Abstract
Free full text

30 years of repeat expansion disorders: What have we learned and what are the remaining challenges?
Abstract
Tandem repeats represent one of the most abundant class of variations in human genomes, which are polymorphic by nature and become highly unstable in a length-dependent manner. The expansion of repeat length across generations is a well-established process that results in human disorders mainly affecting the central nervous system. At least 50 disorders associated with expansion loci have been described to date, with half recognized only in the last ten years, as prior methodological difficulties limited their identification. These limitations still apply to the current widely used molecular diagnostic methods (exome or gene panels) and thus result in missed diagnosis detrimental to affected individuals and their families, especially for disorders that are very rare and/or clinically not recognizable. Most of these disorders have been identified through family-driven approaches and many others likely remain to be identified. The recent development of long-read technologies provides a unique opportunity to systematically investigate the contribution of tandem repeats and repeat expansions to the genetic architecture of human disorders. In this review, we summarize the current and most recent knowledge about the genetics of repeat expansion disorders and the diversity of their pathophysiological mechanisms and outline the perspectives of developing personalized treatments in the future.
Normal and pathological tandem repeats in human genomes
The human genome contains more than one million annotated tandem repeats (TRs).1,2 Due to their repetitive nature, TRs have the highest mutational rate in the genome and are typically polymorphic and multiallelic, with the longest alleles being the most unstable. TRs are mainly scattered in non-coding regions of the genome although repetitions of triplets, usually limited in size, can also be found in coding parts of genes.1 Coding TRs may occasionally become hotspots for harmful frameshift mutations.3
TRs are usually divided into microsatellites (1–9 bp repeats; or short tandem repeats, STRs) and minisatellites (10–99 bp repeats), which together form the variable number of tandem repeats (VNTRs), and satellites (≥100 bp repeats), which mainly constitute heterochromatin and centromeres. STRs have widely been used as markers in genetic linkage analyses and forensic studies, but they are still an enigmatic and understudied class of variation. STRs present in human genomes are recent in terms of evolution and are either human specific or conserved only in very close primate species.4 Most STRs have arisen from other repeated elements, such as long interspersed nuclear element (LINE) and short interspersed nuclear element (SINE), including Alu elements.5 The evolutionary constraints applying to these dynamic, rapidly evolving variations and how TRs have contributed to shape human genomes are still largely unknown although recent evidence suggests that they could play an important role in the regulation of gene expression.6,7
The expansion of TR length across generations is a well-characterized process that results in at least 50 known disorders (Tables 1, ,2,2, and S1). The first expansions, both identified on chromosome (chr) X in 1991, were CGG repeats in the 5′ untranslated region (UTR) of FMR1 (MIM: 309550) and CAG repeats in the first exon of AR (MIM: 313700) as the underlying causes of fragile X syndrome8,9 (FXS [MIM: 300624]) and spinal bulbar muscular atrophy (SBMA [MIM: 313200]),10 respectively (Figure 1). In the next 10 years, a wave of repeat expansion discovery followed, revealing the basis of more than 25 hereditary disorders, most of which primarily affect the central nervous system.
Table 1
List of coding repeat expansion disorders, sorted according to their year of discovery
| ID year | Disordera | Inheritanceb | Chr | Gene | Repeat motif | Normal repeat range | Pathological repeat range | Pathological mechanism(s)c | ID methodd | Ref |
|---|---|---|---|---|---|---|---|---|---|---|
| 1991 | SBMA | XLR | Xq12 | AR | CAG | 9–36 | ≥38–68 | polyQ, RNA? | L, CGA | La Spada et al.10 |
| 1993 | HD | AD | 4p16.3 | HTT | CAG | 6–35 | ≥36–250 | polyQ, RNA? | L, ExScr | The Huntington’s Disease Collaborative Research Group11 |
| 1993 | SCA1 | AD | 6p22 | ATXN1 | CAG | 6–38 | ≥39–88 | polyQ, RNA? | L, ExScr | Orr et al.12 |
| 1994 | DRPLA | AD | 12p13.31 | ATN1 | CAG | 3–35 | ≥48–93 | polyQ, RNA? | L, ExScr | Koide et al.,13 Nagafuchi et al.14 |
| 1994 | SCA3 | AD | 14q32 | ATXN3 | CAG | 12–44 | ≥55–87 | polyQ, RNA (foci) | L, cl | Kawaguchi et al.15 |
| 1996 | SCA2 | AD | 12q24 | ATXN2 | CAG | 13–31 | ≥32–500 | polyQ, RNA? | L, cl | Pulst et al.16 |
| 1996 | SCA7 | AD | 3p21 | ATXN7 | CAG | 4–33 | ≥37–460 | polyQ, RNA? | L, cl | Lindblad et al.17 |
| 1996 | SPD1 | AD | 2q31.1 | HOXD13 | GCG | 15 | 24 | polyA (DN) | L, CGA | Akarsu et al.18 |
| 1997 | SCA6 | AD | 19p13 | CACNA1A | CAG | 4–18 | ≥20–33 | polyQ, RNA? | L, ExScr | Zhuchenko et al.19 |
| 1997 | BCCD | AD | 6p21 | RUNX2 | GCN | 17 | 27 | polyA (DN/LoF?) | L, CGA | Mundlos et al.20 |
| 1998 | OPMD | AD | 14q11.2 | PABPN1 | GCG | 6–10 | ≥12–17 | polyA (DN/LoF?) | L, ExScr | Brais et al.21 |
| 1999 | SCA17 | AD | 6q27 | TBP | CAG (or CAG/CAA) | 25–40 | ≥43–66 | polyQ, RNA? | L, CGA | Koide et al.22 |
| 2000, 2002 | HFGS | AD | 7p15.2 | HOXA13 | tract 1: GCN | 14 | 22 | polyA (DN/LoF?) | L, CGA | Goodman et al.,23 Utsch et al.24 |
| tract 2: GCN | 12 | 18 | ||||||||
| tract 3: GCN | 18 | 24–-30 | ||||||||
| 2001 | HDL2 | AD | 16q24.2 | JPH3 | CAG | 6–28 | ≥41–58 | polyQ, RNA (foci) | L, RED | Margolis et al.25 |
| 2001 | BPES | AD | 3q23 | FOXL2 | GCN | 14 | 19–24 | polyA (LoF) | L, CGA | De Baere et al.26 |
| 2001 | HPE5 | AD | 13q32 | ZIC2 | GCN | 15 | 25 | polyA (LoF) | L, CGA | Brown et al.27 |
| 2002 | EIEE1 | XLR | Xp21.3 | ARX | tract 1: GCN | 16 | 23 | polyA (DN) | L, CGA | Strømme et al.28 |
| tract 2: GCN | 12 | 20 | ||||||||
| 2002 | MRGH | XLR | Xq26.3 | SOX3 | GCN | 15 | 26 | polyA (LoF) | L, CGA | Laumonnier et al.29 |
| 2003 | CCHS | AD | 4p13 | PHOX2B | GCN | 20 | 25–29 | polyA (DN) | L, CGA | Amiel et al.30 |
ID, identification; chr, chromosome; ref, reference(s)
Table 2
List of noncoding repeat expansion disorders involving motifs up to 12 nucleotides, sorted according to their year of discovery
| ID year | Disordera | Inheritanceb | Chr | Gene | Location | Repeat motif | pathogenic motif when different | Normal repeat range | Pathological repeat range | Pathological mechanism(s)c | ID methodd | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1991 | FXS | XLD | Xq27.3 | FMR1 | 5′ UTR | CGG | – | 5–50 | >200 | Me/GS | K, FS, L, Me, cl | Oberlé et al.,8 Verkerk et al.9 |
| 1992 | DM1 | AD | 19q13.32 | DMPK | 3′ UTR | CTG | – | 5–37 | >50–10,000 | RNA (foci, PS), RAN (polyQ) | L, cl | Mahadevan et al.31 |
| 1993 | FRAXE | XLR | Xq28 | AFF2 | 5′ UTR | CCG | – | 4–39 | ≥200–900 | Me/GS | K, L | Knight et al.32 |
| 1996 | FRDA | AR | 9q21.11 | FXN | intron | GAA | – | 5–34 | ≥66–1300 | EGS | L, ExScr | Campuzano et al.33 |
| 1997 | EPM1 | AR | 21q22.3 | CSTB | promoter / 5′ UTR | CCCCGCCCCGCG | – | 2–3 | ≥30–75 | EGS | L, cl, exp | Lalioti et al.34 |
