Abstract
Free full text

Genomic analysis of primary and secondary myelofibrosis redefines the prognostic impact of ASXL1 mutations: a FIM study
Associated Data
Key Points
Mutations of TP53 and high-risk genes (EZH2, CBL, U2AF1, SRSF2, IDH1, IDH2, NRAS or KRAS) are adverse prognostic factors in myelofibrosis.
ASXL1 isolated mutations (ie, without TP53 or high-risk mutations) have no prognostic impact in myelofibrosis.
Abstract
We aimed to study the prognostic impact of the mutational landscape in primary and secondary myelofibrosis. The study included 479 patients with myelofibrosis recruited from 24 French Intergroup of Myeloproliferative Neoplasms (FIM) centers. The molecular landscape was studied by high-throughput sequencing of 77 genes. A Bayesian network allowed the identification of genomic groups whose prognostic impact was studied in a multistate model considering transitions from the 3 conditions: myelofibrosis, acute leukemia, and death. Results were validated using an independent, previously published cohort (n = 276). Four genomic groups were identified: patients with TP53 mutation; patients with ≥1 mutation in EZH2, CBL, U2AF1, SRSF2, IDH1, IDH2, NRAS, or KRAS (high-risk group); patients with ASXL1-only mutation (ie, no associated mutation in TP53 or high-risk genes); and other patients. A multistate model found that both TP53 and high-risk groups were associated with leukemic transformation (hazard ratios [HRs] [95% confidence interval], 8.68 [3.32-22.73] and 3.24 [1.58-6.64], respectively) and death from myelofibrosis (HRs, 3.03 [1.66-5.56] and 1.77 [1.18-2.67], respectively). ASXL1-only mutations had no prognostic value that was confirmed in the validation cohort. However, ASXL1 mutations conferred a worse prognosis when associated with a mutation in TP53 or high-risk genes. This study provides a new definition of adverse mutations in myelofibrosis with the addition of TP53, CBL, NRAS, KRAS, and U2AF1 to previously described genes. Furthermore, our results argue that ASXL1 mutations alone cannot be considered detrimental.
Introduction
The Philadelphia chromosome–negative myeloproliferative neoplasms (MPNs), which include polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF), are acquired clonal hematopoietic stem cell disorders characterized by the abnormal proliferation and accumulation of mature blood cells. Myelofibrosis is characterized by dysregulation of the bone marrow stroma with the development of a reticulin fibrosis. PMF differs from post-PV or post-ET secondary myelofibrosis (SMF), which has an evolution rate of ~10% after 10 years of follow-up.1 The course of myelofibrosis is associated with progressive constitutional symptoms (eg, fatigue, night sweats, and fever), increasing splenomegaly, worsening cytopenia, and a risk of transformation to acute myeloid leukemia (AML). Among MPN, myelofibrosis is associated with the worst prognosis, and its evolution is extremely variable according to prognostic features and treatment.2 Risk-adapted therapies range from no treatment to allogeneic hematopoietic stem cell transplantation.2
More than 90% of myelofibrosis cases harbor somatic mutations in the driver genes JAK2, CALR, or MPL that lead to a constitutive activation of the JAK-STAT5 pathway.3 Other somatic nondriver mutations (so-called additional mutations) have been increasingly detected in MPN with the use of high-throughput sequencing.4 These additional mutations involve genes with various functions, such epigenetics, splicing, signalization, and transcription factors, and are also mutated in other myeloid neoplasms like myelodysplastic syndromes and AML. Since the seminal study by Vannucchi et al,5 the prognostic role of additional mutations in PMF have been confirmed, and new scoring systems have incorporated “high-risk” genes.6,7 Conversely, few data are currently available for SMF. In 2018, Grinfeld et al studied 2035 patients and proposed a prognostic classification for all MPN subtypes, incorporating the mutational landscape to allow personalized prognostic assessment.8 Overall, these results pave the way for the use of genomics in the clinical management of MPN, even if additional data are still needed to specify which genes and types of mutation are definitively associated with adverse prognosis.
In this study, we aimed to characterize the molecular landscape of PMF and SMF in a large multicentric cohort to identify homogeneous genomic groups. The prognosis of these genomic groups was then studied, and the results were validated using an external previously published cohort.
Methods
Design and method of the study were performed according the REMARK guidelines (see supplemental Data).9
Patients and samples
From 2005, patients with a diagnosis of PMF or SMF (ie, post-ET or PV) were registered in the French Intergroup of Myeloproliferative Neoplasms (FIM) observatory of myelofibrosis, according to the World Health Organization 2008 or 2016 classifications.10,11 DNAs from 497 unselected patients with PMF or SMF were centralized in the Angers Hospital for high-throughput molecular analysis. Finally, 479 patients were included in the analysis after exclusion of 10 patients who did not fulfill myelofibrosis diagnostic criteria after central review and 8 patients because of next-generation sequencing (NGS) failure (supplemental Figure 1). All samples were collected at the time of myelofibrosis diagnosis and consisted of whole blood (83%), purified blood granulocytes (14%), or whole bone marrow (3%). The median time between diagnosis and sampling was 17 days (range, 0-88 months). In detail, 77% of samples were collected within 3 months, and 95% of patients were sampled within the first 2 years following diagnosis. As a validation cohort, we used the results of Grinfeld et al on 276 cases of PMF or SMF.8 Details regarding patients, diagnostic review, samples, and the validation cohort are provided in supplemental Data.
NGS
A custom RNA-baits panel was designed to cover all exons of 77 genes involved in myeloid malignancies or previously described in MPN. Bioinformatics tools were used to call and annotate variants (details in supplemental Data). Only exonic or splicing mutations (donor and acceptor sites) with a variant allele frequency ≥2% and not described as common polymorphisms (ie, ≥1% in general population) were retained. Variants were finally classified according to their putative pathogenic effect as pathogenic, likely pathogenic, or variant of unknown significance according to standard guidelines12,13 (details in supplemental Data). Furthermore, mutations of unknown significance with a frequency ≥0.01% in the general population were considered as rare polymorphisms and were removed.
