to MSB with annual royalties paid to UM and D.F.H. Outside the submitted work D.F.H. holds stock options from OncImmune LLC, InBiomotion, and serves on advisory boards for Cepheid, Freenome, CellWorks, Agendia, Salutogenic, EPIC Sciences and L-Nutra and UM receives research funding on his behalf from Merrimack, Eli Lilly, Puma Biotechnology, Pfizer, AstraZeneca. The remaining authors have no conflicts of interest to disclose.
Author contributions: TON contributed to conceptualization and writing—original draft, review and editing. SCYL contributed to conceptualization and writing—original draft, review and editing. DLR contributed to conceptualization and writing—original draft, review and editing. AD contributed to conceptualization and writing—original draft, review and editing. BA contributed to writing—review and editing. SB contributed to writing—review and editing. CD contributed to writing—review and editing. MJE contributed to writing—review and editing. SF contributed to writing—review and editing. MF contributed to writing—review and editing. HHK contributed to writing—review and editing. AL contributed to writing—review and editing. HP contributed to writing—review and editing. FMPL contributed to writing—review and editing. MP contributed to writing—review and editing. RS contributed to writing—review and editing. IES contributed to writing—review and editing. TS contributed to writing—review and editing. JMSB contributed to conceptualization and writing—original draft, review and editing. LMM contributed to conceptualization and writing—original draft, review and editing. MD contributed to conceptualization and writing—original draft review and editing. DFH contributed to conceptualization and writing—original draft, review and editing.
References
Articles from JNCI Journal of the National Cancer Institute are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/jnci/djaa201
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/jnci/article-pdf/113/7/808/40501751/djaa201.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/jnci/djaa201
Article citations
Multimodal apparent diffusion MRI model in noninvasive evaluation of breast cancer and Ki-67 expression.
Cancer Imaging, 24(1):137, 11 Oct 2024
Cited by: 0 articles | PMID: 39394171 | PMCID: PMC11470582
Updates in Systemic Treatment of Hormone Receptor-Positive Early-Stage Breast Cancer.
Curr Treat Options Oncol, 25(10):1323-1334, 03 Oct 2024
Cited by: 0 articles | PMID: 39361142
Review
Precision oncology in patients with breast cancer: towards a 'screen and characterize' approach.
ESMO Open, 9(10):103716, 19 Sep 2024
Cited by: 0 articles | PMID: 39303452 | PMCID: PMC11439525
α-Parvin Expression in Breast Cancer Tissues: Correlation with Clinical Parameters and Prognostic Significance.
Cells, 13(18):1572, 19 Sep 2024
Cited by: 0 articles | PMID: 39329755 | PMCID: PMC11430769
Practical Guidance on Abemaciclib in Combination with Adjuvant Endocrine Therapy for Treating Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative High-Risk Early Breast Cancer.
Breast Cancer (Dove Med Press), 16:517-527, 29 Aug 2024
Cited by: 0 articles | PMID: 39224861 | PMCID: PMC11368096
Review Free full text in Europe PMC
Go to all (244) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Clinical Trials
- (1 citation) ClinicalTrials.gov - NCT01953588
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Ki67 in Breast Cancer Assay: An Ad Hoc Testing Recommendation from the Canadian Association of Pathologists Task Force.
Curr Oncol, 30(3):3079-3090, 06 Mar 2023
Cited by: 2 articles | PMID: 36975446 | PMCID: PMC10047249
Evaluation of the international Ki67 working group cut point recommendations for early breast cancer: comparison with 21-gene assay results in a large integrated health care system.
Breast Cancer Res Treat, 203(2):281-289, 17 Oct 2023
Cited by: 1 article | PMID: 37847456 | PMCID: PMC10787679
The usefulness of CanAssist Breast over Ki67 in breast cancer recurrence risk assessment.
Cancer Med, 12(12):13342-13351, 28 May 2023
Cited by: 1 article | PMID: 37245224
Molecular subclasses of breast cancer: how do we define them? The IMPAKT 2012 Working Group Statement.
Ann Oncol, 23(12):2997-3006, 01 Dec 2012
Cited by: 163 articles | PMID: 23166150
Review
Funding
Funders who supported this work.
Breast Cancer Research Foundation
NCATS NIH HHS (1)
Grant ID: UL1 TR001863






