Abstract
Free full text
Utrophin modulator drugs as potential therapies for Duchenne and Becker muscular dystrophies
Abstract
Utrophin is an autosomal paralogue of dystrophin, a protein whose deficit causes Duchenne and Becker muscular dystrophies (DMD/BMD). Utrophin is naturally overexpressed at the sarcolemma of mature dystrophin‐deficient fibres in DMD and BMD patients as well as in the mdx Duchenne mouse model. Dystrophin and utrophin can co‐localise in human foetal muscle, in the dystrophin‐competent fibres from DMD/BMD carriers, and revertant fibre clusters in biopsies from DMD patients. These findings suggest that utrophin overexpression could act as a surrogate, compensating for the lack of dystrophin, and, as such, it could be used in combination with dystrophin restoration therapies. Different strategies to overexpress utrophin are currently under investigation. In recent years, many compounds have been reported to modulate utrophin expression efficiently in preclinical studies and ameliorate the dystrophic phenotype in animal models of the disease. In this manuscript, we discuss the current knowledge on utrophin protein and the different mechanisms that modulate its expression in skeletal muscle. We also include a comprehensive review of compounds proposed as utrophin regulators and, as such, potential therapeutic candidates for these muscular dystrophies.
INTRODUCTION
Duchenne muscular dystrophy
Duchenne muscular dystrophy (DMD) is a fatal X‐linked disorder that affects approximately one in 5000 live male births worldwide. It is caused by one or more mutations in the DMD gene, which encodes dystrophin protein [1, 2, 3]. This protein provides a structural link between the skeletal muscle cytoskeleton and the extracellular matrix, and it is essential to maintain muscle integrity. DMD patients appear clinically asymptomatic at birth; however, they manifest signs of muscle weakness and walking difficulties during early childhood when they are typically diagnosed. Loss of ambulation and wheelchair dependency ensues around puberty. Thanks to improvements in palliative care, life expectancy and quality of life have improved, and many patients may survive beyond 30 years of age [3]. Becker muscular dystrophy (BMD) is a milder dystrophy form caused by in‐frame mutations in the DMD gene, leading to the expression of a shorter and partially functional dystrophin protein. Individuals with BMD share signs and symptoms with DMD patients, but they present a much later disease onset and a nearly average lifespan [4].
Although the molecular mechanisms of this disease have been extensively investigated, there is still no complete curative treatment available. The current standard of care includes corticoids, such as prednisone or deflazacort, to delay disease progression [5]. Therapies based on dystrophin replacement at the protein or gene level are challenging due to the gene's large size, the wide distribution of the skeletal muscle throughout the body, and the possibility of immune response activation. However, many of these aspects have been overcome, and several micro‐dystrophin gene therapies are currently undergoing clinical trials [6].
In recent years, regulatory agencies have conditionally approved several RNA treatments, based on read‐through (ataluren [7]) or exon‐skipping strategies (eteplirsen [8], golodirsen [9] and viltolarsen [10]). Nevertheless, these therapies are only applicable to a low percentage of DMD patients. Moreover, their delivery to the muscle is challenging [11], and their approval is controversial due to the low efficacy in dystrophin restoration and the limited clinical efficacy demonstrated so far [12].
Alternative strategies to mutation‐specific approaches have been under intense investigation in several laboratories worldwide in order to find a therapy applicable to the Becker and Duchenne community, regardless of their specific mutations. Among them, upregulation of utrophin, a structural and functional paralogue of dystrophin, is one of the most promising therapeutic strategies. Recent studies based on high‐throughput screening have identified small molecules able to induce utrophin upregulation. However, utrophin expression is subject to regulation at multiple steps throughout its synthesis and degradation pathways, which need to be studied in depth to improve pharmacological interventions.
Utrophin vs dystrophin: structure, distribution and function
Dystrophin is a 427 kDa protein encoded by the DMD gene, the largest human gene, localised on the X chromosome. Utrophin, known initially as ‘dystrophin‐related protein’, is a 395 kDa autosomal paralogue of dystrophin encoded by the UTRN gene localised in the human chromosome 6q24 [13]. While four full‐length dystrophin isoforms driven by different promoters have been described, only two full‐length utrophin isoforms have been identified to date, utrophin A and B (Figure 1). These isoforms are transcribed from two different promoters, A and B. The two mRNAs vary at their 5’ ends, resulting in two identical functional proteins with slightly different N‐terminal domains and different expression patterns. While utrophin A is expressed in a variety of structures, including neuromuscular junctions, choroid plexus, pia mater and renal glomerulus [14], utrophin B remains restricted to the endothelial cells [15]. Interestingly, five novel 5’ utrophin isoforms (A’, B’, C, D and F) have been recently identified in human adult and embryonic tissues, but they remain to be fully characterised [16]. Both the DMD and the UTRN gene also encode for shorter dystrophin and utrophin transcripts. Shorter dystrophin isoforms, including Dp260, Dp140, Dp116 and Dp71, have been identified in different non‐muscle tissues such as the brain [17] and retina [18] (Figure 1A,C). Utrophin's internal promoters produce shorter transcripts such as Up71, Up140 and G‐utrophin, which are expressed in many tissues with functions not fully understood [19] (Figure 1B, C).