Statistics
A Bayesian network analysis combined with hierarchical clustering analysis was performed to characterize homogeneous clusters of genes. To describe the natural history of myelofibrosis a multistate model was used, allowing variables associated with each transition to be identified. Correction of P values was performed for multiple testing using a Benjamini-Hochberg procedure. Only pathogenic and likely pathogenic mutations were taking into account for statistical analysis of prognosis. Detailed methodology is described in supplemental Data (pages 8-12).
Results
The mutational landscape of myelofibrosis
The whole cohort included 305 PMF and 174 SMF (70 post-PV and 104 post-ET). The characteristics of patients are summarized in supplemental Table 1. Driver mutations were distributed as follows: 309 JAK2V617F (65%), 110 CALR (23%), 25 MPLW515 (5%), and 8 patients with a double mutation (4 JAK2V617F/MPLW515, 2 CALR/MPLW515, 1 JAK2V617F/CALR, and 1 JAK2V617F/JAK2exon12). For patients with 2 driver mutations, the mutation with the highest allele burden was considered the driver for further analyses. JAK2V617F allele burden was higher in SMF than in PMF (supplemental Figure 2). Of note, 27 patients had no driver mutation detected (ie, triple negative).
A total of 1385 additional variants were detected with the following distribution: 861 pathogenic variants, 179 likely pathogenic variants, and 345 variants of unknown significance. We found a median (range) of 3 (0-11) additional variants per patient and 2 (0-10) additional mutations when considering only pathogenic and likely pathogenic variants. The most frequently mutated genes were ASXL1, TET2, SRSF2, U2AF1 and EZH2 (Figure 1A; supplemental Figure 3). Mutations in the SRSF2 and U2AF1 genes were significantly more frequent in PMF, whereas mutations in NFE2 were more frequent in SMF (Figure 1B). SRSF2 and U2AF1 mutations were more frequently encountered in JAK2-mutated than in CALR-mutated patients (Figure 1C). Mutations in ATM, KMT2C, KMT2D, and NOTCH1 were mainly variants of unknown significance. The allele burden distribution of additional mutations harbored different profiles depending on the genes: (1) an allele burden centered on 40% to 50% (eg, SRSF2 and U2AF1); (2) an allele burden with values ranging from 0% to 100% (eg, EZH2 and TP53); and (3) an allele burden with a bimodal distribution, with some values at a low allele burden and others centered on 40% to 50% (eg, ASXL1 andTET2) (Figure 1D). In further analyses, we considered only pathogenic and likely pathogenic mutations. Indeed, the function of “variants of unknown significance” is unclear, and most of these are likely rare polymorphisms. No difference in the number of additional mutations according to the sex was found (P = .08), but older patients more frequently had ≥1 additional mutation (P = .0009; supplemental Figure 4).
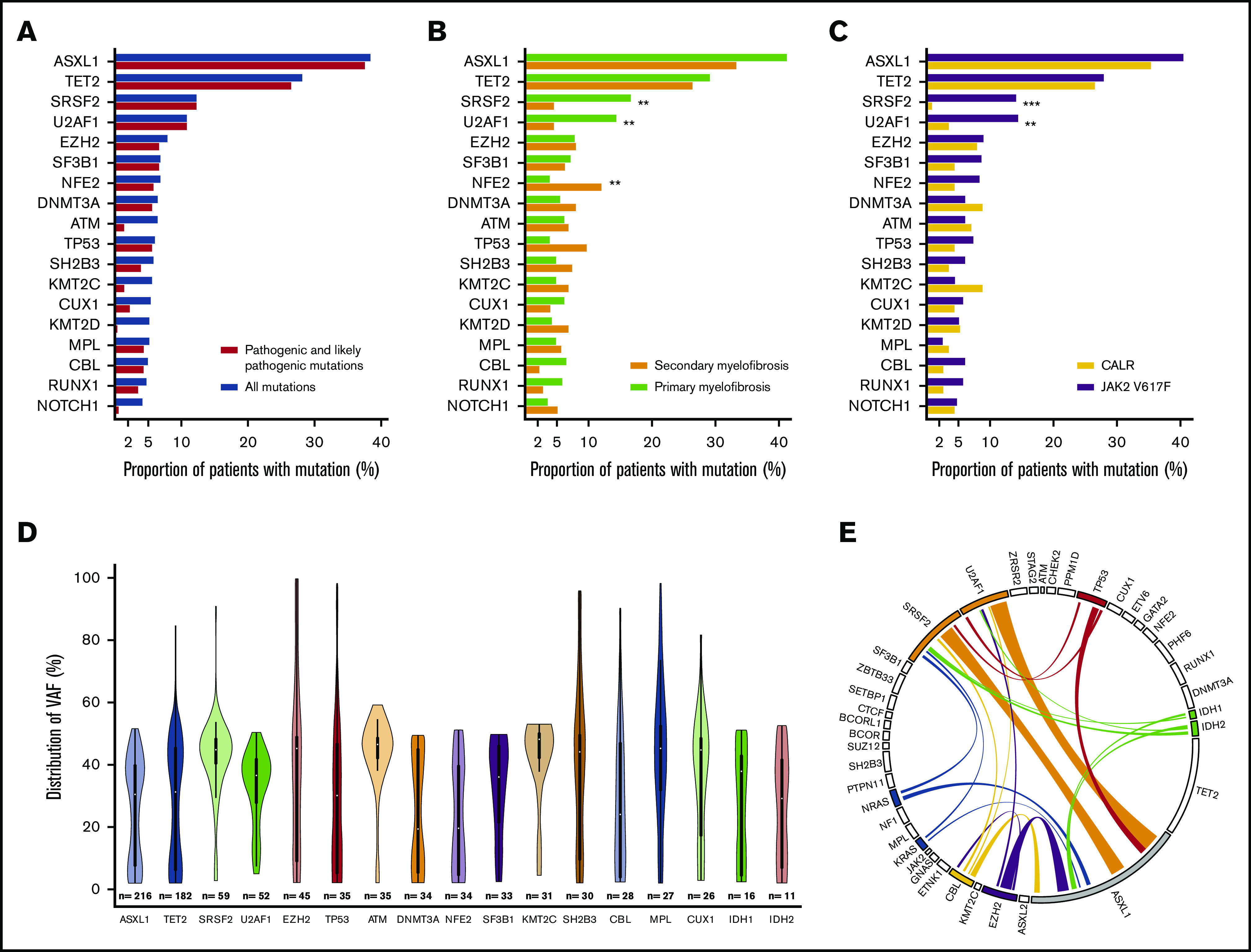
Mutational landscape of the entire cohort. Proportion of patients with ≥1 mutation per gene according to the classification of mutations (A), the type of myelofibrosis (B), and the driver mutation (JAK2 or CALR) (C). The distribution of allelic burden of additional mutations is represented by violin plots (D), and the association between mutations on the genes of the 4-tier classification is shown in a circos plot (E). *P < .05; **P < .01; ***P < .001. Only pathogenic and likely pathogenic mutations were retained for the circos plot. VAF, variant allele frequency.