| 1998 | FXPOI | XL | Xq27.3 | FMR1 | 5′ UTR | CGG | – | 5–50 | 55–200 | RNA (foci), RAN | Fam, ExScr | Conway et al.35 |
| 1999 | SCA8 | AD | 13q21 | ATXN8 / ATXN8OS | 3′ UTR | CAG/CTG | – | 15–50 | >74–250 | RNA (foci), RAN | L, cl | Koob et al.36 |
| 1999 | SCA12 | AD | 5q31 | PPP2R2B | 5′ UTR | CAG | – | 4–32 | ≥43–78 | RAN (polyG)? | L, RED | Holmes et al.37 |
| 2000 | SCA10 | AD | 22q13 | ATXN10 | intron | ATTCT / ATTGT | – | 10–32 | >280–4,500 | RNA (foci) | L, ExScr | Matsuura et al.38 |
| 2001 | DM2 | AD | 3q21.3 | CNBP | intron | CCTG / CAGG | – | 11–30 | >50−11,000 | RNA (foci) | L, ExScr | Liquori et al.39 |
| 2001 | FXTAS | XLR | Xq27.3 | FMR1 | 5′ UTR | CGG | – | 5–50 | 55–200 | RNA (foci), RAN (polyG) | Fam | Hagerman et al.40 |
| 2009 | SCA31 | AD | 16q22 | BEAN1 / TK2 | intron | TAAAA | TGGAA (+TAGAA) | variable | ≥110–760 | RNA (foci, PS), RAN? | L | Sato et al.41 |
| 2011 | SCA36 | AD | 20p13 | NOP56 | intron | GGCCTG | – | 5–14 | ≥650–2,500 | RNA (foci) | L, ExScr | Kobayashi et al.42 |
| 2011 | FTD/ALS | AD | 9p21.2 | C9ORF72 | 5′ UTR / intron | GGGGCC | – | 3–25 | >30 | RNA (foci), RAN (polyG) | L, WGS | Renton et al.,43 DeJesus-Hernandez et al.44 |
| 2015 | FECD3 | AD | 18q21.2 | TCF4 | intron | CTG | – | 5–31 | >50 | unknown | L, GWAS | Mootha et al.45,46 |
| 2017 | XDP | XLR | Xq13.1 | TAF1 | intron (retrotransposon) | CCCTCT | – | none | 30–55 | unknown | L, exp | Bragg et al.47 |
| 2017 | SCA37 | AD | 1p32 | DAB1 | intron | ATTTT | ATTTC | 7-400 (ATTTT) | ≥31–75 ATTTC | RNA | L, ExScr | Seixas et al.48 |
| 2018 | FAME1 | AD | 8q24 | SAMD12 | intron | ATTTT | ATTTC | 7-exp (ATTTT) | ≥440–3,680 ATTTC | RNA? | L, ExScr | Ishiura et al.49 |
| 2018 | FAME6 | AD | 16p12.1 | TNRC6A | intron | ATTTT | ATTTC | N/A | N/A | RNA? | TRhist | Ishiura et al.49 |
| 2018 | FAME7 | AD | 4q32.1 | RAPGEF2 | intron | ATTTT | ATTTC | N/A | N/A | RNA? | TRhist | Ishiura et al.49 |
| 2019 | BSS | AR | 16p12.3 | XYLT1 | promoter | CGG | – | 9–20 | 120–800 | Me/GS | ADO, Me | LaCroix et al.50 |
| 2019 | CANVAS | AR | 4p14 | RFC1 | intron | AAAAG / AAAGG / AAGAG / AGAGG | AAGGG | variable | ≥400–2,000 | unknown | L, WGS | Cortese et al.,51 Rafehi et al.52 |
| 2019 | GDPAG | AR | 2q32.2 | GLS | 5′ UTR | GCA | – | 8–16 | ≥680–1,400 | EGS | CGA | van Kuilenburg et al.53 |
| 2019 | NIID | AD | 1q21.2 | NOTCH2NLC | 5′ UTR / exon 1 | CGG | – | 7–60 | ≥61–500 | Me/GS, RAN (polyG)? | TRhist, L, WGS | Ishiura et al.,54 Sone et al.,55 Tian et al.56 |
| 2019 | OPDM1 | AD | 8q22.3 | LRP12 | 5′ UTR | CGG | – | 13–45 | 90–130 | RAN (polyG)? | TRhist | Ishiura et al.54 |
| 2019 | OPML1 | AD | 10q22.3 | NUTM2B-AS1 | noncoding RNA gene | CGG/CCG | – | 3–16 | 40–60 | RAN (polyG)? | TRhist | Ishiura et al.54 |
| 2019 | FAME4 | AD | 3q27.1 | YEATS2 | intron | ATTTT | ATTTC | 7–400 (ATTTT) | N/A | RNA? | L, ExScr | Yeetong et al.57 |
| 2019 | FAME2 | AD | 2q11.2 | STARD7 | intron | ATTTT | ATTTC | 9–20 | ≥661–735 | RNA? | L, ExScr | Corbett et al.58 |
| 2019 | FAME3 | AD | 5p15.2 | MARCHF6 | intron | ATTTT | ATTTC | 10–30 | ≥660–2,800 | RNA? | L, ExScr | Florian et al.59 |
| 2020 | OPDM2 | AD | 19p13.12 | GIPC1 | 5′ UTR | CGG | – | 12–32 | ≥97–120 | unknown | L, WGS, LRS | Deng et al.60 |
ID, identification; chr, chromosome; N/A, not available; ref, reference(s); UTR, untranslated region
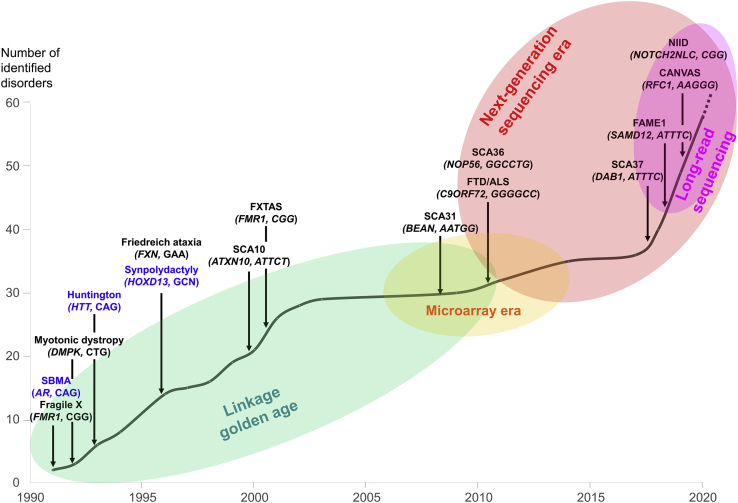
Timeline of repeat expansion discovery in human disorders
During the first twenty years, linkage analyses were the gold standard allowing to map the genomic region containing repeat expansions. The development of next-generation sequencing, and more particularly long-read sequencing, has marked a new milestone, making their identification at a genome-wide scale possible, and has allowed a new wave of repeat expansion discovery.
As illustrated by the initial discoveries, two main types of STR expansions exist: expansions affecting coding regions, mainly leading to abnormally long stretches of polyglutamine (polyQ, mostly encoded by CAG codons) or polyalanine (polyA, encoded by GCN codons) within proteins (Table 1), and expansions altering non-coding regions of genes (Figure 2).61 CAG triplet expansions, including those found in Huntington disease (HD [MIM: 143100]), spinocerebellar ataxias type 1 (SCA1 [MIM: 164400]), SCA2 (MIM: 183090), SCA3 (Machado-Joseph disease [MIM: 109150]), SCA6 (MIM: 183086), SCA7 (MIM: 164500), SCA17 (MIM: 607136), and dentatorubral-pallidoluysian atrophy (DRPLA [MIM: 125370]) are typically associated with neuronal intranuclear protein inclusions that contain the protein (or part of it) exhibiting the expanded polyQ stretch. These inclusions result from the propensity of these abnormal polyQ chains to form β sheet structures leading to intermolecular cross-beta oligomerization and to the formation of insoluble fibrillar aggregates, a mechanism usually described as a toxic gain-of-function at the protein level.62 PolyA peptides can also form β sheet structures, but polyA expansions, usually much more limited in size compared to their polyQ counterpart, have variable effects and can lead to either gain- and/or loss-of-function depending on the gene and protein where they occur.63
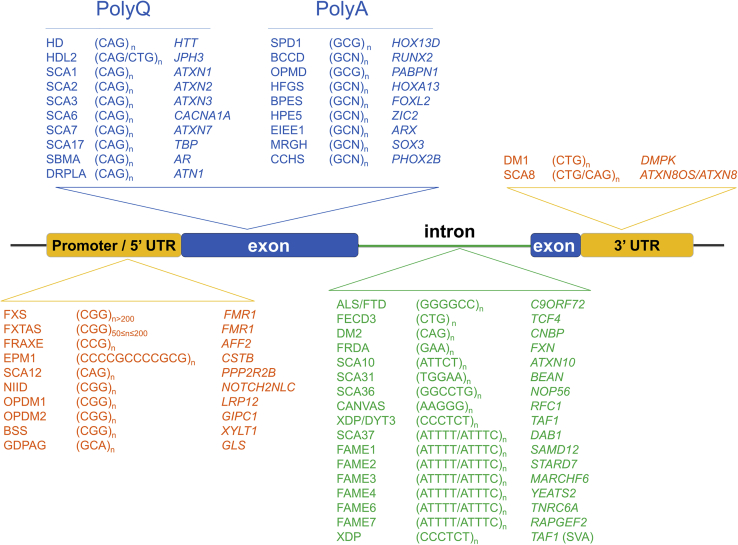
Location of repeat expansions within genes
Repeat expansions may affect coding regions (mainly encoding polyglutamine or polyalanine tracts) or noncoding regions of genes. Noncoding repeat expansions are mainly located in promoters or 5′ untranslated regions (UTRs) or introns. Noncoding repeats in 5′ regions are often GC-rich and their expansion usually affects gene expression through methylation of other epigenetic processes. Intronic expansions and expansions affecting 3′ UTRs more often lead to RNA toxicity or polypeptide synthesis via repeat-associated non-AUG (RAN) translation and subsequent formation of cellular/nuclear aggregates. BCCD, brachydactyly and cleidocranial dysplasia; BSS, Baratela-Scott syndrome; CANVAS, cerebellar ataxia, neuropathy and vestibular areflexia syndrome; CCHS, congenital central hypoventilation syndrome; DM1, myotonic dystrophy type 1; DM2, myotonic dystrophy type 2; DRPLA, dentatorubral-pallidoluysian atrophy; EIEE1, early infantile epileptic encephalopathy type 1; EPM1, progressive myoclonus epilepsy type 1 (Unverricht-Lundborg disease); FAME, familial adult myoclonic epilepsy; FECD3, Fuchs endothelial corneal dystrophy type 3; FRAXE, fragile XE syndrome; FRDA, Friedreich ataxia; FTD/ALS, frontotemporal dementia / amyotrophic lateral sclerosis; FXS, fragile X syndrome; FXTAS, fragile X-associated tremor ataxia syndrome; GDPAG, global developmental delay, progressive ataxia, and elevated glutamine; HD, Huntington disease; HDL2, Huntington disease-like 2; HFGS, hand-foot-genital syndrome; HPE5, holoprosencephaly type 5; MRGH, mental retardation with isolated growth hormone deficiency; OPMD, oculopharyngeal muscular dystrophy; NIID, neuronal intranuclear inclusion disease; OPDM1, oculopharyngodistal myopathy type 1; OPDM2, oculopharyngeal muscular dystrophy type 2; OPML1, oculopharyngeal myopathy with leukoencephalopathy type 1; SBMA, spinal and bulbar muscular atrophy; SPD1, synpolydactyly type 1; SCA, spinocerebellar ataxia; SVA, SINE-VNTR-Alu retrotransposon; XDP, X-linked dystonia parkinsonism.
