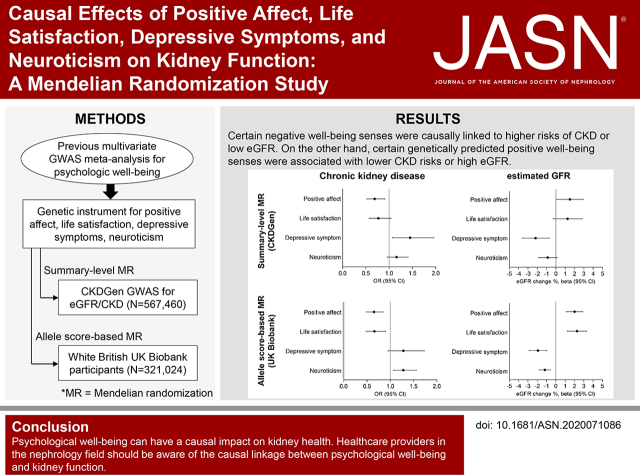
Abstract
Background
Further investigation of the causal effects of psychologic wellbeing on kidney function is warranted.Methods
In this Mendelian randomization (MR) study, genetic instruments for positive affect, life satisfaction, depressive symptoms, and neuroticism were introduced from a previous genome-wide association study meta-analysis of European individuals. Summary-level MR was performed using the CKDGen data of European ancestry (n=567,460), and additional allele score-based MR was performed in the individual-level data of White British UK Biobank participants (n=321,024).Results
In summary-level MR with the CKDGen data, depressive symptoms were a significant causative factor for kidney function impairment (CKD OR, 1.45; 95% confidence interval, 1.07 to 1.96; eGFR change [%] beta -2.18; 95% confidence interval, -3.61 to -0.72) and pleiotropy-robust sensitivity analysis results supported the causal estimates. A genetic predisposition for positive affect was significantly associated with better kidney function (CKD OR, 0.69; 95% confidence interval, 0.52 to 0.91), eGFR change [%] beta 1.50; 95% confidence interval, 0.09 to 2.93) and sensitivity MR analysis results supported the finding for CKD outcome, but was nonsignificant for eGFR. Life satisfaction and neuroticism exposures showed nonsignificant causal estimates. In the UK Biobank with covariate-adjusted allele score MR analysis, allele scores for positive affect and life satisfaction were causally associated with reduced risk of CKD and higher eGFR. In contrast, neuroticism allele score was associated with increased risk of CKD and lower eGFR, and depressive symptoms allele score was associated with lower eGFR, but showed nonsignificant association with CKD.Conclusions
Health care providers in the nephrology field should be aware of the causal linkage between psychologic wellbeing and kidney function.Free full text

Causal Effects of Positive Affect, Life Satisfaction, Depressive Symptoms, and Neuroticism on Kidney Function: A Mendelian Randomization Study
Significance Statement
Poor psychologic wellbeing is prevalent in people with kidney function impairment. A Mendelian randomization investigation identified “causal” effects from psychologic wellbeing on kidney function. The analysis demonstrated that genetic predisposition for certain positive wellbeing senses causally decreases the risk of kidney function impairment. In contrast, genetically predicted negative wellbeing senses were causally linked to a higher risk of CKD, or a lower eGFR. Therefore, this study suggests health care providers in the nephrology field should be aware of the causal linkage between psychologic wellbeing and kidney function.
Visual Abstract
Abstract
Background
Further investigation of the causal effects of psychologic wellbeing on kidney function is warranted.
Methods
In this Mendelian randomization (MR) study, genetic instruments for positive affect, life satisfaction, depressive symptoms, and neuroticism were introduced from a previous genome-wide association study meta-analysis of European individuals. Summary-level MR was performed using the CKDGen data of European ancestry (n=567,460), and additional allele score–based MR was performed in the individual-level data of White British UK Biobank participants (n=321,024).
Results
In summary-level MR with the CKDGen data, depressive symptoms were a significant causative factor for kidney function impairment (CKD OR, 1.45; 95% confidence interval, 1.07 to 1.96; eGFR change [%] beta −2.18; 95% confidence interval, −3.61 to −0.72) and pleiotropy-robust sensitivity analysis results supported the causal estimates. A genetic predisposition for positive affect was significantly associated with better kidney function (CKD OR, 0.69; 95% confidence interval, 0.52 to 0.91), eGFR change [%] beta 1.50; 95% confidence interval, 0.09 to 2.93) and sensitivity MR analysis results supported the finding for CKD outcome, but was nonsignificant for eGFR. Life satisfaction and neuroticism exposures showed nonsignificant causal estimates. In the UK Biobank with covariate-adjusted allele score MR analysis, allele scores for positive affect and life satisfaction were causally associated with reduced risk of CKD and higher eGFR. In contrast, neuroticism allele score was associated with increased risk of CKD and lower eGFR, and depressive symptoms allele score was associated with lower eGFR, but showed nonsignificant association with CKD.
Conclusions
Health care providers in the nephrology field should be aware of the causal linkage between psychologic wellbeing and kidney function.
CKD is one of the major comorbidities related to a large socioeconomic burden.1,2 The prevalence of CKD is increasing worldwide, particularly with the global aging trend. Previous studies have revealed important interventions to reduce the risk of CKD development, particularly focusing on controlling metabolic risk factors. Nevertheless, as the global burden of CKD increases, identifying additional causal factors for kidney function impairment is an important medical issue.
Psychologic wellbeing is related to various health and social outcomes.3,4 In individuals with kidney function impairment, poor psychologic wellbeing has been reported to be prevalent, and studies have focused on the management of poor quality of life.5,6 Furthermore, depression has been reported as a risk factor for incident CKD or progression of CKD by clinical observations.7,8 In the field of cardiology, major depressive disorder is now considered a causal factor for coronary artery disease, as revealed by a recent Mendelian randomization (MR) analysis.9 However, whether such causal effects of negative psychologic wellbeing are present for risk of CKD needs additional investigation. In addition, whether a positive dimension of psychologic wellbeing, including positive affect or life satisfaction, is a causative factor for better kidney function has rarely been described. The evidence for a causal linkage between dimensions of psychologic wellbeing and CKD risk would be essential because it would direct healthcare providers to pay attention to subjective wellbeing, both positive or negative, when managing individuals at risk of kidney function impairment.
The abovementioned MR analysis can reveal the causal effect of a modifiable exposure on complex diseases, because the analysis utilizes inborn-determined genetic instruments minimally affected by clinical confounders or reverse causation.10 MR has been popularized in recent medical literature and revealed important causal factors for certain health outcomes, particularly after the availability of large-scale genetic databases. MR has also been introduced to the nephrology field, assessing the significance of potential risk factors for CKD.11–13
In this study, we aimed to identify the causal effects of dimensions of psychologic wellbeing, including positive affect, life satisfaction, depressive symptoms, and neuroticism, on kidney function by MR analysis. We also performed observational analysis of the population-scale United Kingdom (UK) Biobank cohort to demonstrate a supportive observational association between psychologic wellbeing and CKD. We hypothesized that positive wellbeing senses would be causally linked to better kidney function, whereas negative wellbeing senses would have causal effect on risk of kidney function impairment.
Methods
Ethics Considerations
The study was performed in accordance with the Declaration of Helsinki. The study was approved by the institutional review boards of Seoul National University Hospital (E-2006–043–1131) and the UK Biobank consortium (application 53799). The genetic instrument implemented in this study has been previously published, and summary-level data were utilized. The summary statistics for the CKDGen genome-wide association study (GWAS) meta-analysis for kidney function traits are in the public domain (https://ckdgen.imbi.uni-freiburg.de/). The requirement for informed consent was waived because the study analyzed anonymous databases or summary statistics.