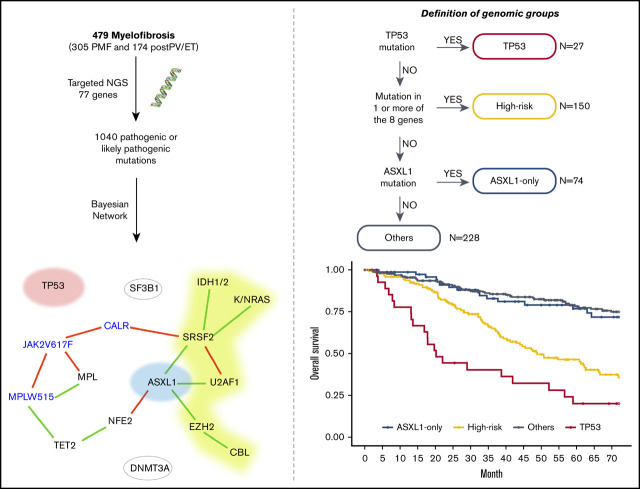
We visualized the molecular structure of the cohort using a Bayesian network (Figure 2). Based on this network and on a hierarchical clustering analysis, we defined 7 genomic groups. Since some of these 7 genomic groups harbored similar outcomes, we then merged some of the genomic groups to generate 4 prognostic groups according to their potent respective impact on overall survival (OS) (supplemental Figure 5; Figure 3A). The 4 groups were created in the following sequence: (1) TP53-mutated patients (n = 27); (2) patients with ≥1 mutation in EZH2, CBL, U2AF1, SRSF2, IDH1, IDH2, NRAS, or KRAS (high-risk group, n = 150); (3) ASXL1-only mutation (ie, no associated mutation in TP53 or in high-risk genes, n = 74); and (4) “other” patients, with the main mutations being in NFE2, DNMT3A, TET2, and SF3B1 (n = 228) (Figure 2).
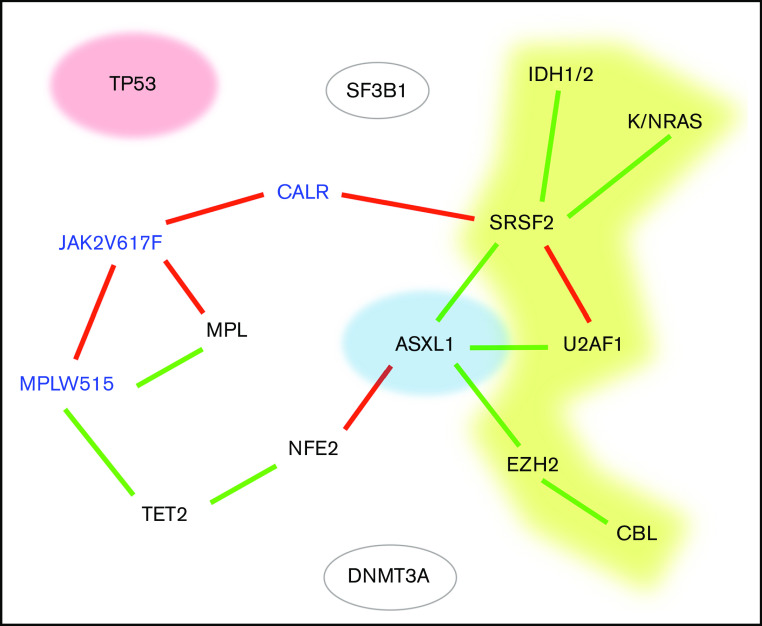
Bayesian network of the molecular landscape. Driver genes are in blue. A green link represents a positive association, and a red one represents 2 mutually exclusive genes. Red, yellow, and blue genes represent the genomic groups identified in the following order: TP53 mutations, high-risk mutation (without TP53 mutation), and ASXL1-only mutation (ie, without TP53 or high-risk mutation).