Non-coding repeat expansions are even more diverse and their impact strongly depends on the type, length, and location of the repeat motif within genes (Table 2).61 They can occur in either the 5′ UTRs, introns, or 3′ UTRs of genes (Figure 2). Repeat expansions located in the 5′ UTRs, promoter, or other regulatory regions are usually GC rich, like triplet (CGG) expansions in FMR1, dodecamer (CCCCGCCCCGCG) expansions in CSTB (MIM: 601145),34 and hexanucleotide (GGGGCC) expansions in C9ORF72 (MIM: 614260).43,44 Large, usually GC-rich expansions overlapping 5′ regulatory regions are frequently—but not always—associated with hypermethylation of the corresponding allele and gene silencing, as exemplified by full mutations (>200 repeats) causing FXS.64 Expanded repeats falling into introns correspond to the majority of recently described expansion disorders and involve motifs of different lengths (mainly tri, tetra, penta, or hexa-nucleotides) and GC content. Only two repeat expansions in 3′ UTRs have been described to date, in myotonic dystrophy 1 (DM1 [MIM: 160900]; CTG expansion in the 3′ UTR of DMPK [MIM: 605377]) and SCA8 (MIM: 608768) (CTG/CAG expansion in the 3′ UTR of ATXN8OS [MIM: 603680]/ATXN8 [MIM: 613289]). Remarkably, non-coding, and more particularly intronic repeat expansions act through a variety of different mechanisms, most of which are associated with dominant inheritance although a few conditions are recessive, generally resulting from the loss-of-function of the gene as a result of the expansion.
Anticipation
A characteristic of dominant repeat expansion disorders is anticipation, a process by which the clinical manifestation appears at an earlier age and/or becomes more severe as the disorder is transmitted from one generation to the next. Strong correlations between the size of the expansion and the age at onset and/or the severity of the disorder have been established for many repeat expansion disorders, including all polyQ as well as many of the non-coding expansions. The larger the expansion is, the earlier the age at onset and the more severe the phenotype becomes. Moreover, expanded repeats, and especially trinucleotide repeats, tend to dynamically expand further during meiosis. Therefore, the offsprings of an affected carrier receiving the pathogenic allele are at higher risk of presenting a more severe or early form of the disorder. In extreme cases, including DM1, HD, and certain types of SCAs, the disorder can occur in infancy or childhood and be fatal in a few years whereas the transmitting parent has a milder adult-onset form of the same disorder.65 The risk of expansion during meiosis generally increases with the number of repeats. Contractions of repeat numbers during meiosis have also been reported but are relatively unfrequent compared to expansions.66 The instability of repeat expansions during meiosis and transmission of larger alleles to offprings is influenced by multiple factors including the size of the repeat, its structure, and the sex and age of the transmitting parent.66,67 Interrupted repeats (for instance AGG interruptions in CGG repeats, or CAA interruptions in CAG repeats) are much more stable than pure ones. Contractions may be more frequent in the male germline, accounting for the exclusive maternal transmission of congenital DM1, or of full FMR1 expansions from premutated mothers, but not from premutated fathers. Males with FXS expansions have spermatozoa with premutation, suggesting that very long repeats may be subject to negative selection during spermatogenesis, by affecting completion of replication.68 Conversely, CAG repeat expansions associated with juvenile HD forms arise only from paternal transmission, and somatic variability can be observed in sperm of HD expansion carriers.
Somatic instability
Repeat expansions are also mitotically unstable, each cell division being associated with a risk of error during DNA replication. This process has been initially attributed to the slippage of DNA polymerase while replicating repetitive DNA sequences and is further amplified by the ability of some repeats to form stable folded structures such as hairpins, stem-loops, cruciform, and/or tetraplexes that favor DNA polymerase pausing.68, 69, 70 However, the observations of expansions and contractions in non-dividing cells have indicated that other mechanisms are at play, and there is increasing evidence from mouse models, and very recently from affected individuals, that somatic stability or instability of repeat expansions largely depends on DNA mismatch repair processes and may impact progression of the disease.68,69,71 Many repeat expansions are thus associated with somatic mosaicism, which degree is usually correlated with expansion size and also age, but can show large variation in different tissues, with a pattern that differs in the various expansion disorders.72, 73, 74, 75 Interruptions by alternative repeat motifs tend to stabilize expansions, making them less prone to alterations during DNA replication or repair.76, 77, 78 Moreover, interrupted expansions are associated with milder and/or different phenotypes compared to uninterrupted expansions, as shown for DM1, SCA2, or SCA10 (MIM: 603516).79,80
Founder effects
Many repeat expansions, especially larger ones, are associated with founder effects, where the expanded allele is associated with a single or predominant haplotype, and result in geographical clustering of the corresponding disorder or highly variable prevalences depending on the population. For instance, DM1 and Friedreich ataxia (FRDA [MIM: 229300]) are, respectively, very rare in Africa and Japan.81,82 This suggests that unexpanded alleles, contrary to their expanded counterparts, are relatively stable. Furthermore, risk haplotypes are in linkage disequilibrium with the large normal alleles at the cognate repeat loci, suggesting a multistep evolution process where an initial historical event is a mutation that generates a large normal allele (protomutation) that then acts as a reservoir for further gradual expansions that will finally result in pathological alleles.82 Another possibility for a rare founder event is the loss of stabilizing interruption by a variant motif in the target normal repeat (such as a loss of AGG interruptions in the CGG repeat at the fragile X locus).83 Examples of repeat expansions for which founder effects have been demonstrated include DM1, FRDA, FXS, SCA3, SCA10, C9ORF72-related frontotemporal dementia and/or amyotrophic lateral sclerosis (FTD/ALS [MIM: 105550]), and myoclonic epilepsy of Unverricht and Lundborg (EPM1 [MIM: 254800]).84, 85, 86, 87
Identification strategies: Remaining challenges
Although repeat expansions were identified as an important source of human disorders in the years 1991–2003, no additional expansion disorder was discovered in the next 5 years (Figure 1). In 2011, the identification of dominant hexanucleotide expansions in C9ORF7243,44 at a locus mapped by linkage in multiple large families in 2006 revived some interest in the search for repeat expansion diseases. But attempts to identify repeat expansions remained limited by exome sequencing, which was the prevalent disease gene identification strategy.
Indeed, owing to their repetitive nature and their abundance in the human genome, TRs are very difficult to study at a genome-wide scale and have been for a long time masked by bioinformatic tools such as RepeatMasker. Short-read encompassing repeats typically map to multiple genomic regions, are clipped off, or are discarded. Furthermore, there is a computational challenge in accurately calling genotypes of polymorphic alleles composed of multiple copies of the same repeats. Detection of TRs is thus generally absent from standard NGS pipelines and requires specific tools in addition to standard SNP, indel, and structural/copy number variant pipelines. Bioinformatics tools specifically assessing repeat numbers from genome (or exome) data have been developed, including LobSTR,88 HipSTR,89 TREDPARSE,90 ExpansionHunter91 STRetch,92 GangSTR,93 and exSTRa.94 However, until recently, most of these tools were only able to call genotypes at specific loci for a given motif. This has recently changed by the development of TRhist95 and ExpansionHunter DeNovo,96 which assess the presence of repeat expansions at a genome-wide scale. Nevertheless, the detection of expansions without linkage data or prior hypotheses on the expanded motif remains a challenge due to the huge number of possible motifs, the abundance of TRs in the human genome, and the difficulty in clearly distinguishing pathogenic expansions from normal polymorphic alleles based on short-read sequences.90 TR expansions thus likely constitute an enormous unexplored reservoir of pathogenic variations. A recent study, which investigated tandem repeats in 17,231 individuals with autism spectrum disorders, suggested that repeat expansions at more than 2,500 loci may represent a collective contribution of 2.6% to autism risk, but the statistic analysis could not distinguish between potential highly penetrant expansions or those associated as polygenic low penetrance risk factors.97
An alternative way to detect TR expansions more accurately is to use long-read technologies, including Oxford Nanopore Technology (ONT) and Pacific Biosciences (PacBio) Single Molecule Real-Time (SMRT) sequencing.98 These technologies allow researchers to obtain single read >10 kb in size, sometimes reaching up to 1–2 Mb, but specific DNA extractions from fresh blood samples or living cells are required to obtain good results. Repeat expansions are detected by pipeline calling structural variants although specific tools, like NanoSatellite99 and tandem-genotypes,100 have been developed to specifically study repeats present in long-read data. Remarkably, these technologies detect thousands of structural variants per individual that predominantly correspond to repeated regions. Because databases such as gnomAD are based on short-read sequencing data, most of the detected structural variants have not yet been described in large control population datasets and filtering of pathogenic variants amid this mass of information is not an easy task.