Study Setting
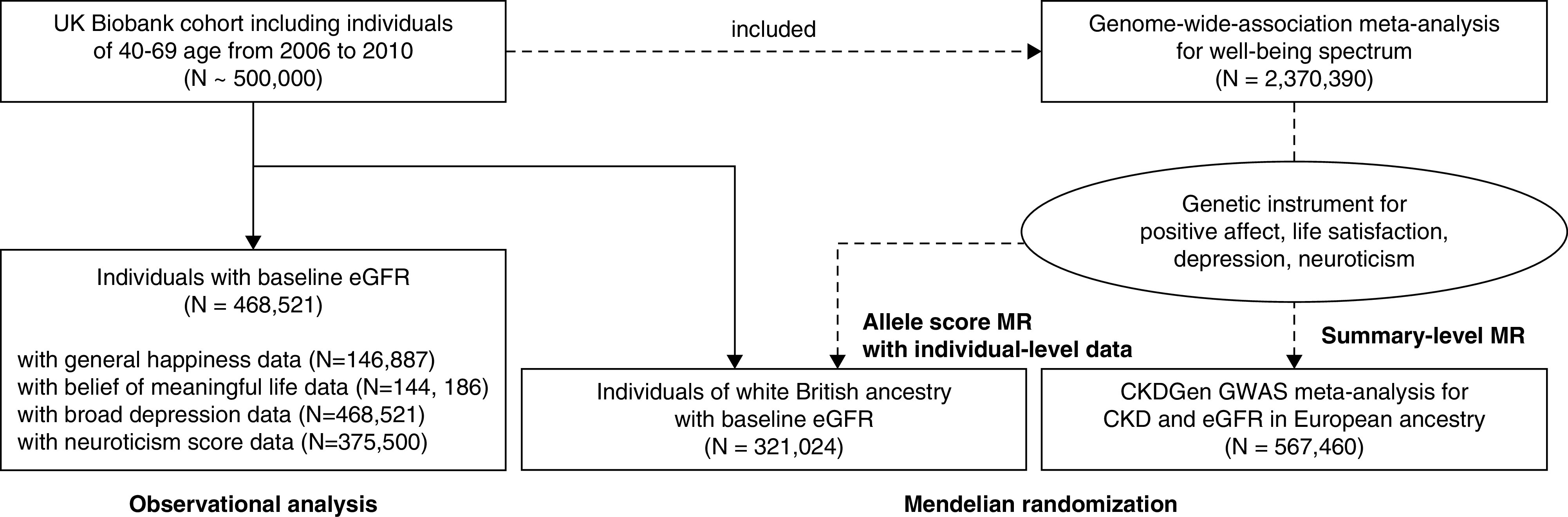
The study consisted of three parts: an observational cohort study with the UK Biobank data, a summary-level MR analysis using the previously published genetic instrument and outcome GWAS summary statistics for kidney function traits using CKDGen, and an allele score–based MR within the UK Biobank (Figure 1). The aim of the first observational analysis was to yield a supportive individual-level observational association between psychologic wellbeing and CKD. The causal effects of dimensions of psychologic wellbeing and risk of CKD were investigated in the summary-level MR analysis. Allele score–based MR was performed to test the causal effects in an independent cohort.
UK Biobank and the Observational Study Data
The UK Biobank is a population-scale prospective cohort including participants aged 40–69 years. The study recruited >500,000 participants from 2006 to 2010 from 22 assessment centers in the UK. The individuals included in the UK Biobank cohort were healthier than the general population in the UK, had a low prevalence of CKD, and had some differences in characteristics, when compared with the general population. The other details of the UK Biobank project have been published previously.14,15
For the observational investigation, among the 502,505 UK Biobank participants available in this study, we included 468,521 individuals with identifiable baseline eGFR values, calculated by the CKD Epidemiology Collaboration equation on the basis of serum creatinine–cystatin C values, measured by the standardized enzymatic method.
Phenotypic Exposures of the Observational Investigation
The exposures of the observational analysis were selected to reflect similar dimensions of psychologic wellbeing as the available genetic instruments: general happiness (to reflect positive affect in the genetic instrument), belief their own life is meaningful (to reflect life satisfaction in the genetic instrument), broad depression (to reflect depressive symptoms in the genetic instrument),16,17 and neuroticism (to reflect neuroticism in the genetic instrument). Responses to questions about general happiness (graded using six possible responses from “extremely happy” to “extremely unhappy”) and belief that one’s own life is meaningful (graded using five possible responses from “an extreme amount” to “not at all”) were collected using a touchscreen questionnaire at the baseline visit. The presence of broad depression was determined by a previous history of medical visits for psychiatric symptoms and has been utilized in the literature on depression in the UK Biobank, because it has been reported to be the most genetically trackable phenotype in the database. The neuroticism score was on the basis of 12 questions related to neurotic symptoms (e.g., mood swings, nervous feelings, tenseness), and the number of “yes” answers consisted of the sum score. The number of individuals reporting the exposures of the observational investigation were different according to the implemented dimensions of psychologic wellbeing in the observational investigation: general happiness (n=146,887), belief their own life is meaningful (n=144,186), broad depression (n=468,521), and neuroticism score (n=375,500). Those without exposure information were not included in the according analysis with the exposure. The details of the collected data, including covariates and missing information, are described in the Supplemental Methods.
CKD Outcome in the UK Biobank Data
Considering the relatively low prevalence of CKD compared with the general population in the UK Biobank,14,18 we implemented a broad definition to define CKD stage 3–5 in the UK Biobank data. The broad definition was to avoid possible underestimation of CKD events because baseline eGFR values could not capture CKD events that occur after the baseline visit, although both prevalent and incident events can be assessed as outcomes in an MR analysis.19 The CKD event was determined by presence of one or more of the following histories: baseline eGFR <60 ml/min per 1.73 m2 by creatinine-, cystatin C–, or creatinine-cystatin C–based CKD Epidemiology Collaboration equation; a prevalent/incident event of ESKD; or an International Classification of Diseases 10th revision diagnostic code indicative of CKD stage 3–5, dialysis, or transplantation.
Statistical Methods for Observational Investigation
The observational association between exposures and CKD stage 3–5 was investigated by the logistic regression model. The first model was age and sex adjusted, and the second multivariable model was additionally adjusted for important confounders, including hypertension, diabetes, body mass index, and the number of household members. A more stringently adjusted model was not constructed, as observed psychologic wellbeing senses would be a sum of various medical and social factors. All analyses were performed by complete-case methods. All statistical analyses were performed using R (version 3.6.2, the R Foundation), and two-sided P values <0.05 were considered significant for the observational investigations and in the genetic analysis, unless otherwise specified.
Genetic Instrument for MR Analysis
The genetic instrument was developed from a novel model-averaging genome-wide association meta-analysis revealing independent (r 2<0.1 in 250 kb window) single nucleotide polymorphisms (SNPs) with genome-wide significance (P<5×10−8) for standardized phenotypes of positive affect (191 SNPs), life satisfaction (148 SNPs), depressive symptoms (239 SNPs), and neuroticism (263 SNPs) from a previous study.20 The study analyzed genetic information from the Social Science Genetic Association Consortium (SSGAC),21 Understanding Society,22 UK Biobank, 23andMe,23,24 and CHARGE25 cohorts of European ancestry, the details of which are presented in Supplemental Table 1. The model-averaging genome-wide association meta-analysis was performed with the published summary statistics, and the joint model averaged effect sizes were calculated, combining the multiple phenotypes as z scores setting the variance of each phenotype in the summary statistics to 1. Polygenic prediction showed that the identified SNPs explained certain variance for positive affect (r 2=1.1%), life satisfaction (r 2=0.9%), depressive symptoms (r 2=1.6%), and neuroticism (r 2=1.6%) in the subsamples. The study reported that related genes were differentially expressed in the subiculum and GABAergic interneurons. We redirected the effects of the genetic instrument toward positive wellbeing sense, and the effect sizes for depressive symptoms/neuroticism were negatively correlated with those for positive affect/life satisfaction.