Prognostic impact of genomic alterations
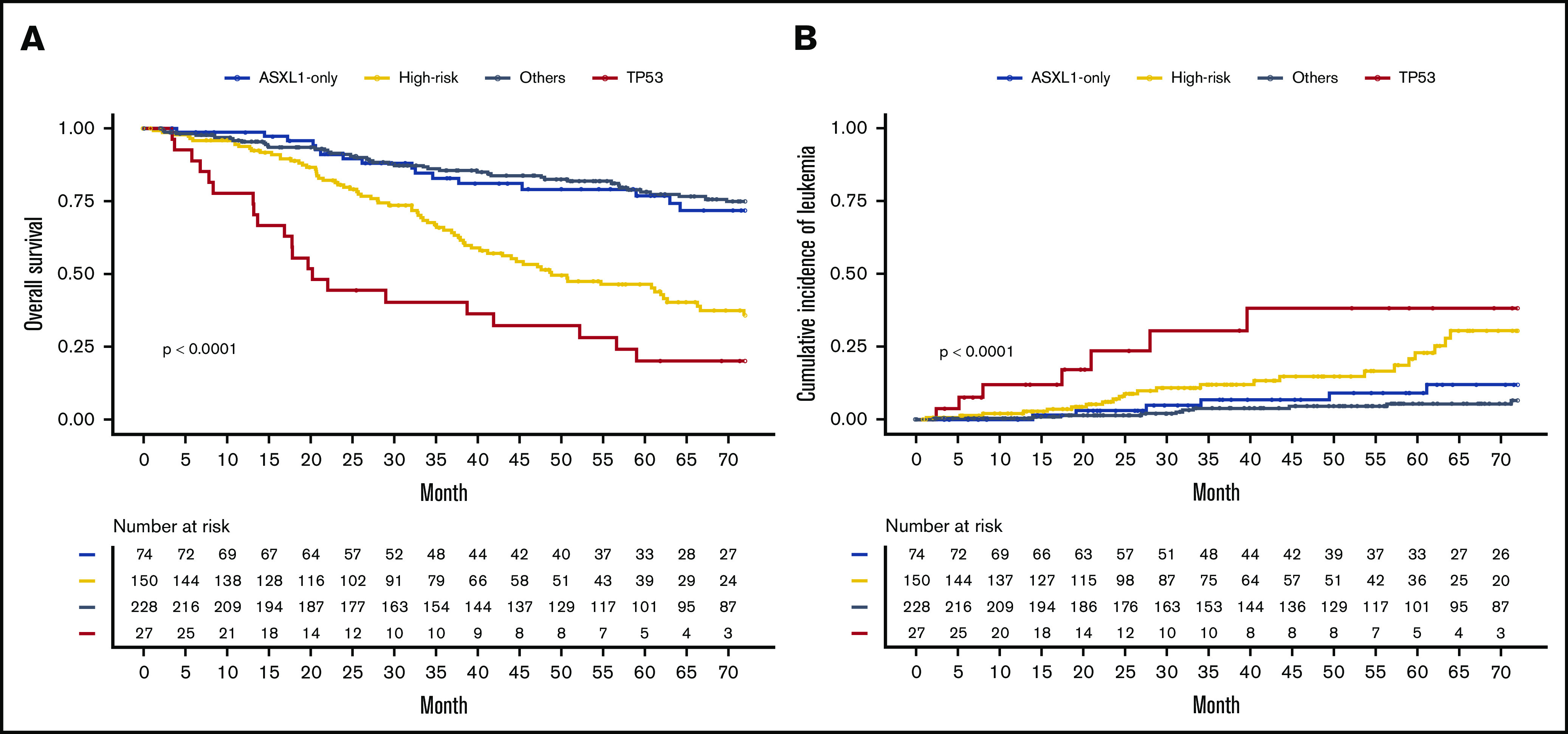
The prognostic impact of the 4 molecular groups on OS and cumulative incidence of leukemic transformation highlighted a worst prognostic for TP53 and high-risk groups (median OS, 20 and 49 months, respectively; median time to leukemic transformation not reached for TP53 and 110 months for high risk) than in patients with ASXL1-only and the “other patients” group (median OS, 89 and 116 months, respectively; time to leukemic transformation not reached) (Figure 3; P < .0001 for OS and leukemic transformation). This prognostic impact was also found on leukemia-free survival (supplemental Figure 6). Then, we studied the prognostic value of these 4 groups using multistate modeling, which included the following variables: sex; age at diagnosis; hemoglobin, platelet, and leukocyte counts; type of driver mutation; type of myelofibrosis; and the 4-tier genomic classification. Univariate analyses of each transition are summarized in supplemental Table 2, and the final model is summarized in Table 1. TP53 and high-risk mutations groups were associated with an independent and significantly higher risk of both leukemic transformation (hazard ratio [HR] [95% confidence interval], 8.68 [3.32-22.73], P < .0001 and HR, 3.24 [1.58-6.64], P = .0013, respectively) and risk of death without AML (HR, 3.03 [1.66-5.56], P = .0003 and HR, 1.77 [1.18-2.67], P = .006, respectively). Patients with ASXL1-only mutation did not have an adverse prognosis either in terms of leukemic transformation (HR, 2.45 [0.95-6.29], P = .063) or OS (HR, 1.17 [0.68-2.01], P = .579). Moreover, we confirm that well-known factors like age at diagnosis, anemia, and leukocytosis were associated with a higher risk of death.2,14 Thrombocytopenia was associated with a higher risk of leukemic transformation, whereas CALR-mutated patients presented with a lower risk of AML transformation. Two sensitivity analyses were performed considering either (1) a censor at the time of bone marrow transplantation for the 50 allografted patients and (2) a time 0 as the time of DNA sampling, and both analyses confirmed our findings (supplemental Tables 3 and 4).
Table 1.
Multistage model for AML and death events
| Transitions | ||||||
|---|---|---|---|---|---|---|
| Variable | Myelofibrosis to AML | Myelofibrosis to death | AML to death | |||
| HR [95% CI] | P | HR [95% CI] | P | HR [95% CI] | P | |
| Age at diagnosis | — | — | 1.05 [1.03-1.07] | <.0001 | — | — |
| Sex (male) | — | — | 1.41 [0.96-2.06] | .077 | — | — |
| Hemoglobin | — | — | 0.81 [0.74-0.89] | <.0001 | — | — |
| Leukocyte count | — | — | 1.04 [1.02-1.05] | <.0001 | — | — |
| Platelet count | 0.99 [0.99-0.99] | .012 | — | — | — | — |
| Driver mutation (JAK2 as reference): CALR | 0.21 [0.06-0.70] | .011 | — | — | — | — |
| Genomic groups (others as reference) | ||||||
 TP53 TP53 | 8.68 [3.32-22.73] | <.0001 | 3.03 [1.66-5.56] | .0003 | 1.21 [0.31-4.72] | .784 |
 High risk High risk | 3.24 [1.58-6.64] | .0013 | 1.77 [1.18-2.67] | .006 | 0.88 [0.33-2.36] | .801 |
 ASXL1 only ASXL1 only | 2.45 [0.95-6.29] | .063 | 1.17 [0.68-2.01] | .579 | 1.22 [0.33-4.47] | .767 |
CI, confidence interval.
Finally, we applied the multistate model to the validation cohort of 276 cases of myelofibrosis published by Grinfeld et al8 and found very similar results (supplemental Table 5). In particular the prognostic impact of TP53 mutations on leukemic transformation and high-risk mutations on leukemic transformation and death were confirmed with HRs close to those found in our cohort. Of note, 2 results cannot be reproduced in the validation cohort: the protective effect of CALR mutations on leukemic transformation and reduced OS for TP53 mutations. However, the absence of prognostic value of the ASXL1-only group was also supported by results obtained in the validation cohort.