Overview of recently identified repeat expansion disorders
Despite the technical and computational difficulties, the last few years have clearly witnessed a second wave of repeat expansion discovery. While no expansions have been identified between 2003 and 2008, the genetic basis of more than 20 repeat expansion disorders have been unraveled since 2009, with 17 identified in the last 3 years (Figure 1 and Tables 1 and and2).2). In almost all studies, the identification of pathogenic expansions has taken many years and was only possible because of a large number of families that allowed reducing the candidate linkage intervals to regions containing only one or very few genes. This suggests that many more rarer expansion disorders remain to be identified. Comprehensive reviews have already covered many aspects of repeat expansion disorders identified before 2017.61,101 We review in the following sections the clinical and genetic characteristics of novel disorders and recent advances made in the field since 2017.
Dominant expansions
Spinocerebellar ataxia 37 (SCA37)
SCAs are a clinically and genetically heterogeneous group of autosomal-dominant disorders, characterized by gait imbalance as the result of the progressive loss of cerebellar neurons. SCA37 (MIM: 615945) was first mapped to 1p32 in a large Spanish family, and the interval was confirmed and further refined to 2.8 Mb in three Portuguese families.48 The systematic search for repeat expansions in this interval led to the identification of an intronic expansion composed of ATTTT repeats with an insertion of ATTTC motifs in DAB1 (MIM: 603448).48 Pathogenic expansions all contain the ATTTC insertion, ranging from 31 to 75 repeats, flanked by variable stretches of ATTTT repeats, while larger pure ATTTT expansions (up to 400 repeats) are present in healthy individuals.48 Furthermore, an inverse correlation exists between the number of ATTTC repeats detected in the blood and the age at disease onset.48 DAB1 encodes a downstream effector of the reelin signaling pathway, contributing to the correct positioning of neurons in the developing brain, with highest expression in the cerebellum. A study on post-mortem brains of SCA37 expansion carriers revealed that expansions increase DAB1 expression and trigger alternative splicing events favoring the inclusion of two exons absent from isoforms normally present in the brain.102 Overexpression of ATTTC repeats led to RNA foci formation in vitro, and injection of ATTTC repeats in vivo during zebrafish development was associated with malformations and lethality, suggesting a toxic effect of RNA molecules containing AUUUC repeats.48
Familial adult myoclonic epilepsy (FAME)
FAME is an autosomal-dominant disorder characterized by myoclonus (cortical myoclonic tremor) and epilepsy.103,104 This familial condition was first described in Japan as benign adult familial myoclonic epilepsy (BAFME) and was independently referred to as autosomal-dominant cortical myoclonus and epilepsy (ADCME) and familial cortical myoclonic tremor with epilepsy (FCMTE).104 Myoclonic tremor is usually the first symptom to appear at a variable age, ranging from 10 to 60 years, with an average of 30 years.104 At least half of affected individuals develop epilepsy, mainly generalized tonic-clonic seizures, concomitantly to tremor or later in life.104 Myoclonus and seizures are exacerbated by photic stimulation, alcohol, sleep deprivation, or emotional stress. At an advanced age, tremors tend to worsen and be associated with gait instability and difficulties to walk.
Four loci on chromosomes 2, 3, 5, and 8 had been described between 1999 and 2013,103 but the genetic cause of FAME had remained elusive for 20 years despite extensive sequencing of the candidate intervals. In 2018, the candidate interval on chr 8 was reduced to a single gene, allowing the identification of intronic repeat expansions in SAMD12 (MIM: 618073) as the cause of BAFME (FAME1 [MIM: 601068]). Similar to SCA37 expansions, SAMD12 expansions are composed of both ATTTT and ATTTC motifs.49 In one family and one sporadic case without SAMD12 expansion, the same expanded repeats were detected in TNRC6A (MIM: 610739) and RAPGEF2 (MIM: 609530).49 This discovery rapidly unlocked the other FAME loci. Similar ATTTT/ATTTC expansions were identified in STARD7 (MIM: 616712) on chr 2,58 YEATS2 (MIM: 613373) on chr 3,57 and MARCHF6 (MIM: 613297) (formerly MARCH6) on chr 5.59 In all six genes, FAME expansions occur at a polymorphic STR site initially composed of ATTTT repeats and contain a ATTTC insertion generally located at the 3′ end of the expansion. As previously observed for SCA37, SAMD12 expansions composed of pure ATTTT repeats exist in healthy individuals, confirming that inserted ATTTC repeats are also the pathogenic part of FAME expansions.49 The size of SAMD12 expansions varied from 2.2 to 18.4 kb (i.e., 440–3,680 repeats),49 while MARCHF6 expansions ranged from 3.3 to 14 kb (i.e., 660–2,800 repeats) on average.59 However, sequencing and staining of single alleles at the MARCHF6 locus using molecular combing revealed an important somatic mosaicism of the expansion, both in terms of length and structure in blood cells. Interestingly, micro-rearrangements occurring at the expansion site were observed in individuals with very large (>10 kb) expansions in 20% of the cells, suggesting that expansions are prone to chromosomal breakages and could constitute fragile sites.59 The mean expansion size was inversely correlated with the age at epilepsy onset49,59 and this effect was mainly driven by the size of ATTTC repeats.59 However, other pathogenic motifs than ATTTC repeats could exist, as suggested by the identification of SAMD12 expansions composed of ATTTG instead of ATTTC repeats in a large Chinese family affected by FAME.105 SAMD12 expansions have been reported mainly in families from Japan, China, India, and Sri Lanka,49,106 while STARD7 and MARCHF6 expansions seem to be restricted to families from European ancestry.58,59 This geographical distribution results from ancient founder effects originating from Asia or Europe, respectively, and may be useful for initial genetic testing prioritization.
Contrary to SCA37 expansions, which alter the expression and splicing of DAB1, a gene with an established role in cerebellum development and function, the six genes harboring FAME expansions have completely different functions and expression profiles. MARCHF6 encodes a ubiquitous E3 ubiquitin ligase mediating the degradation of misfolded or damaged proteins in the endoplasmic reticulum. RAPGEF2 encodes a neuron-specific GTPase activating the RAS signaling pathways in response to the activation of cell membrane receptors, such as dopaminergic receptors. STARD7 encodes a ubiquitous protein involved in lipid transport and metabolism. TNRC6A encodes a subunit of a cytoplasmic ribonucleoprotein complex regulating mRNA silencing, stability, and translation, highly expressed in the cerebellum. YEATS2 encodes a ubiquitous subunit of the ADA2A-containing histone acetyltransferase complex. The function of SAMD12 is unknown. This strongly suggests that the ATTTC repeats are pathogenic independently of the recipient gene and its function and that the gene is only a template for repeat expression. Reads filled with AUUUU/AUUUC repeats and RNA foci associated with abortive transcription following SAMD12 expansions have been observed in postmortem brains of expansion carriers from Japan, suggesting that RNA containing AUUUC repeats could form aggregates able to sequestrate specific RNA-binding proteins.49 On the other hand, despite MARCHF6 and STARD7 ubiquitous expression, reads filled with AUUUU or AUUUC repeats could not be detected in lymphoblasts and fibroblasts of individuals with MARCHF6 or STARD7 expansions and expression and splicing of MARCHF6 and STARD7 were unaltered in these tissues.58,59 These findings possibly indicate that abnormal transcription and accumulation of AUUUC repeats observed in SAMD12 expansion carriers could be restricted to neuronal tissues or be absent in other FAME subtypes, and additional studies are required to determine whether expansions lead to RNA toxicity in a neuron-specific manner.
Neuronal intranuclear inclusion disease (NIID)
NIID (MIM: 603472), also known as neuronal intranuclear hyaline inclusion disease or intranuclear inclusion body disease, is an autosomal-dominant, slowly progressive neurodegenerative disorder first described in 1968. As its name indicates, the diagnosis of this condition was initially based on post-mortem neuropathological investigation showing eosinophilic intranuclear inclusions in neurons, but also in other cell types, such as glial cells, fibroblasts, and muscles. NIID can present as a wide and variable range of clinical manifestations, including pyramidal and extrapyramidal symptoms, cerebellar ataxia, cognitive decline, peripheral neuropathy, and autonomic dysfunction.107 Furthermore, the age at onset is extremely variable, ranging from infancy to late adulthood, though most affected subjects show symptoms after the 3rd decade. Brain MRI shows characteristic white matter abnormalities including T2-weighted hyperintensity signals in the middle cerebellar peduncles and high-intensity signals in the corticomedullary junction on diffusion-weighted imaging. Skin biopsy combined with brain imaging is therefore useful for establishing an ante-mortem diagnosis.