To construct the genetic instruments for psychologic wellbeing sense and perform MR investigation, we considered the three assumptions of MR analysis required to demonstrate valid causal effects.10 First, the relevance assumption means the genetic instrument is strongly associated with the exposure of interest. The previous GWAS meta-analysis results showed SNPs were strongly associated with wellbeing exposure, and we applied a more conservative approach, including SNPs with a Bonferroni-adjusted significance level (P<0.05/5,000,000 [1×10−8]) of association, with each wellbeing dimension serving as the main genetic instrument. To assess the second assumption, the independence assumption, we trimmed the genetic instruments by excluding SNPs that showed a strong association with possible major confounders, including hypertension, diabetes, obesity, income grade, and number of household members. For this, the UK Biobank dataset for the genetic analysis, described below, was utilized because the data included information on the possible confounders. A GWAS was performed for the possible confounders with SNPs included in the genetic instrument, with adjustment for age, sex, age×sex, age2, and the first 20 principal components, and the SNPs with an association with any of the possible confounders that reached a P value <1×10−5 were excluded from the genetic instrument.11 Analytically, additional sensitivity MR analyses were performed to investigate pleiotropy-robust causal estimates. The third assumption, the exclusion restriction assumption, means the genetic effect should be through the exposure of interest. Although this assumption cannot be formally tested, we performed median-based sensitivity MR analysis, which can relax this assumption in certain portions of the instruments.26
Furthermore, we again pruned the genetic variants within a 500 kb window to include independent signals (r 2<0.1) by the genome data of the UK Biobank. Steiger filtering was performed to ensure the direction of the genetic effects was due to the exposure of interest toward the outcome.27 Last, the SNPs present in both summary statistics for the genetic instrument and for the CKDGen results were included, and the SNPs that were palindromic with intermediate allele frequencies were disregarded.
CKDGen GWAS Meta-analysis for Summary-level MR Analysis
The CKDGen GWAS meta-analysis is the largest GWAS for kidney function outcomes to date.28 We implemented the summary statistics for log-transformed eGFR (calculated from creatinine values, log ml/min per 1.73 m2) and CKD (eGFR <60 ml/min per 1.73 m2) from individuals of European ancestry (n=567,460). The individuals of European ancestry in the CKDGen had a median age of 54 years old, and 50% of them were male, with a median eGFR of 91.4 ml/min per 1.73 m2 and a prevalence of CKD of 9% (Supplemental Table 2). There was possible partial overlap in the samples in the CKDGen data with the GWAS that provided the genetic instrument (e.g., Atherosclerosis Risk in Communities Study).
Summary-level MR Methods
First, as we stated above, we utilized the SNPs associated with psychologic wellbeing exposures that reached a Bonferroni-corrected significance level (P<1×10−8). Next, we filtered SNPs with stronger associations (P<1×10−10) with the exposures to decrease the possibility of remaining horizontal pleiotropy, and repeated the analysis. The main summary-level MR method was the multiplicative random-effects inverse variance weighted method.29 As the level of heterogeneity of the utilized variants was significant, the method yielded the same results as the random-effects inverse variance weighted method, which allows balanced pleiotropy. For the sensitivity MR analysis, MR-Egger regression, which yields pleiotropy-robust causal estimates, was performed.30 The analysis was accompanied by the MR-Egger test for directional pleiotropy, also known as the MR-Egger intercept, and a P value <0.1 was considered to indicate the possibility of directional pleiotropy, namely, a nonzero MR-Egger intercept. Nonsignificant evidence for directional pleiotropy or a significant result (P<0.05) from MR-Egger regression in patients with suspected pleiotropy was necessary to indicate a significant causal estimate, along with a significant result from the inverse variance weighted method. Another sensitivity analysis was performed by Mendelian Randomization Pleiotropy ReSidual Sum and Outlier (MR-PRESSO).31 MR-PRESSO can detect and correct the effects caused by outliers and was performed when the MR-PRESSO global test indicated the presence of significant outliers. In addition, median-based methods were implemented, which have strength when invalid instruments are present. The simple median method, providing valid causal estimates while allowing up to 50% invalid instruments, and the weighted median method, allowing up to 50% of invalid weights contributed by the genetic instruments, were utilized.26 Cochran’s Q statistics P values were calculated to assess the presence of heterogeneity among the implemented genetic instruments. Regarding eGFR outcomes, the log-transformed eGFR unit was changed to a % change unit. The summary-level MR analysis was performed by the TwoSampleMR package in R (version 3.6.2, the R foundation,32), and statistical codes to call the causal estimates by the package are provided in the Supplemental Methods.
Sensitivity Analysis for Summary-level MR
It has been reported that the presence of overlapping samples between the data for genetic instrument generation and outcome assessment may cause bias toward observational associations. Thus, we performed an additional sensitivity analysis, which had been previously suggested,33 by reconstructing the genetic instrument including SNPs with different association strengths with the exposure trait of interest. This method has been suggested to inspect whether the effects from overlapping samples may alter the main findings by testing the effects of changing the instrumental power of genetic instruments. We filtered SNPs within a range of thresholds (P<5×10−8 to <1×10−10), repetitively performed summary-level MR analysis by the inverse variance weighted method, and performed the MR-Egger test to detect directional pleiotropy.
UK Biobank Participants for Allele Score–based MR
For allele score–based MR,34 337,138 participants of White British ancestry passed the quality control filter. Those who were outliers in terms of heterozygosity or had a missing rate, those with sex chromosome aneuploidy and those with excess kinship (≥10 third-degree relatives) were excluded on the basis of the information determined by the UK Biobank consortium, and the samples included in the genetic principal component calculation were from unrelated individuals.35 In addition, those with missing creatinine or cystatin C values were excluded, because the information was necessary to determine the study outcome, and the remaining 321,024 participants were finally included in the investigation.
Statistical Methods for Allele Score–based MR
The UK Biobank data were the largest single cohort used to validate the findings of summary-level MR with individual-level data. Yet, a possible bias from clinical confounders was present, as the UK Biobank sample overlapped with the samples included in the study for genetic instrument development.33 However, assessment of causal estimates in different individual-level data has been performed in previous MR studies for validation purposes, and the analysis had strength as additional consideration of individual-level covariates was possible.19,36,37
We calculated allele scores for the exposures by multiplying the gene dosage matrix with the effect sizes of the main genetic instrument for positive affect, life satisfaction, depressive symptoms, and neuroticism, respectively, using PLINK 2.0 (version alpha 2.3).38 As allele scores were calculated from summing the effect size betas, one increase in the allele score reflected one z score increase in the previous model-averaging genome-wide association meta-analysis results setting phenotype variance to 1 as the summary-level MR.37 The association between genetically predicted psychologic wellbeing sense and log-transformed creatinine–cystatin C eGFR or CKD was tested by linear or logistic regression with adjustment for age, sex, genotype measurement batch, and the first 10 principal components. We additionally performed an analysis by adding phenotypical hypertension, diabetes mellitus, body mass index, number of household members, and income grades in the regression model.37 The effect sizes for the eGFR outcome were transformed into % change units as the summary-level MR analysis.
