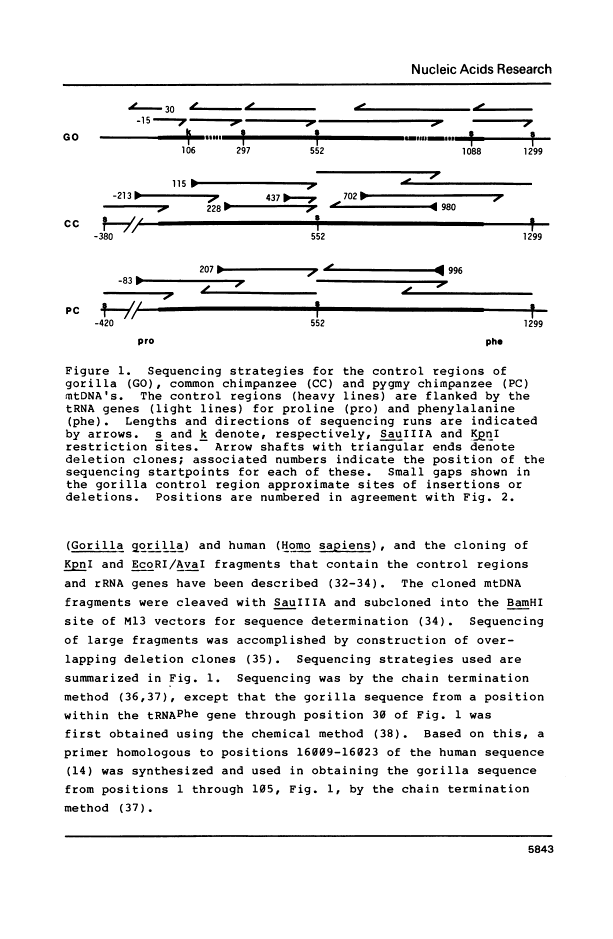
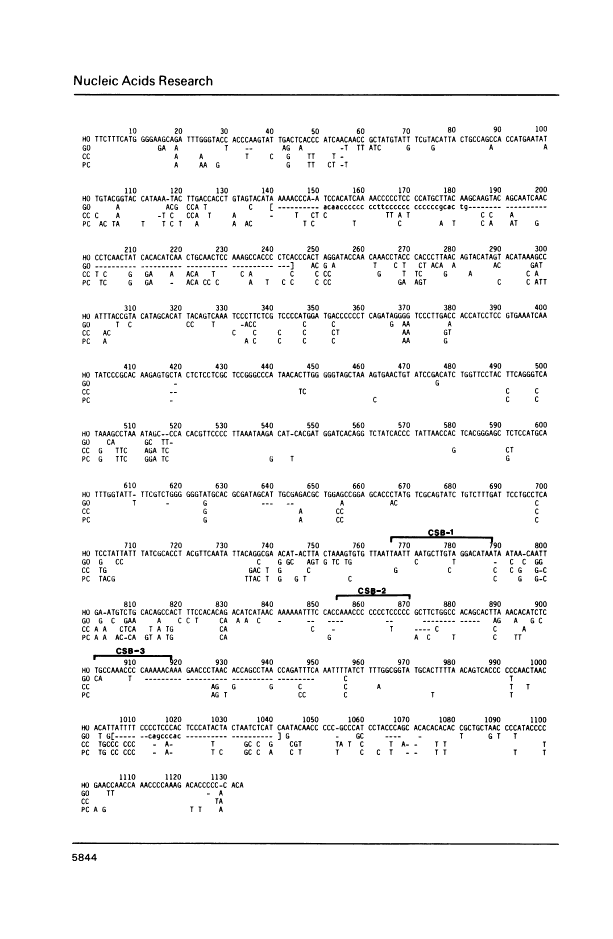
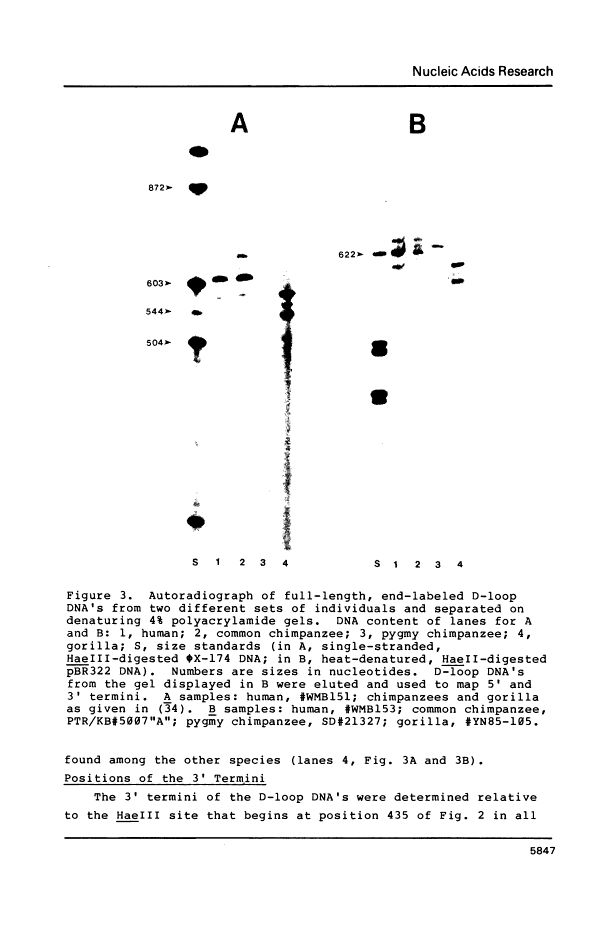
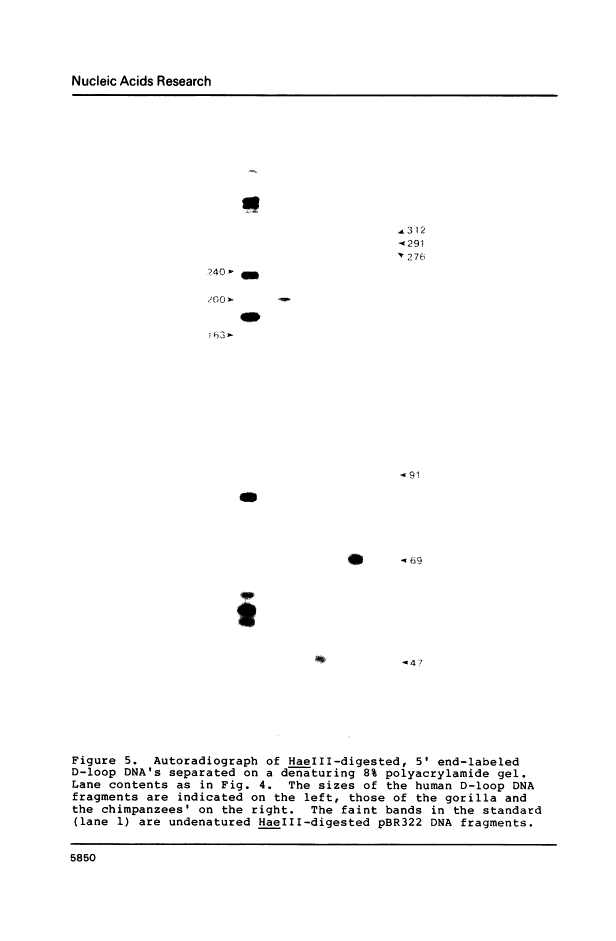
Abstract
Free full text

Comparisons of ape and human sequences that regulate mitochondrial DNA transcription and D-loop DNA synthesis.