White matter abnormalities observed in NIID are reminiscent of those occasionally observed in fragile X-associated tremor ataxia syndrome (FXTAS [MIM: 300623]), a disease caused by intermediate CGG expansions (55–200 repeats) in FMR1.54 Taken together with the presence of neuronal intranuclear inclusions, these findings were suggestive of a repeat-expansion disorder. TRhist combined with SMRT sequencing identified CGG expansions mapping to the 5′ UTR/first exon of NOTCH2NLC (MIM: 618025) (previously NBPF19).54 In two parallel studies, genome-wide linkage analysis first identified overlapping intervals on chromosomes 1p22.1–q21.3 and 1p13.3–23.1 and the same expansion in NOTCH2NLC was revealed by long-read sequencing.55,56 Screening of this expansion in additional neurological disorders led to expand associated phenotypes to essential tremor (ETM6 [MIM: 618866]),108,109 FTD and Azheimer-like dementias,110 and multiple system atrophy.111 Many families with NOTCH2NLC expansions are from Japan and China, suggesting a founder effect in these populations. However, expansions can also occur de novo in sporadic cases112 and can be present in individuals from other geographic origins although their frequency in Europe appears lower than in Asia.113
NOTCH2NLC is one of five genes (NOTCH2 [MIM: 600275], NOTCH2NLA [MIM: 618023], NOTCH2NLB [MIM: 618024], NOTCH2NLC, and NOTCH2NLR [MIM: 618026]) sharing a high degree (>99%) of DNA homology in their 5′ region. The NOTCH2NL copies are distributed as tandem duplicates on each side of the chr 1 centromere and are human specific. They have arisen from evolutionary recent duplications of NOTCH2 that likely contributed significantly to human brain evolution, by increasing neurogenesis and cortical size.114,115 NOTCH2NL genes are correctly annotated only in the hg38 version of the human reference genome and the presence of these multiple, almost identical copies complicates the molecular diagnosis of NIID, which is already challenging due to the GC-rich nature of these expansions. The number of pathological CGG repeats was estimated to range from 61 to 500 repeats whereas in control individuals this number varies from 6 to 60 and CGG repeats can be interrupted by AGG motifs.54,55,116 CGG expansions associated with NIID are not consistently associated with DNA hypermethylation and they do not alter the expression of NOTCH2NLC, but anti-sense transcripts are specifically produced in affected individuals.54,55 This suggests pathophysiological mechanisms possibly similar to FXTAS, with a probable gain of toxic function at the RNA level and the eventual existence of repeat-associated non-AUG (RAN) translation.
Oculopharyngeal and oculopharyngodistal myopathies
In a family with four affected individuals presenting with oculopharyngeal myopathy, limb weakness, ataxia, ptosis, and white matter abnormalities similar to those seen in NIID (OPML1 [MIM: 618637]), Ishiura et al. used TRhist to identify CGG repeats in a long non-coding RNA gene, LOC642361, which overlaps on chr 10q22.3 with an antisense transcript, NUTM2B-AS1 (MIM: 618639).54 This expansion segregated with the disorder in the family and was highly unstable, but additional families with similar phenotypes related to this expansion are needed to confirm the pathogenicity of this expansion.
The same strategy also revealed a CGG expansion in the 5′ UTR of LRP12 (MIM: 618299) in a family with oculopharyngodistal myopathy (OPDM1 [MIM: 164310]).54 Oculopharyngodistal myopathy is an adult-onset neuromuscular disorder characterized by progressive leg and arm weakness associated with external ophtalmoplegia, dysphagia, and ptosis. The CGG expansion in LRP12 was further identified in eight families and 13 sporadic cases with OPDM or milder symptoms, including ptosis and extraocular and pharyngeal weakness.54
Independently, Deng et al. used a combination of whole-genome sequencing and long-read sequencing to identify CGG repeat expansions in the 5′ UTR of GIPC1 (MIM: 605072) in four families and three sporadic cases with OPDM2 (MIM: 618940) of Chinese origin.60 This expansion was further confirmed by repeat-primed PCR in 15 additional families or sporadic cases of Chinese or Japanese origins.60 Expansions increased GIPC1 mRNA expression without altering protein levels. Like ATTTC expansions in FAME, CGG expansions seem to lead to OPDM irrespective of the gene where they occur.
Recessive expansions
Cerebellar ataxia, neuropathy, and vestibular areflexia syndrome (CANVAS)
CANVAS (MIM: 614575) is an adult-onset, slowly progressive disorder characterized by cerebellar ataxia, sensory neuropathy, bilateral vestibulopathy, and autonomic dysfunction, first described in 1991 but recognized as a a specific disease entity in 2011. The symptoms usually start around the 6th decade although the age at onset can vary between 39 and 71 years. Furthermore, CANVAS is possibly a relatively common disorder although its exact prevalence remains to be determined. The description of affected siblings and additional pedigrees pointed to a recessive disorder.117
The identification of repeat expansions in CANVAS was possible through genome-wide linkage analysis in multiple small families, which identified a single locus on chromosome 4p14.51,52 In this candidate interval, bi-allelic repeat expansions composed of AAGGG repeats were identified in intron 2 of RFC1 (MIM: 102579), replacing the 11 AAAAG repeats present in the reference genome. Analysis of control individuals revealed a striking variability of repeat motifs and sizes at this locus. Non-expanded AAAAG repeats correspond to the ancestral and most frequent allele but nonpathogenic expanded AAAAG repeats are quite common in healthy individuals. Additional motifs present at this locus include AAAGG, AAGAG, and AGAGG.118 The relevance of these motifs in human pathology is not yet fully understood, but the observation of an affected individual with bi-allelic ACAGG expansion suggests that other motifs than AAGGG could also be pathogenic.119 Expansion size varied from 400 to 2,000 repeats (i.e., 2–10 kb) but no association between the number of AAGGG repeat unit and the age at onset was found.51 AAGGG expansions are more frequent in the European population and associated with a founder effect,51 although expanded alleles also exist in other populations.119 Sensory neuropathy is present in nearly 100% of individuals with RFC1 expansions, and a dry spasmodic cough is often the first manifestation, long before walking difficulty.120 More rarely, bi-allelic RFC1 expansions can be detected in individuals with multiple system atrophy.121
Remarkably, although AAGGG expansions are recessive, suggesting a loss of function of some kind, no detectable impact was observed on RCF1 gene expression or splicing in peripheral tissues or post-mortem brain samples (including cortex and cerebellum).51 In addition, no RNA molecule with expanded AAGGG or CCCUU repeats corresponding to RFC1 pre-mRNA transcript and no RNA foci could be detected in any of the tissues examined.51 RFC1 encodes the large subunit of replication factor C, a ubiquitous DNA polymerase accessory protein required for DNA replication and DNA repair. However, the function of this complex is unaltered by expansions as fibroblasts from affected individuals showed a normal response to DNA damage.51 Therefore, the mechanisms by which AAGGG expansions lead to CANVAS remains mysterious so far.
Recessive expansions causing gene silencing
Baratela-Scott syndrome (BSS) is a recessive skeletal dysplasia associated with developmental delay, short stature, and facial dysmorphy that clinically overlaps with Desbuquois dysplasia 2 (DBQD2 [MIM: 615777]), another skeletal dysplasia caused by bi-allelic loss-of-function variants in XYLT1 (MIM: 608124). Using genome sequencing, microarray analysis, and Sanger sequencing, Lacroix and collaborators identified homozygous or compound heterozygous pathogenic variants or deletions altering XYLT1 in a few individuals with BSS while other affected individuals had only one heterozygous or no XYLT1 variant. Based on possible allelic drop-out, they looked for DNA methylation defects and showed that alleles without identified variant or deletion were hypermethymated. This hypermethylation was secondary to a CGG expansion occurring in the 5′ UTR of XYLT1 and was associated with silencing of the corresponding allele.50 These results show that BBS and DBQD2 are allelic disorders and that the most frequent loss-of-function variant in XYLT1 is a CGG expansion in its 5′ UTR.
Recessive expansions leading to a loss of function of GLS (MIM: 138280) were identified thanks to their associated biochemical defect. The discovery started with the identification of heterozygous variants (one missense and one nonsense) in GLS in two of three unrelated individuals with global developmental delay, progressive ataxia, and elevated plasma glutamine (GDPAG [MIM: 618412]). GLS encodes glutaminase, the enzyme catalyzing the first reaction of glutamine catabolism. Glutaminase activity was strongly impaired in all three subjects, suggesting the presence of additional variants refractory to exome detection. Using ExpansionHunter and repeat-primed PCR, the authors identified a GCA repeat expansion in the 5′ UTR of GLS, which was either homozygous or compound heterozygous with the previously identified variants. The number of repeats ranged from 400 to 1,500 whereas the normal range in control individuals was 8–16. There was no evidence of DNA methylation associated with this expansion, but histone modifications associated with transcriptionally active regions (H3ac and H3K4m3) were reduced while histone marks of transcriptionally silent regions were increased. These findings suggest that the repeat expansion in GLS leads to a change in chromatin configuration that subsequently results in reduced transcription.53
X-linked expansion related to the insertion of a retrotransposon
X-linked dystonia-parkinsonism (XDP or “Lubag” syndrome [MIM: 314250]) is a neurodegenerative disorder characterized by severe and progressive torsion dystonia associated with extra-pyramidal signs including tremors, bradykinesia, and rigidity (parkinsonism) described in 1976 (see GeneReviews by Evidente in web resources). XDP mainly affects males and more rarely females, and is almost exclusively limited to individuals of Filipino descent originating from Panay Island, Philippines, where its prevalence is above 5 in 100,000. Age at onset varies from 12 to 79 years.122 Autopsy of XDP-affected brains has shown lesions in the striatum resembling those seen in individuals with HD.123
Genome-wide linkage in XDP-affected families had identified the DYT3 locus on Xq13.1.124 Using shotgun sequencing from BAC clones from a subject with XDP, Makino et al.122 identified a SINE-VNTR-Alu (SVA) retrotransposon inserted in an intron of TAF1 (MIM: 313650), encoding the largest component of the transcription factor IID complex. Gene expression analysis in a XDP postmortem brain showed a decrease of a neuron-specific TAF1 isoform associated with increased DNA methylation at the site of SVA insertion.122
The inserted SVA is associated with an identical founder haplotype spanning 294 kb and including five single nucleotide substitutions and a 48-bp deletion.122 Further analysis of the SVA sequence revealed a polymorphic hexanucleotide (CCCTCT) tandem repeat located at the 5′ end of the SVA. Interestingly, the length of this repeat insertion is inversely correlated with the age at onset and its sequence is able to modulate the expression of a reporter gene, suggesting that this repeat expansion contributes to the pathogenesis of XDP.47,125
Expansions of minisatellites
A 99-bp coding expansion in PLIN4 underlies a rare type of myopathy
In a large family with autosomal-dominant progressive myopathy with rimmed ubiquitin-positive autophagic vacuolation (MRUPAV), Ruggieri et al.126 identified a linkage interval of 5.12 Mb on chr 19p13.3 (Table 3). The authors performed a combination of exome, genome, and RNA sequencing from skeletal muscle biopsies, which was unproductive. They then performed a proteome analysis of microdissected vacuoles that revealed a 20-fold increase in periculin-4, which gene (PLIN4 [MIM: 613247]) was contained within the linked interval. PLIN4, like other periculin genes, contains a domain composed of 27 to 31 repetitions of 33-amino acids (99 bp). In PLIN4, these 33-amino acid repeats are formed of imperfect 11-mer tracts that each form three amphipathic helices able to anchor the protein to the phospholipid monolayer of lipid droplets. A reanalysis of genome and transcriptome data showed an unusually high coverage of exon 3, encoding the repeats. Long-range PCR revealed a 1,000 pb higher band, and nanopore sequencing showed that this band corresponds to a repetition of 40 × 33-mer instead of the 27–31 normal repeats. This expansion was associated with increased aggregation of periculin-4 and aggrephagy.126 This example illustrates the complexity of identifying repeat expansions involving minisatellites, due to a computational challenge in mapping and accurately calling repeats of several dozens of nucleotides.