Summary-level MR for Causal Estimates from Kidney Function to Psychologic Wellbeing
We performed additional summary-level MR to assess the causality from kidney function to subjective wellbeing traits in the utilized data. The 256 independent SNPs that showed genome-wide significant (P<5×10−8) associations with log-transformed eGFR in the CKDGen dataset of European individuals were utilized as potential genetic instruments. To meet the independence assumption, confounder-associated SNPs were disregarded following the abovementioned methods. In addition, to generate a genetic instrument for kidney function that was associated with not only creatinine-based eGFR values but also an alternative kidney function biomarker, namely, BUN, the summary statistics of the GWAS for BUN were implemented as previously suggested. The SNPs with effect directions that were reversed for eGFR and BUN and had stronger association strength than a Bonferroni-corrected level (<0.05/256) with BUN were filtered in. The summary-level MR was performed for summary statistics of the model-averaging GWAS meta-analysis for positive affect, life satisfaction, depressive symptoms, and neuroticism from the study, which provided the genetic instruments for psychologic wellbeing.20 The outcome summary statistics did not include the results from the 23andMe data due to data restrictions.
Results
Clinical Characteristics of the Study Population for the Observational Analysis
The characteristics of the 468,521 study subjects for observational investigation are described in Supplemental Table 3. The median age of the study subjects was 58 years old, with 54.2% females and 46.7% males. Most of the individuals reported moderately happy or very happy status, with more than half of the subjects considering life “very much” or “extremely” meaningful. A total of 34.4% reported broad depression and a history of medical visits for depression-related psychiatric symptoms, which was higher in females. Their median eGFR was 91.2 ml/min per 1.73 m2, with 6.1% CKD stage 3–5 events identified in the UK Biobank data.
Observational Association Between Psychologic Wellbeing and CKD
Patients’ age and sex were strongly associated with their psychologic wellbeing (Supplemental Table 4). In the age- and sex-adjusted model (Table 1), those who reported general happiness status showed approximately 35% lower odds for CKD stage 3–5 when compared with those who were unhappy. The results remained similar even after adjusting for body mass index, hypertension, diabetes, and the number of household members. Those who considered their own life to be “very” or “extremely” meaningful showed approximately 17% lower odds for CKD in the multivariable model than the others. Those who reported broad depression or neuroticism scores ≥5 had approximately 11% or 13% higher adjusted odds for CKD in the multivariable model than those with better psychologic wellbeing.
Table 1.
Observational association between psychologic wellbeing and odds of CKD stage 3–5 in the UK Biobank
| Exposure | N with Exposure | N with CKD and Proportion | Age,- Sex-Adjusted Model | Multivariable Model | ||
|---|---|---|---|---|---|---|
| Adjusted OR (95% CI) | P | adjusted OR (95% CI) | P | |||
| General happiness | ||||||
 Moderately, very, or extremely happy Moderately, very, or extremely happy | 138,686 | 5404 (3.9%) | 0.65 (0.58 to 0.73) | <0.001 | 0.71 (0.63 to 0.80) | <0.001 |
 Moderately, very, or extremely unhappy Moderately, very, or extremely unhappy | 8201 | 347 (4.2%) | Reference | reference | ||
| Belief that own life is meaningful | ||||||
 Very or an extreme amount Very or an extreme amount | 94,812 | 3601 (3.8%) | 0.83 (0.78 to 0.88) | <0.001 | 0.87 (0.82 to 0.92) | <0.001 |
 A moderate amount, a little, or not at all A moderate amount, a little, or not at all | 49,374 | 2034 (4.1%) | reference | reference | ||
| Broad depression | ||||||
 Presence of broad depression Presence of broad depression | 161,349 | 10,484 (6.5%) | 1.22 (1.19 to 1.26) | <0.001 | 1.11 (1.09 to 1.15) | <0.001 |
 Absence of broad depression Absence of broad depression | 307,172 | 18,090 (5.9%) | reference | reference | ||
| Neuroticism | ||||||
 Neuroticism score ≥5 Neuroticism score ≥5 | 153,918 | 9137 (5.9%) | 1.21 (1.17 to 1.24) | <0.001 | 1.13 (1.10 to 1.16) | <0.001 |
 Neuroticism score <5 Neuroticism score <5 | 221,582 | 12,882 (5.8%) | reference | reference | ||
The multivariable model was adjusted for age, sex, body mass index, hypertension, diabetes mellitus, and number of household members. The complete-case multivariable analysis was performed for 145,443 individuals with general happiness exposure (5663 patients with CKD), 142,784 individuals with meaningful life exposure (5549 patients with CKD), 458,965 individuals with broad depression exposure (27,631 patients with CKD), and 369,575 individuals with neuroticism exposure (21,363 patients with CKD), respectively, without any missing information in the covariates. OR, odds ratio; CI, confidence interval.
Genetic Instrument for Psychologic Wellbeing Spectra
After excluding possible confounder-associated SNPs (Supplemental Table 5), the independent SNPs (500-kb window, r 2 < 0.1) with Bonferroni-corrected significant level association (P<1×10−8) with each psychologic wellbeing dimension were filtered in. The summary statistics of the main genetic instrument for positive affect (102 SNPs), life satisfaction (76 SNPs), depressive symptoms (125 SNPs), and neuroticism (133 SNPs) are presented in Supplemental Table 6.
Summary-level MR Results
In the summary-level MR (Figure 2, Table 2, Supplemental Figure 1) conducted with the inverse variance weighted method, genetically predicted positive affect was associated with a lower risk of CKD. In contrast, depressive symptoms showed significant causal estimates toward a higher risk for CKD. With the SNPs and Bonferroni-corrected association level significance (P<1×10−8), the causal estimates from depressive symptoms to CKD risk were supported by the utilized sensitivity analysis results, including the MR-Egger, weighted median, simple median, and MR-PRESSO results. However, only the MR-PRESSO analysis yielded significant causal estimates from positive affect to a lower risk for CKD. Otherwise, the causal estimates from life satisfaction or neuroticism were found to be nonsignificant by the inverse variance weighted method.