Abstract
The mitochondrial DNA (mtDNA) control regions for common chimpanzee, pygmy chimpanzee and gorilla were sequenced and the lengths and termini of their D-loop DNA's characterized. In these and all other species for which there are data, 5' termini map to sequences that contain the trinucleotide YAY. 3' termini are 25-51 nucleotides downstream from a sequence that is moderately conserved among vertebrates. Substitutions were greater than 1.5 times more frequent in the control region than in regions encoding structural genes. Additions and deletions were also frequent, especially in gorilla. Sequences of promoters and of two of four transcription factor binding sites were highly conserved. Comparisons of sequence similarity and transition/transversion ratios suggest that human and chimpanzees may be more closely related to each other than either is to gorilla, if substitution rates are approximately equal among these species.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.9M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982 Apr;28(4):693–705. [Abstract] [Google Scholar]
- Clayton DA. Transcription of the mammalian mitochondrial genome. Annu Rev Biochem. 1984;53:573–594. [Abstract] [Google Scholar]
- Brown WM, Shine J, Goodman HM. Human mitochondrial DNA: analysis of 7S DNA from the origin of replication. Proc Natl Acad Sci U S A. 1978 Feb;75(2):735–739. [Europe PMC free article] [Abstract] [Google Scholar]
- Densmore LD, Wright JW, Brown WM. Length variation and heteroplasmy are frequent in mitochondrial DNA from parthenogenetic and bisexual lizards (genus Cnemidophorus). Genetics. 1985 Aug;110(4):689–707. [Europe PMC free article] [Abstract] [Google Scholar]
- Dunon-Bluteau DC, Brun GM. Mapping at the nucleotide level of Xenopus laevis mitochondrial D-loop H strand: structural features of the 3' region. Biochem Int. 1987 Apr;14(4):643–657. [Abstract] [Google Scholar]
- Gillum AM, Clayton DA. Displacement-loop replication initiation sequence in animal mitochondrial DNA exists as a family of discrete lengths. Proc Natl Acad Sci U S A. 1978 Feb;75(2):677–681. [Europe PMC free article] [Abstract] [Google Scholar]
- King TC, Low RL. Mapping of control elements in the displacement loop region of bovine mitochondrial DNA. J Biol Chem. 1987 May 5;262(13):6204–6213. [Abstract] [Google Scholar]
- King TC, Low RL. Mitochondrial DNA displacement loop structure depends on growth state in bovine cells. J Biol Chem. 1987 May 5;262(13):6214–6220. [Abstract] [Google Scholar]
- MacKay SL, Olivo PD, Laipis PJ, Hauswirth WW. Template-directed arrest of mammalian mitochondrial DNA synthesis. Mol Cell Biol. 1986 Apr;6(4):1261–1267. [Europe PMC free article] [Abstract] [Google Scholar]
- Tapper DP, Clayton DA. Mechanism of replication of human mitochondrial DNA. Localization of the 5' ends of nascent daughter strands. J Biol Chem. 1981 May 25;256(10):5109–5115. [Abstract] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. [Abstract] [Google Scholar]
- Anderson S, de Bruijn MH, Coulson AR, Eperon IC, Sanger F, Young IG. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J Mol Biol. 1982 Apr 25;156(4):683–717. [Abstract] [Google Scholar]
- Brown GG, Gadaleta G, Pepe G, Saccone C, Sbisà E. Structural conservation and variation in the D-loop-containing region of vertebrate mitochondrial DNA. J Mol Biol. 1986 Dec 5;192(3):503–511. [Abstract] [Google Scholar]
- Cairns SS, Bogenhagen DF. Mapping of the displacement loop within the nucleotide sequence of Xenopus laevis mitochondrial DNA. J Biol Chem. 1986 Jun 25;261(18):8481–8487. [Abstract] [Google Scholar]
- Sekiya T, Kobayashi M, Seki T, Koike K. Nucleotide sequence of a cloned fragment of rat mitochondrial DNA containing the replication origin. Gene. 1980 Oct;11(1-2):53–62. [Abstract] [Google Scholar]