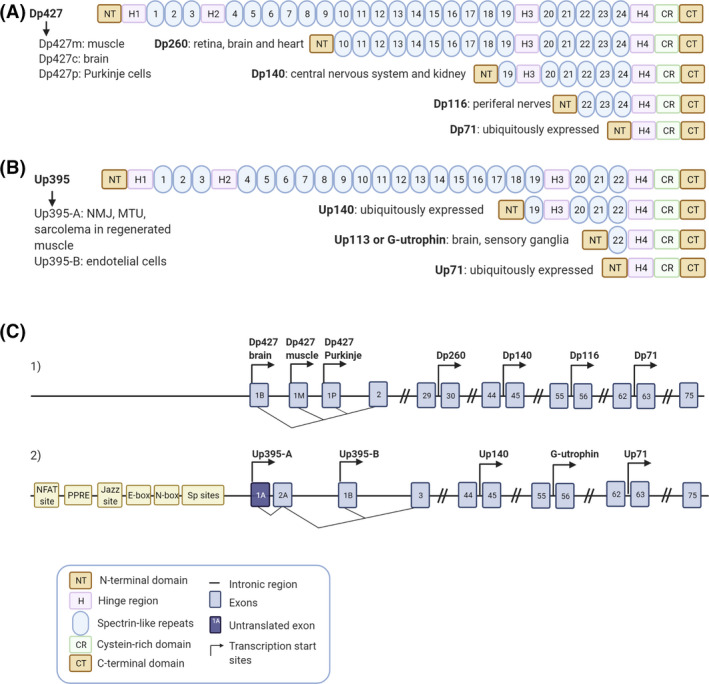
Dystrophin and utrophin isoforms. Schematic representation of the full length and truncated dystrophin (A) and utrophin (B) protein isoforms including their most representative expression in tissues. (C1 and C2) Blue boxes show the specific exons, the black line represents the intronic regions and the transcription start sites of the different promoters are indicated by arrows within the dystrophin (C1) and the utrophin (C2) gene. (C1) Full‐length dystrophin expression is driven by three promoters Dp427 brain, muscle and Purkinje and the smaller isoforms are produced from four internal promoters, Dp260, Dp140, Dp116 and Dp71. (C2) Full‐length utrophin expression is driven by two promoters Up395‐A and Up395‐B and the smaller isoforms are produced from three internal promoters Up140, G‐utrophin and Up71. Different elements of the utrophin A promoter are also specified in the panel. Created with BioRender.com
In the adult skeletal muscle, dystrophin is an essential structural protein that links the extracellular matrix to the actin cytoskeleton through assembly to the dystrophin–glycoprotein complex (DGC) (Figure 2A). This large multi‐protein complex is critical for maintaining the fibre's structural integrity, the stability of the neuromuscular synapse, and the muscle fibre's strength and flexibility while protecting the membrane from contraction‐induced damage [20]. In DMD patients, loss of dystrophin leads to destabilisation and deterioration of the whole complex. Mutations in genes encoding different components of the DGC result in a variety of muscular dystrophies, which highlights the importance of this complex [21].

Schematic representation of dystrophin and utrophin glycoprotein complexes (DGC/UGC). (A) Dystrophin glycoprotein complex (DGC) and (B) utrophin glycoprotein complex (UGC) consist of dystrophin (or utrophin), syntrophins, dystrobrevins, sarcoglycans, sarcospan and dystroglycans distributed in cytoplasmic, transmembrane and extracellular protein complex. The cytoplasmic part includes α1 and β1 syntrophin isoforms and α‐dystrobrevin; transmembrane part includes the sarcoglycan (α, β, γ, δ) and sarcospan complex. Dystroglycan complex consists in the extracellular component, α‐dystroglycan (α‐DG) which binds to agrin and laminin in the extracellular matrix and the transmembrane isoform β‐dystroglycan (β‐DG). Biglycan is another extracellular matrix component of the DGC/UGC that binds to α‐dystroglycan and α‐ and γ‐sarcoglycan [22]. Finally, β‐DG binds to dystrophin or utrophin, completing the link between the actin‐based cytoskeleton and the extracellular matrix [23]. Furthermore, utrophin is associated with large acetylcholine receptors (AChR) clusters at the crests of post‐junctional folds in neuromuscular junctions (NMJs) [24]. Notice that the main differences between dystrophin and utrophin are their lateral interactions with actin and the impossibility of the UGC to recruit nNOS. Created with BioRender.com
Dystrophin has four main domains: an N‐terminal actin‐binding domain (NTD), a central spectrin‐like repeat region, a cysteine‐rich domain (CR), which binds the DGC, and a C‐terminal domain (CTD) (Figure 2A). Utrophin shares those domains with dystrophin, but there are some structural and mechanical differences. Both proteins differ in their lateral interactions with actin [20] (Figure 2B), utrophin containing fewer spectrin‐like repeats, and sharing only a 35% homology in the central domain with dystrophin. Moreover, a significant difference in the mechanical behaviour between spectrin repeats has been recently demonstrated [25]. Crucially, they also differ in their capacity to recruit neuronal nitric oxide synthase (nNOS), which cannot be recruited by utrophin [26]. nNOS, a signalling protein associated with the DGC that produces nitric oxide (NO), is considerably reduced in dystrophic muscle fibres, leading to functional ischaemia due to decreased contraction‐induced vasodilation.