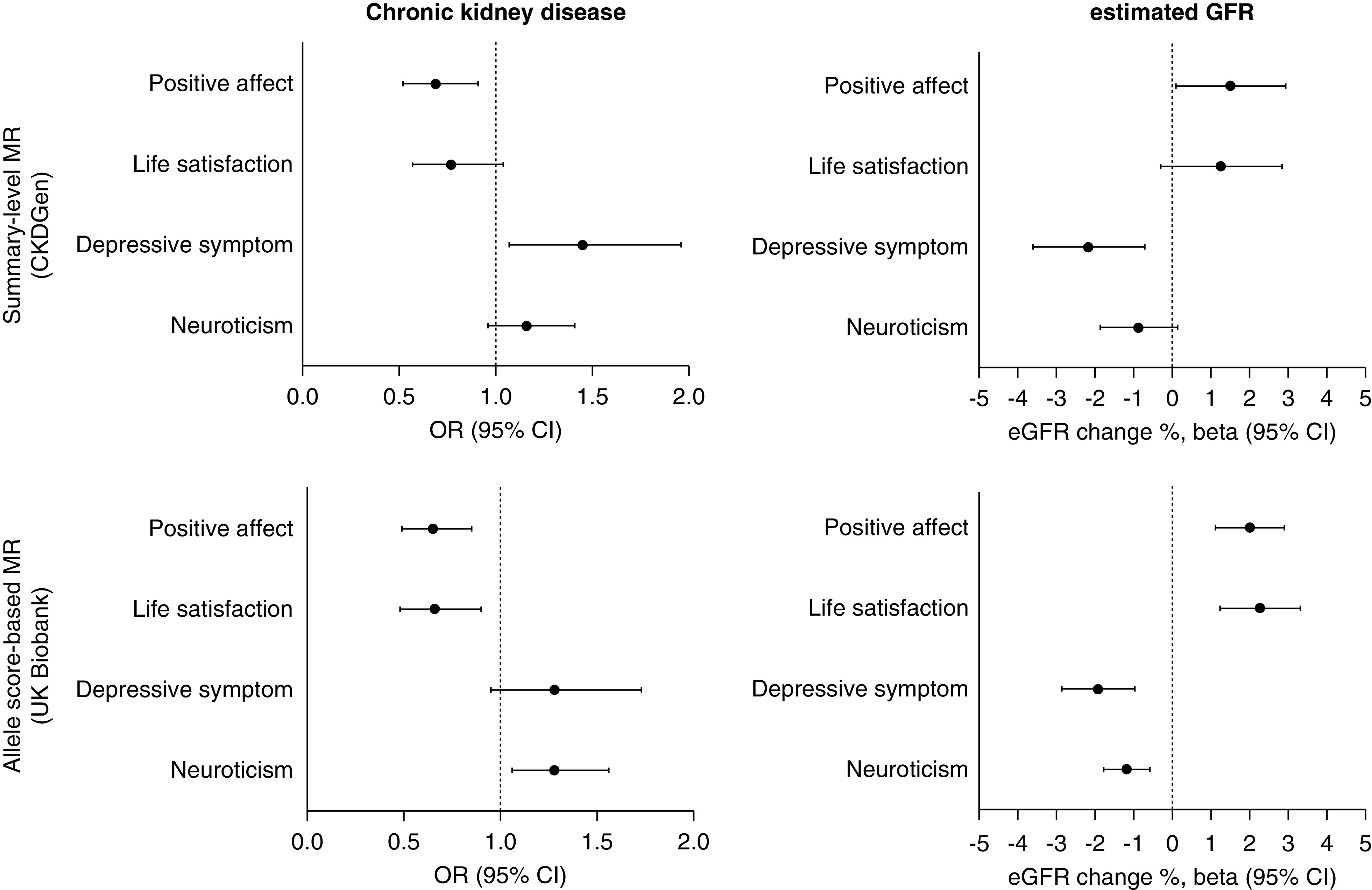
MR analysis results indicated the causal linkage between dimensions of psychologic wellbeing and kidney function. The summary statistics for eGFR and CKD outcomes were introduced from the CKDGen GWAS meta-analysis. The effect sizes in the summary-level MR were from the genetic predisposition to a z score in the previous model-averaging genome-wide association meta-analysis setting phenotype variance to 1. The presented causal estimates from the summary-level MR analysis are from multiplicative random-effect inverse variance weighted method. The effect sizes of the allele score–based MR was similarly scaled to sum of the betas with the unit of a z score in the previous model-averaging genome-wide association meta-analysis setting phenotype variance to 1. The allele score–based MR analysis results are from the covariate-adjusted logistic regression model including age, sex, the first 10 genetic principal components, body mass index, hypertension, diabetes mellitus, income grade, and number of household members. The unit of the eGFR outcome was % change, calculated from the log-transformed eGFR values. Cr, creatinine; CI, confidence interval; OR, odds ratio.
Table 2.
Summary-level MR results for the causal estimates from psychologic wellbeing on risk of CKD in the CKDGen data
| Genetically Predicted Exposure | N of Instrumented SNPs | Cochran’s Q Statistics P Value | MR-Egger Pleiotropy Test P Value | MR Method | OR (CKD) (95% Confidence Interval) a | P |
|---|---|---|---|---|---|---|
| Positive affect | 102 | 0.001 | 0.23 | Multiplicative random-effect IVW | 0.69 (0.52 to 0.91) | 0.008 |
| MR-Egger | 0.34 (0.10 to 1.11) | 0.08 | ||||
| Weighted median | 0.81 (0.57 to 1.16) | 0.25 | ||||
| Simple median | 0.73 (0.52 to 1.03) | 0.07 | ||||
| MR-PRESSO (identified 1 outlier SNPs) | 0.72 (0.55 to 0.94) | 0.02 | ||||
| Life satisfaction | 76 | 0.01 | 0.05 | Multiplicative random-effect IVW | 0.77 (0.57 to 1.04) | 0.09 |
| MR-Egger | 0.16 (0.03 to 0.79) | 0.03 | ||||
| Weighted median | 0.84 (0.57 to 1.22) | 0.36 | ||||
| Simple median | 0.90 (0.62 to 1.29) | 0.56 | ||||
| MR-PRESSO (identified 1 outlier SNPs) | 0.82 (0.60 to 1.13) | 0.18 | ||||
| Depressive symptom | 125 | <0.001 | 0.07 | Multiplicative random-effect IVW | 1.45 (1.07 to 1.96) | 0.02 |
| MR-Egger | 5.21 (1.26 to 21.62) | 0.02 | ||||
| Weighted median | 1.54 (1.07 to 2.22) | 0.02 | ||||
| Simple median | 1.50 (1.03 to 2.18) | 0.03 | ||||
| MR-PRESSO (identified 0 outlier SNPs) | NA | NA | ||||
| Neuroticism | 133 | <0.001 | 0.11 | Multiplicative random-effect IVW | 1.16 (0.96 to 1.41) | 0.12 |
| MR-Egger | 2.46 (0.97 to 6.26) | 0.06 | ||||
| Weighted median | 1.15 (0.91 to 1.47) | 0.24 | ||||
| Simple median | 1.08 (0.85 to 1.37) | 0.53 | ||||
| MR-PRESSO (identified 1 outlier SNP) | 1.13 (0.94 to 1.36) | 0.20 |
IVW, inverse variance weighted.
When we filtered the SNPs with stronger associations (P<1×10−10) with the exposures (Supplemental Table 7), the inverse variance weighted method again yielded significant causal estimates from positive affect to a lower risk for CKD, and from depressive symptoms to a higher risk for CKD. The simple-median, weighted-median, and MR-PRESSO results were significant, as the main findings for genetically predicted positive affect being associated with a lower risk for CKD. Although the causal estimates by MR-Egger regression did not reach a statistically significant level, the MR-Egger intercept P value indicated the absence of significant directional pleiotropy, supporting the causal estimates from the inverse variance weighted method. In this analysis, causal estimates from depressive symptoms to a higher risk for CKD were supported by median-based methods, with the results of the MR-Egger test for directional pleiotropy indicating the absence of a significant pleiotropic effect.
For eGFR outcome (Figure 2, Table 3, and Supplemental Figure 1), the inverse variance weighted method showed genetically predicted positive affect to be associated with higher eGFR values. However, the pleiotropy-robust sensitivity MR analysis results did not support the finding as the MR-PRESSO results did. Genetically predicted depressive symptoms were significantly associated with lower eGFR values by the inverse variance weighted, MR-Egger regression, and MR-PRESSO methods. Again, the causal estimates from life satisfaction or neuroticism did not reach a significant level.
Table 3.