Deciphering the spectrum of ASXL1 mutations
We then examined in detail the spectrum of ASXL1 mutations and found that they were frequently associated with TP53 or high-risk genes mutations (Figure 1E). Indeed, an ASXL1 mutation was found in 60% of cases in these 2 genomic groups and was associated with an increased risk of death (HR, 1.5 [1.01-2.29], P = .044; supplemental Figure 7).
The allele burden of ASXL1 mutations was higher when associated with high-risk mutations (supplemental Figure 8). In the ASXL1-only group, the allele burden of ASXL1 mutation was not associated with acute leukemia or death (P = .293 and P = .763, respectively, with a threshold of 20%). A sensitivity analysis of the multistate model considering only additional mutations with >15% allele burden was performed and previous results were confirmed, showing the lack of prognostic significance for ASXL1-only mutations (supplemental Table 6).
Comparison of prognostic performances
Finally, we aimed to evaluate the additional value of the genomic groups for prognosis assessment as compared with other prognostic classifications. For this purpose, the performance (C-index and time-dependent area under the curve [AUC])15,16 and the accuracy (Brier score)17 to predict death and leukemic transformation were evaluated for standard prognostic scoring systems (ie, International Prognostic Scoring System [IPSS]2 for PMF and Myelofibrosis Secondary to PV and ET–Prognostic Model [MYSEC-PM]18 for SMF), the present 4-tier genomic classification alone, and the combination of our 4-tier genomic with previous scores. The more recent prognostic scores MIPSS706 and MIPSS70+v27 were also evaluated. Results are summarized in Table 2 and supplemental Table 7. The 4-tier genomic classification alone harbored equal or slightly inferior C-index performance as compared with IPSS or MYSEC-PM for death and was superior to IPSS but inferior to MYSEC-PM for leukemic transformation. It is worth noting that the association of genomic groups with standard prognostic scoring systems improved the prediction and accuracy of prognosis for both short-term and long-term events, as illustrated in Figure 4 for death and in supplemental Figure 9 for leukemic transformation. MIPSS70 and MIPS70+v2 had good predictive values in particular for death in PMF and SMF and for leukemic transformation in PMF. However, the combination of our 4-tier genomic classification combined with IPSS or MYSEC-PM remained superior to MIPSS70 and MIPS70+v2.
Table 2.
Prognostic performance comparison
| C-index | Events at 24 mo | Events at 48 mo | Events at 72 mo | ||||
|---|---|---|---|---|---|---|---|
| Brier score | AUC | Brier score | AUC | Brier score | AUC | ||
| OS | |||||||
 PMF (n = 305) PMF (n = 305) | |||||||
  NGS NGS | 0.66 | 0.052 | 66 | 0.101 | 72 | 0.13 | 73 |
  IPSS IPSS | 0.68 | 0.051 | 71 | 0.098 | 72 | 0.129 | 74 |
  MIPSS70 MIPSS70 | 0.67 | 0.05 | 70 | 0.095 | 73 | 0.122 | 71 |
  NGS-IPSS NGS-IPSS | 0.73 | 0.05 | 74 | 0.093 | 79 | 0.118 | 82 |
 SMF (n = 174) SMF (n = 174) | |||||||
  NGS NGS | 0.66 | 0.057 | 68 | 0.117 | 61 | 0.147 | 68 |
  MYSEC-PM MYSEC-PM | 0.71 | 0.061 | 74 | 0.11 | 76 | 0.133 | 80 |
  MIPSS70* MIPSS70* | 0.68 | 0.06 | 73 | 0.111 | 71 | 0.135 | 72 |
  NGS-MYSEC-PM NGS-MYSEC-PM | 0.77 | 0.054 | 82 | 0.102 | 79 | 0.124 | 86 |
| Leukemic transformation | |||||||
 PMF (n = 305) PMF (n = 305) | |||||||
  NGS NGS | 0.71 | 0.017 | 76 | 0.041 | 74 | 0.058 | 79 |
  IPSS IPSS | 0.59 | 0.017 | 60 | 0.04 | 61 | 0.062 | 61 |
  MIPSS70 MIPSS70 | 0.72 | 0.017 | 76 | 0.038 | 78 | 0.056 | 73 |
  NGS-IPSS NGS-IPSS | 0.73 | 0.016 | 78 | 0.039 | 77 | 0.056 | 79 |
 SMF (n = 174) SMF (n = 174) | |||||||
  NGS NGS | 0.67 | 0.021 | 70 | 0.047 | 65 | 0.062 | 72 |
  MYSEC-PM MYSEC-PM | 0.75 | 0.022 | 76 | 0.046 | 82 | 0.059 | 85 |
  MIPSS70* MIPSS70* | 0.66 | 0.022 | 64 | 0.048 | 71 | 0.062 | 73 |
  NGS-MYSEC-PM NGS-MYSEC-PM | 0.76 | 0.02 | 81 | 0.042 | 83 | 0.054 | 87 |
Comparisons were performed between classical prognosis score systems and the 4-tier genomic classification (NGS) alone or in combination with a clinical score. C-index and AUC evaluated the performance of the model, and Brier score reflected the accuracy of the prediction (ie, the rate of error). Bold indicates the best values.