Table 3
List of repeat expansions involving motifs larger than 12 nucleotides (minisatellites)
| ID year | Disorder | Inheritance | Chrom | Gene | Location | Repeat motif unit | Normal repeat range | Pathological repeat range | Pathological mechanism(s) | Method of identification | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2018 | SCHIZO / BD | AD (susceptibility factor) | 12p13.33 | CACNA1C | intron | 30 bp | variable | N/A | specific 30-mer sequences associated with disease risk | GWAS, CGA | Song et al.129 |
| 2018 | Alzheimer disease | AD (susceptibility factor) | 19p13.3 | ABCA7 | intron | 25 bp | 12–427 | >200 | association of longer repeat length with AD | GWAS, WES, CGA | De Roeck et al.132 |
| 2020 | MRUPAV | AD | 19p13.3 | PLIN4 | exon | 99 bp | 27–31 | 40 | protein aggregation | L, WGS, proteomics | Ruggieri et al.126 |
| 2020 | ALS | AD (susceptibility factor) | 18q21.31 | WDR7 | intron | 69 bp | variable | N/A | association of longer repeat length with ALS | ExpScr, CGA | Course et al.134 |
AD, autosomal dominant; ALS, amyotrophic lateral sclerosis; MRUPAV, myopathy with rimmed ubiquitin-positive autophagic vacuolation; SCHIZO / BD, schizophrenia / bipolar disorders; N/A, not available; CGA, candidate gene analysis; GWAS, genome-wide association study; ExScr, expansion screening; WES, whole-exome sequencing; WGS, whole-genome sequencing.
30-bp repeat array in CACNA1C associated with schizophrenia and bipolar disorder
Schizophrenia and bipolar disorder are highly heritable psychiatric conditions that are part of a clinical continuum. Genome-wide association studies (GWASs) on thousands of affected and control individuals have identified more than 150 loci contributing to these disorders.127 One of the most reliable association signals lies on chr 12p13.33 within CACNA1C (MIM: 114205), the gene encoding the main subunit of the neuronal Cav1.2 calcium channel, and consists of SNPs in linkage disequilibrium within a 100-kb interval.128 In an attempt to identify the variants underlying this association, Song et al. reexamined the corresponding region and identified an intronic 30-mer repeat array that has specifically expanded in humans.129 Indeed, while chimpanzees have a single copy of the 30-mer, human individuals have multiple copies, ranging from 100 to 3,000 copies. They show evidence that the human reference genome, which contains 10 repeat copies at this position, was wrongly annotated due to a repeat contraction subsequent to the BAC cloning method originally used to sequence the human genome. Using long-read sequencing they demonstrate that these 30-mer repeats are highly variable not only in size but also in their sequence, some positions being nearly invariant whereas others show significant variation. Interestingly, specific sequence variants are associated with a higher risk of developing a psychiatric disorder while others are on the contrary protective. Besides, these repeats are able to modulate the expression of a luciferase reporter in a neural progenitor cell line, indicating that these repeat arrays have variable enhancer activities that could contribute to a differential expression of CACNA1C during brain development.129 This study shows that VNTRs are far more polymorphic than the reference genome suggests and that deciphering their contribution to gene expression and human pathology is of crucial importance but only possible using long-read technologies.
25-bp repeat in ABCA7 associated with Alzheimer disease (AD)
AD is a frequent neurodegenerative disorder characterized by progressive dementia affecting memory, thinking, and behavior and is multifactorial is most cases. GWASs have identified an association with ABCA7 (MIM: 605414) and truncating variants in this gene are five times more frequent in AD-affected individuals versus control individuals (these variants are present in up to 4% of affected individuals).130,131 In order to identify variants underlying the GWAS signal, De Roeck et al. examined repetitive regions in ABCA7 poorly covered by NGS and identified a 25-bp tandem repeat adjacent to the splice donor site of exon 18.132 Using Southern blotting, they observed a high variability in repeat length, ranging from 12 to 427 repeat units. Expanded repeat length (>200 repeats) were significantly associated with AD, with an odd ratio of 4.5 (AD9 [MIM: 608907]). Furthermore, the VNTR length was associated with a decrease in ABCA7 expression, as a result of alternative splicing events involving exons 18 and 19.132
69-bp repeat in WDR7 modifying the risk of amyotrophic lateral sclerosis (ALS)
ALS is a progressive and fatal disorder characterized by the rapid neurodegeneration of motor neurons. Based on the observation that repeat expansions in C9ORF72 contribute to a significant proportion of individuals with ALS and intermediate CAG repeat number in SCA2 are able to modify its progression,133 Course et al. looked for human-specific polymorphic VNTRs in introns and identified 20 repeat-containing regions with unit length >25-mer that mapped to a unique location in human genomes. Among those was a 69-bp repeat located in WDR7 (MIM: 613473), on chr 18q21, within the ALS3 (MIM: 606640) locus and encoding rabconnectin-3, a protein enriched in synaptic vesicles.134 As previously shown for CACNA1C repeats, WRD7 repeats are specifically expanded in humans and highly polymorphic in size. In addition, these repeats are complementary and predicted to form unique secondary structures corresponding to hairpins. By comparing the repeat lengths in 376 individuals with ALS, 531 individuals with Parkinson disease, and 639 control subjects, the authors suggested a significant association of longer repeat lengths with ALS, although the distribution was variable in all three groups, ranging from 51 to 86 repeats. Furthermore, the repeat sequences are variable, and individual alleles are composed of reproducible patterns. The authors suggest that WDR7 repeats can be transcribed in microRNAs in vitro, although there was no evidence of their presence in neuronal RNA-seq datasets, and that ectopically expressed repeats form aggregates.134
Pathophysiological mechanisms
Epigenetic gene silencing
Loss-of-function by gene silencing is one possible consequence of tandem repeat expansions (Figure 3A). Generally associated with recessive (or X-linked) disorders, these expansions are easily missed. Recessive expansions can be either bi-allelic or associated with a pathogenic point variant in trans and the phenotype associated with expansions and point variants is usually identical. Non-coding repeat expansions can modulate the expression of their recipient gene by directly or indirectly altering chromatin conformation and availability. GC-rich expansions may create new or strengthen existing CpG islands, leading to persistent DNA hypermethylation, as observed in FXS (FMR1)135 and BSS (XYLT1).50 However, some GC-rich expansions seem to lead to epigenetic gene silencing independently of DNA methylation, like expansions in CSTB (EPM1)136 and more recently in GLS (GDPAG).53 The recent example of GDPAG has shown that histone acetylation and methylation marks can be altered without any association with DNA hypermethylation. This example is reminiscent of the effect of GAA expansions in FXN (MIM: 606829) (causing FRDA), which alters transcription of Frataxin by creating secondary DNA/RNA structures (R-loops) that block RNA polymerase and are associated with repressive histone marks.137,138 Of note, hypermethylation and inhibition of gene transcription can be only one of several consequences of the repeat expansions, as observed for C9ORF72-FTD/ALS, one of the few dominant disorders associated with hypermethylation.101 Although DNA methylation inversely correlates with repeat size and age at disease onset,139 loss of function of C9ORF72 is likely insufficient to lead to FTD/ALS as no truncating mutations have been yet reported in this gene, but recent evidence suggests that it could contribute to disease pathogenesis by worsening the repeat-dependent gain-of-function mechanisms.140
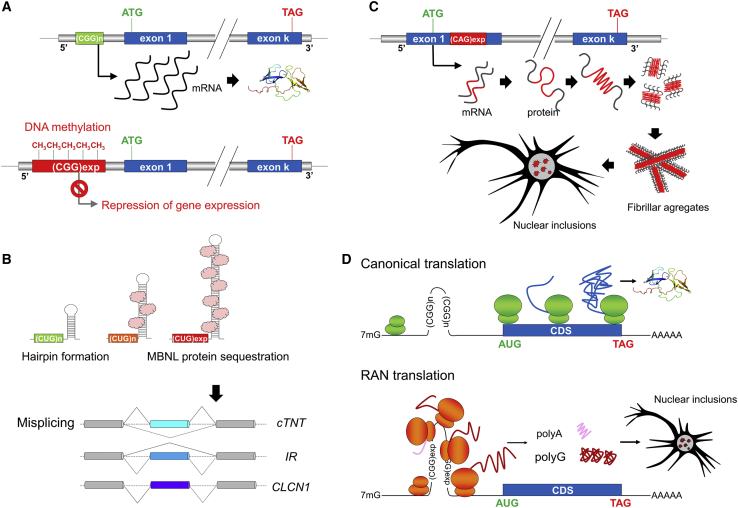
Main mechanisms associated with repeat expansions
(A) Epigenetic gene silencing. Large, usually GC-rich expansions located in promoters and/or 5′ UTRs are frequently associated with DNA hypermethylation at CpG sites. Methylated expanded alleles are locked in a chromatin configuration preventing gene expression, and therefore leading to a loss-of-function of the expanded allele. Examples of this mechanisms include full expansions in FMR1 (fragile X syndrome) and expansions in XYLT1 (Scott-Baratela syndrome).