Summary-level MR results for the causal estimates from psychologic wellbeing on eGFR change (%) in the CKDGen data
| Genetically Predicted Exposure | N of SNPs of the Genetic Instrument | Cochran’s Q Statistics P Value | MR-Egger Pleiotropy Test P Value | MR Method | Beta (eGFR change, %) (95% Confidence Interval) a | P |
|---|---|---|---|---|---|---|
| Positive affect | 102 | <0.001 | 0.83 | Multiplicative random-effect IVW | 1.50 (0.09 to 2.93) | 0.04 |
| MR-Egger | 2.18 (−3.86 to 8.59) | 0.49 | ||||
| Weighted median | −0.03 (−1.45 to 1.41) | 0.97 | ||||
| Simple median | 0.61 (−0.86 to 2.11) | 0.42 | ||||
| MR-PRESSO (identified 1 outlier SNPs) | 1.35 (0.01 to 2.71) | 0.05 | ||||
| Life satisfaction | 76 | <0.001 | 0.04 | Multiplicative random-effect IVW | 1.25 (−0.30 to 2.83) | 0.11 |
| MR-Egger | 10.14 (1.54 to 19.46) | 0.02 | ||||
| Weighted median | 0.77 (−0.74 to 2.31) | 0.32 | ||||
| Simple median | 0.74 (−0.70 to 2.21) | 0.32 | ||||
| MR-PRESSO (identified 1 outlier SNPs) | 1.12 (−0.23 to 2.49) | 0.11 | ||||
| Depressive symptom | 125 | <0.001 | 0.001 | Multiplicative random-effect IVW | −2.18 (−3.61 to −0.72) | 0.003 |
| MR-Egger | −12.33 (−18.06 to −6.20) | <0.001 | ||||
| Weighted median | −1.27 (−2.80 to 0.29) | 0.11 | ||||
| Simple median | −1.35 (−2.84 to 0.17) | 0.08 | ||||
| MR-PRESSO (identified 0 outlier SNPs) | −1.78 (−3.09 to −0.45) | 0.01 | ||||
| Neuroticism | 133 | <0.001 | 0.02 | Multiplicative random-effect IVW | −0.88 (−1.87 to 0.13) | 0.09 |
| MR-Egger | −6.48 (−10.87 to −1.87) | 0.007 | ||||
| Weighted median | −0.71 (−1.68 to 0.27) | 0.15 | ||||
| Simple median | −0.67 (−1.65 to 0.32) | 0.18 | ||||
| MR-PRESSO (identified 1 outlier SNP) | −0.85 (−1.73 to 0.05) | 0.07 |
IVW, inverse variance weighted.
When the SNPs with higher association strength (P<1×10−10) were utilized (Supplemental Table 8), the causal estimates from depressive symptoms to a lower eGFR values were significant according to the inverse variance weighted method, as supported by the median-based and MR-PRESSO results. Although the MR-Egger regression result did not reach a significant level, the absence of directional pleiotropy was supported by the MR-Egger intercept P value. For the SNPs with P<1×10−10 for the association for each type of wellbeing, genetically predicted positive affect was no longer significantly associated with a higher eGFR, and the findings for life satisfaction or neuroticism exposure remained to be nonsignificant.
Additional Sensitivity Analysis for Summary-level MR
In an additional sensitivity analysis, the causal estimates made by the summary-level MR showed similar findings as the main results, even after ranges of P value thresholds were applied for wellbeing sense to define the genetic instruments (Supplemental Table 9). The findings suggested that genetically predicted depressive symptoms were associated with a higher risk of CKD or lower eGFR, and genetically predicted positive symptoms were associated with a lower risk of CKD when the conventional genome-wide significance level (P<5×10−8) or a different threshold P<1×10−9 was applied and the number of variants utilized as the genetic instruments was changed.
Allele Score–based MR Results
In the allele score–based MR in the UK Biobank, genetically predicted positive affect or life satisfaction was significantly associated with higher eGFR change (Table 4 and Figure 2). In contrast, genetically predicted depressive symptoms or neuroticism were significantly associated with eGFR decrease. The results remained significant even when we additionally adjusted clinical covariates including hypertension, diabetes, body mass index, income grades, and number of household members.
Table 4.
Causal estimates from individual-level data from allele score–based MR in the UK Biobank
| Outcome | Genetically Predicted Exposure | Age, Sex, and Principal Components Adjusted a | Additional Covariate Adjusted b | ||
|---|---|---|---|---|---|
| OR (CKD) or Beta (eGFR) (95% CI) c | P | OR (CKD) or Beta (eGFR) (95% CI) c | P | ||
| CKD | Positive affect | 0.71 (0.56 to 0.90) | 0.004 | 0.65 (0.49 to 0.85) | 0.002 |
| Life satisfaction | 0.83 (0.63 to 1.10) | 0.19 | 0.66 (0.48 to 0.90) | 0.01 | |
| Depressive symptoms | 1.16 (0.90 to 1.51) | 0.26 | 1.28 (0.95 to 1.73) | 0.11 | |
| Neuroticism | 1.14 (0.97 to 1.35) | 0.11 | 1.28 (1.06 to 1.56) | 0.01 | |
| eGFR (% change) | Positive affect | 1.59 (0.73 to 2.46) | <0.001 | 2.01 (1.12 to 2.91) | <0.001 |
| Life satisfaction | 1.32 (0.33 to 2.33) | 0.009 | 2.27 (1.24 to 3.32) | <0.001 | |
| Depressive symptoms | −1.24 (−2.16 to −0.30) | 0.01 | −1.92 (−2.86 to −0.97) | <0.001 | |
| Neuroticism | −0.86 (−1.44 to −0.28) | 0.004 | −1.18 (−1.77 to −0.58) | <0.001 | |
For CKD outcome, genetically predicted positive affect was significantly associated with lower risk of CKD, whereas other three wellbeing senses did not show significant association. After adjusting clinical covariates, genetically predicted positive affect and life satisfaction was significantly associated with lower risk of CKD, whereas higher allele scores for neuroticism were associated with higher risks of CKD. In contrast, genetically predicted depressive symptoms did not show a significant association with CKD risks, although the direction of the association from a higher allele score for depressive symptoms was toward a higher risk of CKD.
Causal Estimates from Kidney Function to Psychologic Wellbeing
The genetic instrument for kidney function included 30 SNPs present in the summary statistics for psychologic wellbeing with a genome-wide significant association with eGFR, and with a strong and relevant association with BUN levels but not with possible confounders (Supplemental Table 10). The summary-level MR did not yield significant causal estimates from kidney function to dimensions of psychologic wellbeing, and these findings were similar to the results from the utilized sensitivity MR methods (Table 5).
Table 5.
Summary-level MR results for the causal estimates of kidney function on psychologic wellbeing spectra
| Outcome | N of SNPs of the Genetic Instrument | Cochran’s Q Statistics P Value | MR-Egger Pleiotropy Test P Value | MR Method | Beta (95% Confidence Interval) a | P |
|---|---|---|---|---|---|---|
| Positive affect | 30 | 0.03 | 0.10 | Multiplicative random-effect IVW | 0.02 (−0.15 to 0.20) | 0.79 |
| MR-Egger | 0.49 (−0.07 to 1.05) | 0.10 | ||||
| Weighted median | 0.08 (−0.12 to 0.29) | 0.41 | ||||
| Simple median | 0.02 (−0.19 to 0.24) | 0.83 | ||||
| MR-PRESSO (identified 0 outlier SNPs) | NA | NA | ||||
| Life satisfaction | 30 | 0.24 | 0.16 | Multiplicative random-effect IVW | 0.07 (−0.12 to 0.26) | 0.45 |
| MR-Egger | 0.49 (−0.10 to 1.08) | 0.12 | ||||
| Weighted median | 0.13 (−0.11 to 0.37) | 0.29 | ||||
| Simple median | 0.06 (−0.18 to 0.31) | 0.62 | ||||
| MR-PRESSO (identified 0 outlier SNPs) | NA | NA | ||||
| Depressive symptom | 30 | 0.02 | 0.06 | Multiplicative random-effect IVW | −0.02 (−0.15 to 0.12) | 0.82 |
| MR-Egger | −0.43 (−0.87 to 0.01) | 0.07 | ||||
| Weighted median | −0.08 (−0.25 to 0.09) | 0.37 | ||||
| Simple median | −0.02 (−0.18 to 0.14) | 0.34 | ||||
| MR-PRESSO (identified 1 outlier SNPs) | −0.04 (−0.16 to 0.09) | 0.54 | ||||
| Neuroticism | 30 | 0.02 | 0.08 | Multiplicative random-effect IVW | −0.03 (−0.25 to 0.18) | 0.76 |
| MR-Egger | −0.66 (−1.36 to 0.05) | 0.08 | ||||
| Weighted median | −0.11 (−0.36 to 0.14) | 0.39 | ||||
| Simple median | −0.03 (−0.28 to 0.23) | 0.82 | ||||
| MR-PRESSO (identified 1 outlier SNPs) | −0.07 (−0.27 to 0.13) | 0.49 |
Discussion
In summary-level MR with the CKDGen data, depressive symptom was a significant causative factor for kidney function impairment and the causal estimates were supported by pleiotropy-robust sensitivity analysis results. A genetic predisposition for positive affect was significantly associated with better kidney function and sensitivity MR analysis results supported the finding to CKD outcome, but was nonsignificant to eGFR. Other psychologic wellbeing senses observationally associated with CKD (e.g., life satisfaction or neuroticism) did not show significant causal estimates for CKD or eGFR in the CKDGen data. In the UK Biobank with covariate-adjusted allele score MR analysis, allele scores for positive affect and life satisfaction were causally associated with reduced risk of CKD and higher eGFR. In contrast, neuroticism allele score was associated with increased risk of CKD and lower eGFR, and depressive symptom allele score was associated with lower eGFR, but showed nonsignificant association with CKD. Overall, our study results demonstrated that psychologic wellbeing sense is causally linked to kidney function.