- Walberg MW, Clayton DA. Sequence and properties of the human KB cell and mouse L cell D-loop regions of mitochondrial DNA. Nucleic Acids Res. 1981 Oct 24;9(20):5411–5421. [Europe PMC free article] [Abstract] [Google Scholar]
- Wong JF, Ma DP, Wilson RK, Roe BA. DNA sequence of the Xenopus laevis mitochondrial heavy and light strand replication origins and flanking tRNA genes. Nucleic Acids Res. 1983 Jul 25;11(14):4977–4995. [Europe PMC free article] [Abstract] [Google Scholar]
- Fisher RP, Topper JN, Clayton DA. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell. 1987 Jul 17;50(2):247–258. [Abstract] [Google Scholar]
- Chang DD, Clayton DA. Precise identification of individual promoters for transcription of each strand of human mitochondrial DNA. Cell. 1984 Mar;36(3):635–643. [Abstract] [Google Scholar]
- Montoya J, Christianson T, Levens D, Rabinowitz M, Attardi G. Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7195–7199. [Europe PMC free article] [Abstract] [Google Scholar]
- Chang DD, Clayton DA. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc Natl Acad Sci U S A. 1985 Jan;82(2):351–355. [Europe PMC free article] [Abstract] [Google Scholar]
- Chang DD, Clayton DA. A novel endoribonuclease cleaves at a priming site of mouse mitochondrial DNA replication. EMBO J. 1987 Feb;6(2):409–417. [Europe PMC free article] [Abstract] [Google Scholar]
- Chang DD, Hauswirth WW, Clayton DA. Replication priming and transcription initiate from precisely the same site in mouse mitochondrial DNA. EMBO J. 1985 Jun;4(6):1559–1567. [Europe PMC free article] [Abstract] [Google Scholar]
- Doda JN, Wright CT, Clayton DA. Elongation of displacement-loop strands in human and mouse mitochondrial DNA is arrested near specific template sequences. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6116–6120. [Europe PMC free article] [Abstract] [Google Scholar]
- Bogenhagen DF, Yoza BK, Cairns SS. Identification of initiation sites for transcription of Xenopus laevis mitochondrial DNA. J Biol Chem. 1986 Jun 25;261(18):8488–8494. [Abstract] [Google Scholar]
- Aquadro CF, Greenberg BD. Human mitochondrial DNA variation and evolution: analysis of nucleotide sequences from seven individuals. Genetics. 1983 Feb;103(2):287–312. [Europe PMC free article] [Abstract] [Google Scholar]
- Greenberg BD, Newbold JE, Sugino A. Intraspecific nucleotide sequence variability surrounding the origin of replication in human mitochondrial DNA. Gene. 1983 Jan-Feb;21(1-2):33–49. [Abstract] [Google Scholar]
- Brown WM, George M, Jr, Wilson AC. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1967–1971. [Europe PMC free article] [Abstract] [Google Scholar]
- Brown WM, Vinograd J. Restriction endonuclease cleavage maps of animal mitochondrial DNAs. Proc Natl Acad Sci U S A. 1974 Nov;71(11):4617–4621. [Europe PMC free article] [Abstract] [Google Scholar]
- Hixson JE, Brown WM. A comparison of the small ribosomal RNA genes from the mitochondrial DNA of the great apes and humans: sequence, structure, evolution, and phylogenetic implications. Mol Biol Evol. 1986 Jan;3(1):1–18. [Abstract] [Google Scholar]
- Hong GF. A systemic DNA sequencing strategy. J Mol Biol. 1982 Jul 5;158(3):539–549. [Abstract] [Google Scholar]
- Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977 Feb 24;265(5596):687–695. [Abstract] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. [Europe PMC free article] [Abstract] [Google Scholar]
- Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. [Abstract] [Google Scholar]
- Yousaf SI, Carroll AR, Clarke BE. A new and improved method for 3'-end labelling DNA using [alpha-32P]ddATP. Gene. 1984 Mar;27(3):309–313. [Abstract] [Google Scholar]
- Upholt WB, Dawid IB. Mapping of mitochondrial DNA of individual sheep and goats: rapid evolution in the D loop region. Cell. 1977 Jul;11(3):571–583. [Abstract] [Google Scholar]