(B) Sequestration of RNA-binding splicing factors. Specific expanded repeats within RNA molecules are able to form stable secondary structures such as hairpins, which can bind specific RNA-binding splicing factors (e.g., muscleblind-like proteins) with high affinity. These RNA molecules accumulate to form inclusions in the nucleus and sequester bound RNA-binding splicing factors, resulting in misplicing of a subset of transcripts requiring these factors. The illustration of this mechanism is provided by CTG/CAG expansions in DMPK.
(C) Protein misfolding and aggregation. Coding expansions, particularly those encoding polyQ stretches, lead to neuronal intranuclear protein inclusions. This mechanism results from the ability of abnormally long polyQ chains to form β sheet structures prone to form insoluble fibrillar aggregates.
(D) Repeat-associated non-AUG (RAN) translation. RAN translation is a non-canonical protein synthesis process in which peptide synthesis is initiated at the site of the expanded repeats in absence of an AUG codon. This process can theoretically occur in all reading frames on both sense and antisense DNA strands, resulting in up to six different polypeptides, although only some of these peptides are toxic and/or preferentially expressed. For example, intermediate CGG expansions (ranging from 55 to 200 repeats) associated with FXTAS mainly lead to synthesis of polyglycine peptides that are able to accumulate and form protein aggregates. The list of repeat expansion disorders associated with RAN occurs is rapidly increasing and so far includes DM2, FXTAS, FXPOI, HD, C9ORF72-FTD/ALS, SCA8, and SCA31.
RNA toxicity mediated by protein titration
RNA is the molecule that could mediate the pathogenicity of repeat expansion disorder in many cases. Indeed, the vast majority of repeat expansions undergo transcription, and the transcription often even occurs bilaterally, i.e., in forward and reverse senses.101,141 Repeat-containing RNA molecules can act through different mechanisms and alter multiple pathways, as previously reviewed.101 The best-characterized example of these mechanisms has been studied in myotonic dystrophy: DMPK mRNA molecules with 3′ CUG expanded repeats accumulate to form inclusions in muscle and neuron nuclei and sequester specific RNA-binding splicing factors (e.g., muscleblind-like, MBNL protein). The subsequent depletion of these proteins leads to misplicing of a subset of tissue-specific transcripts (Figure 3B).142,143 It is still unclear whether recently identified repeat expansion disorders, including for instance SCA37 and FAME in which RNA foci have been detected, could fall within the category of disorders resulting from the titration of neuronal-specific splicing factors.
Protein misfolding and aggregation in repeat disorders
Protein aggregation has long been a characteristic feature of coding expansions (Figure 3C), in particular polyQ disorders. The recent identification of 99-mer expansions in PLIN4 illustrates that repeat motifs other than trinucleotides can lead to protein aggregation.126 Furthermore, the identification of 5′ UTR CGG expansions in NIID and oculopharyngeal myopathies demonstrates that neuronal nuclear aggregates can also be a consequence of non-coding expansions although the precise mechanisms related to these aggregates remain to be determined. These disorders share many characteristics in common with FXTAS and it is hence possible that pathogenic mechanisms are alike.
Repeat-associated non-AUG (RAN) translation
RAN translation is a non-canonical protein synthesis process, first described SCA8 and DM1 in 2011.144 RAN translation (Figure 3D) initiates at the site of the expanded repeats in the absence of an AUG codon and can theoretically occur in all three reading frames using both sense and antisense directions, resulting in up to six different polypeptides.145,146 For example, non-coding CAG/CTG expansions in SCA8 result in polyserine, polyglutamine, and polyalanine peptides, the two latter being able to accumulate and form aggregates. The list of repeat expansion disorders associated with RAN translation is constantly growing and now also includes myotonic dystrophy type 2 (DM2 [MIM: 602668]) caused by CCTG expansions in CNBP (MIM: 116955), FXTAS, fragile X-associated premature ovarian infertility (FXPOI [MIM: 311360]) in females with intermediate CGG repeat expansion in FMR1, HD (CAG repeat expansions in HTT [MIM: 613004]), C9ORF72-FTD/ALS, and SCA31 (MIM: 117210).145,147, 148, 149, 150, 151 RAN translation of specific reading frames and accumulation of some peptides could occur in some cell types but not in others, providing a possible ground for the tissue specificity of some disorders.147,151 Mechanisms involving toxic RNA and RAN translation are not mutually exclusive, as illustrated by DM1 and C9ORF72-FTD/ALS in which both RNA foci and RAN peptide aggregates have been detected in affected tissues.146
Precision medicine in repeat expansions disorders
Precision medicine (previously referred to as personalized medicine), which consists of developing a treatment entirely adapted to an individual’s disease, taking the genome and environment into account, is increasingly becoming an accessible goal in genetic disorders.152 This concept has also been developed for several years for specific repeat expansion disorders, in particular HD. In this section of the review, we will focus on the recent progress in this field.
A possible way of developing personalized treatments is to understand the pathological consequences of the expansion at the cellular level and to find molecules that are able to either reverse or delay these consequences. For example, in polyQ disorders, the accumulation of polyQ protein aggregates leads to the dysfunction of several pathways, including altered transcription, reduced mitochondrial metabolism, excitotoxicity, authophagy, and apoptosis. Molecules targeting one or several of these deregulated pathways have therefore been suggested as possible treatments.153 However, these approaches are usually limited as they are not able to fully restore the cellular dysfunctions. Yet, some of these strategies can prove useful on the long term in combination with other approaches. For expansion disorders associated with RAN translation, antibodies have been designed to target specifically the toxic peptides produced and block their effect in vivo. This strategy has recently proven successful in a mouse model of C9ORF72-FTD/ALS.154
A promising way to treat dominant disorders associated with a gain of function of the expanded allele is to target and neutralize specifically this allele. This implies that one functional copy of the gene is sufficient and this strategy is not suitable for genes associated with haplointolerance. CRISPR-Cas9 technologies offer the possibility to edit individual genomes but so far they have limitations, including difficulties in delivering the proteins and RNA guide necessary to gene editing in the affected tissues, as well as possible off-target effects, which still restrict their use for clinical purposes.155 Alternative approaches making use of antisense oligonucleotides (ASO) or siRNA delivered intrathecally have proven to be efficient in downregulating the expression of the expanded allele and have already reached phase III in clinical trials.156 The designed oligonucleotides cannot directly target the repeats because of their repetitive nature and their presence in other genes and thus need to be complementary to a specific sequence located in cis of the expansion. In this setting, single-nucleotide polymorphisms (SNPs) are used to make the difference between the expanded and the normal alleles and target specifically the allele carrying the expansion.157,158 This requires researchers to genotype and determine cis associations existing in each affected individual prior to designing the treatment, and this strategy can only be used for subjects with informative coding SNPs in the corresponding gene.
Another recent pioneering approach could become a game changer in HD and other CAG trinucleotide expansion disorders. A small molecule, naphthyridine-azaquinolone, was shown to specifically bind to expanded CAG repeats and induce their contraction in cells from affected individuals and the brains of mice with an expansion in Htt.159 This approach could be used to reduce the number of CAG repeats in the brain of individuals with HD, hopefully leading to a delay in disease onset of several decades or even a lifetime protection.
Conclusion: Future studies and remaining challenges
The identification of tandem repeat expansions in human disorders is still in its infancy and many more disorders, monogenic or complex, likely remain to be uncovered. Furthermore, in the last few years we have witnessed an important widening of phenotypes related to known repeat expansions, as illustrated by the recent identification of HTT expansions in individuals with FTD and ALS.160 Therefore, repeat expansions should be searched for in unsolved disorders of supposedly genetic origin, even in phenotypes not specifically evoking a known expansion.
The more recent studies have highlighted the extreme diversity in repeat motifs susceptible to expand and their location in genes. However, only specific repeat motifs appear to be pathogenic while others can be expanded in human genomes without any harmful consequences. Making the difference between these pathogenic and non-pathogenic motifs is of utmost importance as this information could be used to specifically chase the pathogenic ones among the many repeats contained in our genomes. Interestingly, most of the pentanucleotide repeat expansions recently discovered are composed of pathogenic motifs distinct from the one(s) present in reference and control genomes. Studying how these alternative pathogenic repeat insertions have arisen could help finding new repeat expansion disorders. In the case of SCA37 expansions, it has been hypothesized that the thymidine to cytosine transition has occurred after expansion of ATTTT repeats to ~200 copies, followed by expansion of the mutant ATTTC sequence itself.161 This mechanism is in agreement with previous studies showing the existence of susceptibility haplotypes predisposing to the disorders in specific populations.