Along with previous reports showing a high prevalence of psychologic discomfort and impaired quality of life in patients with CKD,5 an association between psychologic wellbeing and the risk of kidney function impairment has been suggested. A study including patients with diabetes mellitus suggested the presence of depression was a risk factor for incident CKD.8 Another study including patients with CKD showed that depression is associated with a higher risk of accelerated kidney function decline.7 Additional study was necessary to investigate whether actual causality is present in dimensions of psychologic wellbeing on the risk of kidney function impairment, but a clinical trial with interventions for patients’ subjective wellbeing is difficult to perform. With the benefit of recently utilized MR analysis, which can demonstrate “causal” effects by implementing genetic instruments, we showed that certain dimensions of psychologic wellbeing may causally affect kidney function. Thus, our study encourages health care providers to pay attention to psychologic wellbeing when assessing or managing individuals with regard to their risk of kidney function impairment.
MR requires the following assumptions to be met to demonstrate causal effects.10 First, the relevance assumption is that the genetic instrument should be strongly associated with the exposure of interest. We implemented the findings from a previous study that robustly identified SNPs that were strongly related to the psychologic wellbeing dimensions by joint analysis of multiple cohorts reporting genetic information explaining the largest degree of variance for the complex phenotypical wellbeing dimension to date. Second, the independence assumption is that the utilized genetic instrument should be independent from confounders. We used a large number of SNPs to genetically explain the partial variance of the complex psychologic wellbeing exposures; however, such an approach may increase possibility of heterogeneity or horizontal pleiotropy. We made efforts to investigate pleiotropy-robust causal estimates by excluding the genetic variants associated with potential confounders and by performing sensitivity MR analyses that also gave supportive results for depressive symptoms and positive affect exposures in the summary-level MR. Last, the exclusion restriction assumption is that the genetic instrument should affect the risk of the disease of interest through the exposure trait. Although a direct test for this assumption is not possible, the median-based MR sensitivity analysis eases this assumption for up to half of the genetic instruments, and yielded supportive causal estimates in part of the results. Therefore, through MR, our study results suggest the presence of causal effects of certain dimensions of psychologic wellbeing on kidney function.
Whether the risk of kidney function impairment can be improved through modification of psychologic wellbeing should be answered in a future clinical trial, which would further confirm the clinical implications of our findings. Being engaged in favorable health behaviors in those with good psychologic wellbeing has been considered a cause of the health effects of wellbeing.39,40 In addition, a prior study suggested that psychologic symptoms may directly affect one’s sympathetic nervous system41 or metabolic health,42,43 resulting in better health outcomes. Considering the possible mechanisms and our study results, psychologic wellbeing may be a targetable factor for interventions to ameliorate the burden of kidney function impairment. Additionally, screening psychologic wellbeing status may be considered in those at risk of kidney function impairment and with consideration for early intervention. Conversely, clinicians may consider monitoring kidney function in those with depressive symptoms, because the risk of kidney function impairment may be higher in those with such negative psychologic wellbeing. The above suggested clinical implications support the possible benefits of multidisciplinary care, including psychologic assessment and interventions in the nephrology field.44,45
Some of the results from our MR investigation need additional explanations. First, in the summary-level MR, some causal estimates from sensitivity MR methods were nonsignificant. MR-Egger regression yields pleiotropy-robust causal estimates and provides a test for directional pleiotropy by the MR-Egger intercept.30 However, the method has weak statistical power, particularly when the number of instrumented SNPs decreases, and can still be biased, as inflated type 1 error, if the untestable INstrument Strength Independent of Direct Effect (InSIDE) assumption is violated, which occurs when a group of instruments act through pleiotropic pathways.46 The simple or weighted median method can provide consistent causal estimates, allowing up to half of invalid variants or contributed weights, and even when violating the InSIDE assumption or other main MR assumptions.26 In our results, MR-Egger regression indicated significant causal estimates from positive affect or depressive symptoms when the possibility of directional pleiotropy remained, further supporting the presence of causal effects of psychologic wellbeing on kidney function. In patients of nonsignificant causal estimates by MR-Egger regression, which might have been related to the method’s weak statistical power, the MR-Egger test for directional pleiotropy indicated nonsignificant pleiotropy, also supporting the causal estimates by the inverse variance weighted method. However, whether the InSIDE assumption was met could still be questioned, particularly under conditions with nonzero MR-Egger intercepts and with nonsignificant results by the median-based methods. However, the simple or weighted median methods also yielded significant causal estimates in certain investigations, and the inverse variance weighted method showed significant results when reducing the number of SNPs with a decreased possibility of horizontal pleiotropy, supporting that the suggested causal effects are not the findings from type 1 error. The complexity in psychologic wellbeing exposures, which is only partially explained by the genetic information and difficulty in objectively measuring such phenotypes, may be the reason some analyses have shown marginal statistical power with the genetic instrument in several investigations (e.g., weak instrument bias). The issue of the remaining possibility of directional pleiotropy or weak instrument bias in our MR investigation should be noted. Next, along with the abovementioned limitation of the utilized genetic instrument, although the instrument genetically explained the largest degree of psychologic wellbeing to date, the null findings from life satisfaction and neuroticism in the summary-level MR are not conclusive. Considering that types of psychologic wellbeing are closely correlated clinically and genetically,20 the causal effects might have been less apparent in the statistical analyses because of the weak instrument bias.
There were some differences in the results from the CKDGen data and the UK Biobank. The differences in findings potentially originated from the differences in the population’s genetic structure, the proportion of CKD events affected by one’s wellbeing sense, or the population’s socioeconomic or clinical characteristics. In addition, along with the possibility of a false-negative finding in some of the results due to weak instruments, the allele score–based MR results should be interpreted with caution as the possibility of a false-positive finding remains because the analysis cannot detect a directional pleiotropic effect.34
The reverse-direction MR implied that kidney function may not have significant causal effects on psychologic wellbeing in the general population within the utilized data. However, considering a certain diseased state may cause impairment in psychologic wellbeing, the null results may be due to the heterogeneous nature of the phenotyping for the psychologic wellbeing dimension in the utilized summary statistics. Additionally, because the genetic instrument for kidney function in our investigation may hardly capture the signals from profound kidney dysfunction or acute kidney injury, the causal effects from such severe kidney function impairment on psychologic wellbeing cannot be investigated in this study.