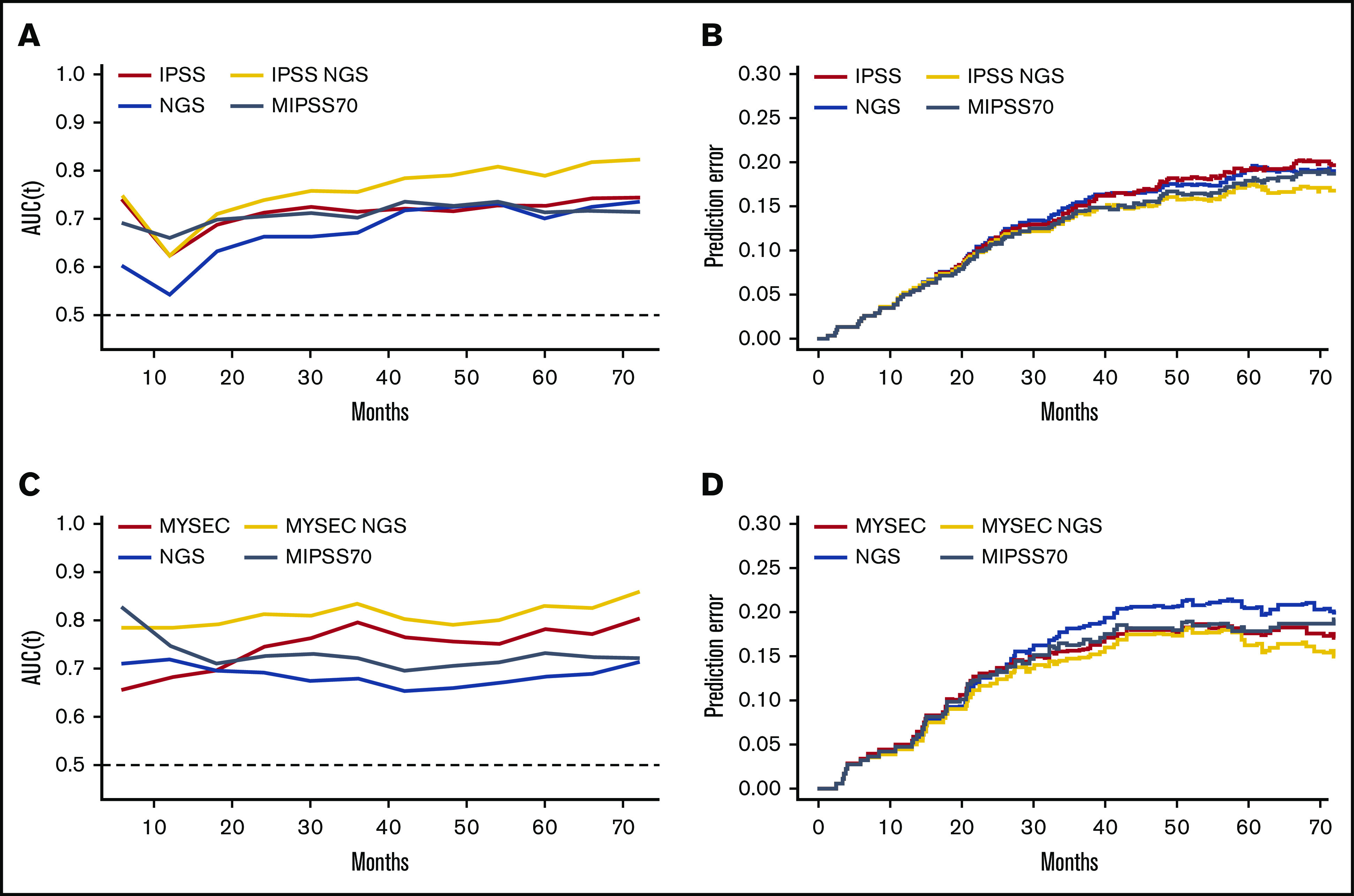
Prognosis performance comparison. The performance (time-AUC; A and C) and accuracy (Brier score; B and D) for death prediction were measured over the time for usual prognosis scoring systems, the 4-tier genomic classification (NGS) alone, or in combination for primary (A-B) and secondary (C-D) myelofibrosis patients.
Discussion
Herein, we analyzed the molecular landscape of a large cohort of patients with PMF and SMF. Gene variants were classified according to their suspected pathogenicity and only pathogenic or likely pathogenic mutations were kept in the statistical analysis for prognosis. The Bayesian network enabled the characterization of the molecular structure of our cohort and it was indeed close to that of the network described by Grinfeld et al.8 Both genomic networks reveal ASXL1 as a central node of the MPN molecular landscape, since most additional mutations arise from this first hit. ASXL1 mutations were found in approximately one-third of myelofibrosis patients as previously described.4,6,19-22 We defined 4 genomic groups, 2 of them, the “TP53” and high-risk (ie, ≥1 mutation in groups EZH2, CBL, U2AF1, SRSF2, IDH1, IDH2, NRAS, or KRAS) groups, were associated with an adverse outcome (transition from myelofibrosis to acute leukemia and from myelofibrosis to death). Furthermore, we investigated the prognostic impact of each gene within the high-risk group by analyzing the performance for the prediction of OS by leaving out the genes one by one. This confirmed that all genes were associated and improved the prognostic evaluation (supplemental Table 8).
Conversely, the ASXL1-only group (ie, without mutation in TP53 or high-risk genes) had no prognostic impact on the risk of death and had only a moderate impact for leukemic transformation. The multivariate analysis also found that age at diagnosis, leukocytosis, and anemia were associated with a higher risk of death, while thrombocytopenia was associated with a risk of leukemic transformation, in agreement with risk factors included in previous prognosis scoring systems, from the Lille score to IPSS and Dynamic International Prognostic Scoring System–Plus (DIPSS-plus).2,14,23 We also found that CALR-mutated patients had a lower risk of leukemic transformation, as previously described.24 Of note, this prognostic advantage was reported as being limited to type 1 and type 1–like mutations of CALR.25,26 In our cohort, 79% of CALR mutations were type 1/type 1–like, and no distinction was made in multivariate analysis, because no difference was found in univariate analysis between type 1/type 1–like and other CALR mutations (P = .77 and .6 for leukemia-free survival and OS, respectively). The 2 major differences of the multistage model in the validation cohort concerned the absence of significance of CALR mutations for leukemic transformation and the TP53 genomic group for OS and may be explained by the limited number of patients with either CALR mutation (n = 46) and TP53 mutations, with 11 mutated cases, of which 5 progressed to acute leukemia and are therefore not considered for myelofibrosis to death transition in the multistate model.