The example of FMR1, in which CGG repeats in 5′ UTR have been associated with three different disorders depending on the repeat size and sex (due to the location of the gene on the X chromosome), has demonstrated that expansions may have different downstream effects depending on the expansion length and that intermediate expansion sizes should also be carefully studied as they could constitute susceptibility factors for more common disorders or be associated with other or late-onset phenotypes more difficult to recognize. So far, most expansion disorders affect the central nervous system and the muscles and there are only a few examples of expansion disorders limited to other tissues. This might be attributed to an increased susceptibility of some cells or tissues to mechanisms related to these expansions, for example somatic mosaicism of the repeats with higher repeat lengths in particular tissues,162 the sequestration of proteins that have a tissue-specific impact (e.g., muscle-blind proteins like), or an increased expression or specific function of the gene where the expansion occurs (e.g., DAB1).102 Neuronal and muscle cells, which are terminally differentiated non-cycling cells, might also be more susceptible to somatic repeat expansions or to toxic gain-of-function mechanisms. However, it is also likely that repeat expansions may affect other tissues, as observed for FXPOI.163
A recent field of research is to understand the impact of normal and pathogenic tandem repeats on gene expression and chromatin conformation. This is now possible thanks to advanced techniques allowing researchers to study gene expression, chromatin conformation, or both at a high resolution, even from single cells. We have just started to fully acknowledge the role of intragenic microsatellites and minisatellites in modulation of gene expression and as a potential source of gene deregulation.6,129 However, the study of minisatellites is associated with even greater challenges, including difficulties in determining sequence variation within repeats and integrating how both repeat number and sequence affect gene expression in a given cell type. This challenge is achievable only through novel long-read technologies. Furthermore, recent studies regarding minisatellites suggest that this category of repeated elements could act more frequently as susceptibility factors and modifiers rather than causing monogenic disorders, and establishing association with common or rare disorders might require large sample sizes. As for the role of satellites in human pathology, this is an almost untouched research topic. To date, only one disorder, facioscapulohumeral muscular dystrophy (FSHD1 [MIM: 158900]), has been linked to the heterozygous contraction of a satellite repeat array at the D4Z4 locus (encoding DUX4 [MIM: 606009]) in the subtelomeric region of chromosome 4q35. Diagnosing this disorder still represents a challenge as only specific techniques, such as Southern blotting, Fiber FISH, or optical genome mapping, can be used to specifically detect the contraction of these very large repeats (approx. 100 kb each) and distinguish them from non-pathogenic contractions of an almost identical repeat array at 10q26 (see GeneReviews by Preston et al. in web resources).
Finally, we have to keep in mind that repeat tracts can be wrongly annotated in the human reference genome. An accumulation of short reads in a region typically suggests that this region is either variable in copy number or wrongly annotated but this observation can be made for many regions. Moreover, there are still genomic regions whose sequence remains unknown because of their high complexity. Most of these regions, including centromeres and short arms of acrocentric chromosomes, actually correspond to repeats. The telomere-to-telomere (T2T) consortium has just started sequencing all the gaps remaining in the human genome reference, a tour de force that will undoubtedly result in clarifying many human disorders in the future. The first sequence, released in 2020, was a complete (i.e., gapless) telomere-to-telomere assembly of the single chr X present in the complete hydatidiform mole CHM13.164 This achievement was possible thanks to a combination of high-coverage, ultra-long-read nanopore sequencing and alternative long-read technologies (SMRT sequencing and optical genome mapping) that improved the accuracy sequencing or facilitated the assembly. The T2T consortium has since announced the completion of the CHM13 genome sequence. The characterization of the variability of these previously unsequenceable regions and distinction between their normal and pathogenic states will require many years but this is obviously the start of a new genomic era where the systematic study of repeats will finally be possible.
Acknowledgments
We thank the editors of the American Journal of Human Genetics and the three anonymous reviewers who made valuable contributions to the revision of this review. We also thank the University Hospital Essen, the Deutsche Forschungsgemeinschaft (DFG), the Tom-Wahlig-Stiftung (TWS), the Deutsche Stiftung Neurologie (DSN), and the University of Strasbourg Institute of Advanced study (USIAS) for their financial support to the research studies conducted by the authors.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.03.011.
Data and code availability
This review is based on a literature search in PubMed and individual knowledge of the authors on the topic. It does not include any unpublished data or code.
Web resources
GeneReviews, Evidente, V.G.H. (1993). X-Linked Dystonia-Parkinsonism. https://pubmed.ncbi.nlm.nih.gov/20301662/
GeneReviews, Preston, M.K., Tawil, R., and Wang, L.H. (1993). Facioscapulohumeral Muscular Dystrophy. https://pubmed.ncbi.nlm.nih.gov/20301616/
OMIM, https://www.omim.org/
Pubmed, https://pubmed.ncbi.nlm.nih.gov/
Supplemental information
References
Articles from American Journal of Human Genetics are provided here courtesy of American Society of Human Genetics
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.ajhg.2021.03.011
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S0002929721000951/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/103188310
Article citations
Methylated GCC repeat expansion in AFF3 associates with intellectual disability.
Nat Genet, 56(11):2302-2303, 01 Nov 2024
Cited by: 0 articles | PMID: 39333768
Molecular diagnostic approach to rare neurological diseases from a clinician viewpoint.
Genomics Inform, 22(1):18, 10 Oct 2024
Cited by: 0 articles | PMID: 39390516 | PMCID: PMC11468364
Review Free full text in Europe PMC
Decoding Nucleotide Repeat Expansion Diseases: Novel Insights from <i>Drosophila melanogaster</i> Studies.
Int J Mol Sci, 25(21):11794, 02 Nov 2024
Cited by: 0 articles | PMID: 39519345 | PMCID: PMC11546515
Review Free full text in Europe PMC
Age-dependent somatic expansion of the ATXN3 CAG repeat in the blood and buccal swab DNA of individuals with spinocerebellar ataxia type 3/Machado-Joseph disease.
Hum Genet, 143(11):1363-1378, 08 Oct 2024
Cited by: 0 articles | PMID: 39375222 | PMCID: PMC11522074
Comprehensive genome analysis and variant detection at scale using DRAGEN.
Nat Biotechnol, 25 Oct 2024
Cited by: 0 articles | PMID: 39455800
Go to all (136) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Diseases (Showing 68 of 68)
- (1 citation) OMIM - 105550
- (1 citation) OMIM - 614260
- (1 citation) OMIM - 618025
- (1 citation) OMIM - 618026
- (1 citation) OMIM - 610739
- (1 citation) OMIM - 618023
- (1 citation) OMIM - 613373
- (1 citation) OMIM - 618024
- (1 citation) OMIM - 183086
- (1 citation) OMIM - 613297
- (1 citation) OMIM - 618866
- (1 citation) OMIM - 618940
- (1 citation) OMIM - 164400
- (1 citation) OMIM - 109150
- (1 citation) OMIM - 183090
- (1 citation) OMIM - 615945
- (1 citation) OMIM - 254800
- (1 citation) OMIM - 605072
- (1 citation) OMIM - 138280
- (1 citation) OMIM - 313700
- (1 citation) OMIM - 143100
- (1 citation) OMIM - 607136
- (1 citation) OMIM - 606640
- (1 citation) OMIM - 606009
- (1 citation) OMIM - 606829
- (1 citation) OMIM - 602668
- (1 citation) OMIM - 603516
- (1 citation) OMIM - 314250
- (1 citation) OMIM - 618299
- (1 citation) OMIM - 614575
- (1 citation) OMIM - 618412
- (1 citation) OMIM - 116955
- (1 citation) OMIM - 613289
- (1 citation) OMIM - 313200
- (1 citation) OMIM - 616712
- (1 citation) OMIM - 613004
- (1 citation) OMIM - 164310
- (1 citation) OMIM - 613247
- (1 citation) OMIM - 117210
- (1 citation) OMIM - 603680
- (1 citation) OMIM - 601145
- (1 citation) OMIM - 601068
- (1 citation) OMIM - 603448
- (1 citation) OMIM - 160900
- (1 citation) OMIM - 158900
- (1 citation) OMIM - 164500
- (1 citation) OMIM - 613473
- (1 citation) OMIM - 114205
- (1 citation) OMIM - 615777
- (1 citation) OMIM - 313650
- (1 citation) OMIM - 608124
- (1 citation) OMIM - 309550
- (1 citation) OMIM - 603472
- (1 citation) OMIM - 609530
- (1 citation) OMIM - 605377
- (1 citation) OMIM - 102579
- (1 citation) OMIM - 605414
- (1 citation) OMIM - 608768
- (1 citation) OMIM - 300623
- (1 citation) OMIM - 608907
- (1 citation) OMIM - 300624
- (1 citation) OMIM - 311360
- (1 citation) OMIM - 618639
- (1 citation) OMIM - 125370
- (1 citation) OMIM - 618637
- (1 citation) OMIM - 229300
- (1 citation) OMIM - 600275
- (1 citation) OMIM - 618073
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Identification and characterization of repeat expansions in neurological disorders: Methodologies, tools, and strategies.
Rev Neurol (Paris), 180(5):383-392, 08 Apr 2024
Cited by: 1 article | PMID: 38594146
Review
Triplet repeat length bias and variation in the human transcriptome.
Proc Natl Acad Sci U S A, 106(40):17095-17100, 17 Sep 2009
Cited by: 22 articles | PMID: 19805156 | PMCID: PMC2746125
The unstable trinucleotide repeat story of major psychosis.
Am J Med Genet, 97(1):77-97, 01 Jan 2000
Cited by: 36 articles | PMID: 10813808
Review
Anticipation, imprinting, trinucleotide repeat expansions and psychoses.
Prog Neuropsychopharmacol Biol Psychiatry, 25(1):167-192, 01 Jan 2001
Cited by: 7 articles | PMID: 11263751
Review
 and
and 