- Brown WM, Prager EM, Wang A, Wilson AC. Mitochondrial DNA sequences of primates: tempo and mode of evolution. J Mol Evol. 1982;18(4):225–239. [Abstract] [Google Scholar]
- Ferris SD, Wilson AC, Brown WM. Evolutionary tree for apes and humans based on cleavage maps of mitochondrial DNA. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2432–2436. [Europe PMC free article] [Abstract] [Google Scholar]
- Karawya EM, Martin RG. Monkey (CV-1) mitochondrial DNA contains a unique triplication of 108 bp in the origin region. Biochim Biophys Acta. 1987 Jun 6;909(1):30–34. [Abstract] [Google Scholar]
- Albertini AM, Hofer M, Calos MP, Miller JH. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. [Abstract] [Google Scholar]
- Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. [Abstract] [Google Scholar]
- Hixson JE, Clayton DA. Initiation of transcription from each of the two human mitochondrial promoters requires unique nucleotides at the transcriptional start sites. Proc Natl Acad Sci U S A. 1985 May;82(9):2660–2664. [Europe PMC free article] [Abstract] [Google Scholar]
- Hauswirth WW, Van de Walle MJ, Laipis PJ, Olivo PD. Heterogeneous mitochondrial DNA D-loop sequences in bovine tissue. Cell. 1984 Jul;37(3):1001–1007. [Abstract] [Google Scholar]
- Baldacci G, Chérif-Zahar B, Bernardi G. The initiation of DNA replication in the mitochondrial genome of yeast. EMBO J. 1984 Sep;3(9):2115–2120. [Europe PMC free article] [Abstract] [Google Scholar]
- Zyskind JW, Smith DW. The bacterial origin of replication, oriC. Cell. 1986 Aug 15;46(4):489–490. [Abstract] [Google Scholar]
Associated Data
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/16.13.5841
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc336833?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/nar/16.13.5841
Article citations
Mitogenome sequences of domestic cats demonstrate lineage expansions and dynamic mutation processes in a mitochondrial minisatellite.
BMC Genomics, 24(1):690, 17 Nov 2023
Cited by: 1 article | PMID: 37978434 | PMCID: PMC10655372
Indian Red Jungle fowl reveals a genetic relationship with South East Asian Red Jungle fowl and Indian native chicken breeds as evidenced through whole mitochondrial genome sequences.
Front Genet, 14:1083976, 09 Aug 2023
Cited by: 4 articles | PMID: 37621706 | PMCID: PMC10445952
Comparative mitogenomics and phylogenetics of the family Carangidae with special emphasis on the mitogenome of the Indian Scad Decapterus russelli.
Sci Rep, 12(1):5642, 04 Apr 2022
Cited by: 2 articles | PMID: 35379869 | PMCID: PMC8980026
Mitochondrial genomes of African pangolins and insights into evolutionary patterns and phylogeny of the family Manidae.
BMC Genomics, 18(1):746, 21 Sep 2017
Cited by: 12 articles | PMID: 28934931 | PMCID: PMC5609056
Novel duplication pattern of the mitochondrial control region in Cantor's Giant softshell turtle Pelochelys cantorii.
Gene, 593(1):242-248, 23 Aug 2016
Cited by: 3 articles | PMID: 27565702
Go to all (77) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Mitochondrial DNA diversity in gorillas.
Mol Phylogenet Evol, 6(1):39-48, 01 Aug 1996
Cited by: 58 articles | PMID: 8812304
Paired chimpanzee hepatitis B virus (ChHBV) and mtDNA sequences suggest different ChHBV genetic variants are found in geographically distinct chimpanzee subspecies.
Virus Res, 79(1-2):103-108, 01 Nov 2001
Cited by: 20 articles | PMID: 11551650
Chimpanzee fetal G gamma and A gamma globin gene nucleotide sequences provide further evidence of gene conversions in hominine evolution.
Mol Biol Evol, 2(5):370-389, 01 Sep 1985
Cited by: 25 articles | PMID: 3870867
Nonneutral mitochondrial DNA variation in humans and chimpanzees.
Genetics, 142(3):953-963, 01 Mar 1996
Cited by: 117 articles | PMID: 8849901 | PMCID: PMC1207032
Funding
Funders who supported this work.
NCRR NIH HHS (1)
Grant ID: RR07050
NIGMS NIH HHS (2)
Grant ID: GM30144
Grant ID: GM07544