In the present study, we identified mutations in TP53, EZH2, SRSF2, IDH1, IDH2, U2AF1, CBL, NRAS, and KRAS genes as adverse prognostic factors in myelofibrosis. The detrimental role of TP53 mutations has been previously described in cohorts that included all subtypes of MPN, and the role of TP53 in leukemic transformation has been shown in mice models.4,8,27,28 Among the other genes, we found a worse prognosis associated with EZH2, SRSF2, IDH1, and IDH2, as previously reported by others for high-risk profiles.5 U2AF1 and N/KRAS have also been described as adverse prognostic factors.7,29,30 CBL protein is a RAS pathway regulator, and previous studies have reported a reduced OS in MPN patients with CBL mutations.22,30,31
Our results about the prognostic value of ASXL1 mutations seem, at a first glance, to contrast those of previous studies in PMF, where they were classified as high molecular risk (HMR).5,6,22 Nevertheless, some other studies found no prognostic impact of ASXL1 mutations in SMF,32 and in ET and in PV in MIPSS-ET and MIPSS-PV did not include ASXL1 mutations.33 Santos et al have recently studied a cohort of 723 patients with myelofibrosis and found a prognostic value of N/KRAS mutations but no impact of HMR mutations using a multivariate model considering sex, platelet count, transfusion dependency, DIPSS classification, karyotype, driver mutation, HMR (ie, ASXL1, EZH2, and IDH1/2), and N/KRAS mutations.30 In this later study, there was a positive association between N/KRAS mutations and ASXL1 mutations. Similarly, we found that 59% of patients with ASXL1 mutations also had either a mutation in TP53 or in a gene from the high-risk genomic group (EZH2, CBL, U2AF1, SRSF2, IDH1, IDH2, NRAS, or KRAS). The selection of genomic groups based on the Bayesian network allowed us to study the prognostic role of isolated ASXL1 mutations. Thus, we hypothesize that the apparent prognostic impact of ASXL1 mutations found in previous studies could be due to either (1) other mutations of adverse prognostic not detected or not included in the NGS panels or in statistical models and frequently associated with ASXL1 or (2) the additional impact of ASXL1 mutations in patients harboring other high-risk mutations. Our results have been validated on an independent cohort. Thus, it is unlikely that recruitment bias explains differences about the impact of ASXL1 mutations.
Our sequencing technology allowed the detection of very low allelic burdens (down to 2%). Such ASXL1 mutations with low allele burden might simply reveal age-related small clones not linked to the MPN (ie, clonal hematopoiesis of indeterminate potential),34 but a sensitivity analysis showed similar results when considering only mutations with an allele burden >15%. The type of ASXL1 mutation may also be important. Indeed, Tefferi et al demonstrated a similar prognosis for frameshift, nonsense, and missense mutations, whereas Kuykendall et al found no apparent impact of missense mutations.35,36 In our cohort, all ASXL1 mutations harbored a truncated C-terminal domain.
Finally, in this study, we have considered the c.1934dupG mutation of ASXL1, whose status as a true mutation or artifact has previously been debated, but it is now considered a bona fide mutation (with 472 occurrences in COSMIC).37,38 A previous study found a worse prognosis for this particular mutation in PMF.39 This led us to conduct 2 sensitivity analyses, one excluding the ASXL1 c.1934dupG mutation and the second dissociating this mutation from the other ASXL1 mutations, and we found no prognosis impact of ASXL1 mutations for either analysis (supplemental Tables 9 and 10).
The role of ASXL1 mutations in myeloid neoplasms development is still unclear. ASXL1 mutations result in a loss of PRC2-mediated H3K27 trimethylation.40 Deletion of Asxl1 in mice induced a myelodysplasia phenotype, and a knockin expression of an Asxl1 mutation with protein truncation led to an age-dependent myeloid skewing with anemia, thrombocytosis, and dysplasia.41,42 Furthermore, mice models have shown a cooperation between Asxl1 and other mutations (eg, Nras and Runx1) in promoting leukemic transformation.40,42 These findings fit with our results showing that ASXL1 mutations had no prognostic impact alone but seemed to confer a worse prognosis when associated with high-risk mutations.
For the evaluation of the prognostic performance, we compiled (1) C-index reflecting the concordance probability of the prognostic model, (2) AUC for different times of follow-up (time-AUC) reflecting the performance of prediction, and (3) Brier score for different times of follow-up reflecting the accuracy of the prediction. All these indicators showed that our 4 genomic group classification allowed a good prognostic performance equivalent to most previous scores.2,6,7,18 Furthermore, the prognostic prediction for both death and leukemic transformation were highly improved by combination of the 4 genomic groups with standard scores.
In conclusion, our results argue for a revision of the prognostic classification of somatic mutations in myelofibrosis with the exclusion of ASXL1 and the inclusion of the TP53, U2AF1, CBL, NRAS, and KRAS genes to the already-recognized SRSF2, EZH2, IDH1, and IDH2 HMR mutations. These results provide a basis for a new molecular classification that will need to be validated in large multicentric cohorts. Indeed, the accurate definition of a high-risk molecular signature is crucial, as it could influence patient management and therapy.43,44
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors are indebted to Rébecca Jouanneau-Courville, who performed sequencing experiments, and Annaëlle Beucher (Angers) and Arnaud Guille (Marseille), who helped with the bioinformatics. They thank the patients, biologists, pathologists, hematologists, and data managers who participated in the FIMBANK project. They especially thank Jean-Marie Chrétien (Délégation à la Recherche Clinique et Innovation, CHU Angers) and Matthieu Resche-Rigon (Université de Paris, ECSTRRA Team U1153, INSERM) for electronic case report form and data coordination and Célia Belkhodja, Kahina Oukherfellah, Eléonore Ourouda-Mbaya, Oryanne Wojtowicz, Alicia Delisle, and Mathilde Favreau for many years of biological and clinical data collection in FIM and FIMBANK electronic case report forms. The authors also thank all participating biobanks (Centres de Ressources Biologiques) for providing high-quality DNA samples, the Plateau de Biologie et de Médecine Moléculaire and the Génomique Onco-Hématologique platform (CHU Angers) for sequencing facilities, and the team of Yves Delneste (Université d’Angers, INSERM, CRCINA, Angers) for scientific discussions. They thank the French Innovative Leukemia Organization, especially Valérie Rolland-Neyret, for continuous support to the FIM. Finally, they thank Moritz Gerstung and Peter Campbell’s groups for sharing their cohort’s data via the MPN-multistage tool.