There are several limitations and unanswered questions in our study. First, the study does not confirm whether directly treating poor wellbeing would lower the risk of CKD (e.g., by treating depression), because psychologic wellbeing is a mixture of health dimensions. Such a question should be answered by a future clinical trial targeting subjective wellbeing, and the mechanism for the benefits of positive wellbeing on kidney function warrants additional study. Second, because MR analyses generally provide qualitative evidence for the directions of the causal effects, the magnitude of the potential clinical effect of subjective wellbeing on kidney function may be different from the genetic effect sizes.47 Moreover, heterogeneous phenotyping methods for wellbeing dimensions additionally limit the quantitative interpretation of the study and might have caused weak instrument bias in some of the sensitivity MR investigations. Third, the exposures tested in this study are subjective terms that may not be similarly applied to certain individuals (e.g., someone feeling a sense of wellbeing from dangerous social or medical behavior). Thus, clinicians should carefully evaluate subjective psychologic wellbeing on the basis of an individual’s condition. Fourth, the study is mainly on the basis of the general population, so whether the findings can be applied to those with severe illness or established CKD is not certain. Last, the study mainly included data from individuals of European ancestry; thus, the generalizability of the findings is not secured for individuals of other ethnicities.
In conclusion, this MR analysis generally supports that psychologic wellbeing can have a causal effect on kidney health. Health care providers in the nephrology field should be aware of the causal linkage between psychologic wellbeing and kidney function.
Disclosures
K. Joo reports receiving research funding from Baxter. All remaining authors have nothing to disclose.
Funding
This work was supported by the Ministry of Trade, Industry & Energy (Korea) through funding from the Industrial Strategic Technology Development Program, Development of Biocore Technology (10077474, Development of early diagnosis technology for acute/chronic renal failure) and a Seoul National University R&DB Foundation grant (800-20190571). The study was performed independently by the authors.
Supplementary Material
Acknowledgments
The study was on the basis of the data provided by the UK Biobank consortium (application 53799). We thank the investigators of the CKDGen, BIOS, and SSGAC consortia who provided the summary statistics for the genetic instrument and the outcome of this study.
The corresponding author attests that all listed authors meet the authorship criteria and that no others meeting the criteria have been omitted. D.K. Kim, H. Lee, K. Kim, K.W. Joo, and S. Park contributed to the conception and design of the study; C.S. Lim, D.K. Kim, J.P. Lee, K.W. Joo, M.W. Kang, S.S. Han, S. Lee, Y. Kim, Y.C. Kim, Y. Lee, and Y.S. Kim provided statistical advice and interpreted the data; K. Kim and S. Park performed the main statistical analysis, assisted by S. Lee and Y. Kim; C.S. Lim, D.K. Kim, H. Lee, J.P. Lee, K.W. Joo, and Y.S. Kim, and provided advice regarding the data interpretation; C.S. Lim, H. Lee, J.P. Lee, K.W. Joo, S.S. Han, Y.C. Kim, and Y.S. Kim provided material support during the study; D.K. Kim and S. Park had full access to all data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis; All authors participated in drafting the manuscript, reviewed the manuscript, and approved the final version to be published.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/10.1681/ASN.2020071086/-/DCSupplemental.
Supplemental Table 1. The study population of the genome-wide association meta-analysis for wellbeing spectrum by Baselmans et al.
Supplemental Table 2. Characteristics of the CKDGen genome-wide association meta-analysis dataset of European ancestry.
Supplemental Table 3. Characteristics of the observational analysis dataset of the UK Biobank.
Supplemental Table 4. Association between psychological wellbeing and age or sex.
Supplemental Table 5. The association between the list of SNPs and potential confounders in the UK Biobank.
Supplemental Table 6. The summary statistics of the final genetic instrument utilized for psychological wellbeing dimensions.
Supplemental Table 7. Summary-level MR results for the causal estimates from psychological wellbeing on risk of CKD from the genetic instrument with stronger association (P<1×10−10) with the wellbeing exposures.
Supplemental Table 8. Summary-level MR results for the causal estimates from psychological wellbeing on eGFR change (%) from the genetic instrument with stronger association (P<1×10−10) with the wellbeing exposures.
Supplemental Table 9. Sensitivity analysis results by including SNPs ranges of association strengths for psychologic wellbeing as the genetic instruments for summary-level MR.
Supplemental Table 10. List of the 30 SNPs utilized as the genetic instrument for kidney function.
Supplemental Figure 1. Scatter plots visualizing the summary-level Mendelian randomization results.
References
Articles from Journal of the American Society of Nephrology : JASN are provided here courtesy of American Society of Nephrology
Full text links
Read article at publisher's site: https://doi.org/10.1681/asn.2020071086
Read article for free, from open access legal sources, via Unpaywall:
https://jasn.asnjournals.org/content/jnephrol/32/6/1484.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/103013181
Article citations
The association between patterns of exposure to adverse life events and the risk of chronic kidney disease: a prospective cohort study of 140,997 individuals.
Transl Psychiatry, 14(1):424, 07 Oct 2024
Cited by: 0 articles | PMID: 39375339 | PMCID: PMC11458756
Recent advances in the application of Mendelian randomization to chronic kidney disease.
Ren Fail, 46(1):2319712, 04 Mar 2024
Cited by: 0 articles | PMID: 38522953 | PMCID: PMC10913720
Review Free full text in Europe PMC
Association between incident depression and clinical outcomes in patients with chronic kidney disease.
Clin Kidney J, 16(11):2243-2253, 26 May 2023
Cited by: 3 articles | PMID: 37915918 | PMCID: PMC10616442
Genetic variations in HMGCR and PCSK9 and kidney function: a Mendelian randomization study.
Kidney Res Clin Pract, 42(4):460-472, 22 May 2023
Cited by: 4 articles | PMID: 37448291 | PMCID: PMC10407636
Genetically Predicted Body Selenium Concentration and estimated GFR: A Mendelian Randomization Study.
Kidney Int Rep, 8(4):851-859, 20 Jan 2023
Cited by: 0 articles | PMID: 37069993 | PMCID: PMC10105058
Go to all (12) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A Mendelian randomization study found causal linkage between telomere attrition and chronic kidney disease.
Kidney Int, 100(5):1063-1070, 30 Jul 2021
Cited by: 32 articles | PMID: 34339747
Causal Effects of Homocysteine, Folate, and Cobalamin on Kidney Function: A Mendelian Randomization Study.
Nutrients, 13(3):906, 11 Mar 2021
Cited by: 7 articles | PMID: 33799553 | PMCID: PMC8001564
Observational or Genetically Predicted Higher Vegetable Intake and Kidney Function Impairment: An Integrated Population-Scale Cross-Sectional Analysis and Mendelian Randomization Study.
J Nutr, 151(5):1167-1174, 01 May 2021
Cited by: 5 articles | PMID: 33693791
Recent advances in the application of Mendelian randomization to chronic kidney disease.
Ren Fail, 46(1):2319712, 04 Mar 2024
Cited by: 0 articles | PMID: 38522953 | PMCID: PMC10913720
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Medical Research Council (2)
UK Biobank (core renewal)
Professor Sir Rory Collins, UK Biobank
Grant ID: MC_PC_17228
UK Biobank
Professor Sir Rory Collins, UK Biobank
Grant ID: MC_QA137853
Ministry of Trade, Industry & Energy (1)
Grant ID: 10077474
Seoul National University (1)
Grant ID: 800-20190571
 3
,
4
,
7
3
,
4
,
7