This study was supported by grants from the Force Hemato foundation (Fonds De Recherche Clinique en Hématologie) (AAP-FH-2016) and the Association Laurette Fugain (ALF 2018/12). This work is also a deliverable of the FIMBANK network, which was founded by the French Institut National du Cancer (INCa BCB 2013).
Footnotes
All data folders and code for figure generation are freely available (https://github.com/Hematology-Lab-of-Angers-Hospital/FIM-Myelofibrosis-NGS).
Authorship
Contribution: D.L.P. and V.U. designed the study; E.V., B.C., A.C., O.M., V.M.-D.M., Y.L.B., F.G., J.-C.C., A.M., P.S., R.B.A., O.K., I.S., O.N., P.M., V.C., and E.L. provided DNA samples; J.-C.I., B.D., F.B., J. Rey, G.E., S.T., F.V., S. Girault, F.G., D.R., P.C.-M., M. Robles, L.R., M.W., B.S., G.D., S. Giraudier, E.L., and J.-J.K. cared for patients; M. Renard developed the bioinformatics tools; D.L.P., E.V., B.C., A.C., O.M., A.M., L.C., I.S., and E.L. analyzed the NGS; B.B. coordinated the histological evaluation; A.W.-P. coordinated the maintenance of the clinical database; N.J. and V.U. coordinated the FIMBANK network; D.L.P. and J. Riou performed statistical analysis; and D.L.P., J. Riou, V.U., G.S., and J.-J.K. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Valérie Ugo, Laboratoire d’Hématologie, Institut de Biologie en Santé, CHU Angers, 4 Rue Larrey, 49933 Angers Cedex 9, France; e-mail: [email protected]; and Damien Luque Paz, Laboratoire d’Hématologie, Institut de Biologie en Santé, CHU Angers, 4 Rue Larrey, 49933 Angers Cedex 9, France; e-mail: [email protected].
A list of the members of the FIM appears in “Appendix.”
Appendix: study group members
Additional members of the FIM include: Fiorenza Barraco (Lyon, France), Yan Beauverd (Geneva, Switzerland), Annaëlle Beucher (Angers, France), Audrey Bidet (Bordeaux, France), Chrystèle Bilhou-Nabera (Paris, France), Odile Blanchet (Angers, France), Anne Bouvier (Angers, France), Marie-Madeleine Coudé (Saint-Ouen L'Aumône, France), Alexandre De Brevern (Paris, France), François Delhommeau (Paris, France), Bernard Drénou (Mulhouse, France), Franck Geneviève (Angers, France), Alban Godon (Angers, France), Alexandre Guy (Bordeaux, France), Pierre Hirsh (Paris, France), Mathilde Hunault (Angers, France), Pascal Hutin (Quimper, France), Norbert Ifrah (Angers, France), Chloé James (Bordeaux, France), Marie-Caroline Le Bousse Kerdiles (Villejuif, France), Benjamin Lebecque (Clermont-Ferrand, France), Laurence Legros (Villejuif, France), Nabih Maslah (Paris, France), Mathieu Meunier (Grenoble, France), Dina Naguib (Caen, France), Corentin Orvain (Angers, France), Florence Pasquier (Villejuif, France), Isabelle Plo (Villejuif, France), Aline Praire (Lyon, France), Emmanuel Raffoux (Paris, France), Bénédicte Ribourtout (Angers, France), Alberto Santagostino (Troyes, France), Isabelle Sudaka-Sammarcelli (Nice, France), Marie-Hélène Schlagetter (Paris, France), Aline Schmidt (Angers, France), Sylvain Thépot (Angers, France), Marie Tuffigo (Angers, France), and Laura Velazquez (Villejuif, France).
References
Articles from Blood Advances are provided here courtesy of The American Society of Hematology
Full text links
Read article at publisher's site: https://doi.org/10.1182/bloodadvances.2020003444
HAL Open Archive
https://www.hal.inserm.fr/inserm-03187575
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/101390014
Article citations
Synergistic effect of concurrent high molecular risk mutations and lower JAK2 mutant variant allele frequencies on prognosis in patients with myelofibrosis-insights from a multicenter study.
Leukemia, 04 Oct 2024
Cited by: 0 articles | PMID: 39367172
Prognostic and Predictive Models in Myelofibrosis.
Curr Hematol Malig Rep, 19(5):223-235, 24 Aug 2024
Cited by: 0 articles | PMID: 39179882 | PMCID: PMC11416430
Review Free full text in Europe PMC
Myelofibrosis and allogeneic transplantation: critical points and challenges.
Front Oncol, 14:1396435, 20 Jun 2024
Cited by: 0 articles | PMID: 38966064 | PMCID: PMC11222377
Review Free full text in Europe PMC
Integrating AIPSS-MF and molecular predictors: A comparative analysis of prognostic models for myelofibrosis.
Hemasphere, 8(3):e60, 20 Mar 2024
Cited by: 1 article | PMID: 38510992 | PMCID: PMC10951878
Myeloproliferative Neoplasms: Contemporary Review and Molecular Landscape.
Int J Mol Sci, 24(24):17383, 12 Dec 2023
Cited by: 0 articles | PMID: 38139212 | PMCID: PMC10744078
Review Free full text in Europe PMC
Go to all (33) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The prognostic contribution of CBL, NRAS, KRAS, RUNX1, and TP53 mutations to mutation-enhanced international prognostic score systems (MIPSS70/plus/plus v2.0) for primary myelofibrosis.
Am J Hematol, 99(1):68-78, 17 Oct 2023
Cited by: 3 articles | PMID: 37846894
Mutations and prognosis in primary myelofibrosis.
Leukemia, 27(9):1861-1869, 26 Apr 2013
Cited by: 452 articles | PMID: 23619563
Clinical impacts of the mutational spectrum in Japanese patients with primary myelofibrosis.
Int J Hematol, 113(4):500-507, 02 Jan 2021
Cited by: 2 articles | PMID: 33389584
Transplant Decisions in Patients with Myelofibrosis: Should Mutations Be the Judge?
Biol Blood Marrow Transplant, 24(4):649-658, 08 Nov 2017
Cited by: 4 articles | PMID: 29128551
Review
 1,2,3, on behalf of the French Intergroup of Myeloproliferative Neoplasms
1,2,3, on behalf of the French Intergroup of Myeloproliferative Neoplasms