While dystrophin is predominantly expressed in muscle and to a lesser extent in the brain, utrophin is widely expressed in several non‐skeletal muscle tissues such as lung, kidney and liver [27]. During foetal muscle development and at early gestational stages, utrophin is present at the sarcolemma of muscle fibres. After birth, utrophin is progressively silenced by the Ets‐2 repressor factor and replaced by dystrophin in adult myofibres. Thereafter, utrophin disappears from the membrane, and its expression is confined to the neuromuscular and myotendinous junctions, where it participates in post‐synaptic membrane maintenance and acetylcholine receptor clustering [28, 29]. However, there is an increase in utrophin expression and redistribution of this protein to the sarcolemma in the dystrophic muscle, in mature dystrophin‐deficient fibres, regenerating fibres and dystrophin‐competent revertant fibres found both in DMD and BMD patients, as well as in mdx mice [30, 31, 32].
Utrophin overexpression in Duchenne muscular dystrophy
Utrophin is naturally increased at the sarcolemma of skeletal muscle samples in DMD and BMD patients compared to healthy individuals [31, 32, 33] by a repair process that also occurs in animal models of the disease, proposed as a compensatory mechanism to mitigate the lack of dystrophin. Moreover, preclinical studies indicate an inverse correlation between utrophin expression and disease severity in DMD, suggesting that utrophin could play a role as a dystrophin surrogate. However, while some human studies report a positive effect of this utrophin expression on disease severity, delaying its progression [34], others do not find any correlation [35].
The most widely used animal model for DMD research is the mdx mouse, carrying a nonsense point mutation (C‐to‐T transition) in exon 23 of the Dmd gene, which completely abolishes dystrophin expression. Despite being dystrophin‐deficient, mdx mice have mild clinical symptoms and a long lifespan, in contrast to DMD patients [36]. Utrophin levels are increased at the sarcolemma of regenerating myofibres in the adult mdx skeletal muscle [37, 38], but this increase may also occur independently of regeneration [39]. Moreover, experimental data suggest that upregulation of utrophin may compensate for dystrophin deficiency. The potential compensatory role of utrophin has been assessed by generating double knockout mice for both dystrophin and utrophin genes (dko). These mice display a much more severe pathology compared to mdx mutants, as well as multiple systemic degenerative changes, in addition to earlier muscle degeneration [40]. On the other hand, the Fiona mouse, a dystrophin‐deficient mdx transgenic mouse that overexpresses utrophin, shows a correction of the dystrophic phenotype [38, 41].
Over the years, preclinical studies have demonstrated that transgenic overexpression and pharmacological modulation of utrophin prevent skeletal muscle pathology in mdx mice. These studies reveal that a 2‐fold increase in sarcolemmal utrophin completely rescues the mechanical function and effectively normalises classical markers of DMD‐related muscle damage [42, 43]. However, even a 1.5 fold increase may be beneficial for mdx mice, given that utrophin localises at the sarcolemma of dystrophic fibres [38]. Utrophin levels also influence mitochondrial pathology that contributes to oxidative stress and propagates muscle damage in DMD. While utrophin deficiency aggravates the pathology, utrophin over‐expression in the dystrophic muscle supports mitochondrial function in mouse models [44]. Interestingly, another study focused on the role of utrophin replacing dystrophin in the male reproductive system discovered that full‐length dystrophin deficiency disturbed the balance between proliferation and apoptosis of germ cells during spermatogenesis. In this case, there is also a utrophin upregulation and relocation as a compensatory response to dystrophin deficiency [45]. Taken together, data in animal models suggest that utrophin can functionally compensate for the lack of dystrophin.
Utrophin overexpression in patients is a promising therapeutic strategy for treating muscle dystrophies, since it targets the primary cause of the disease and would apply to all DMD and BMD patients regardless of their genetic mutation. Several approaches may be used to modulate utrophin levels including direct mechanisms, such as gene or protein replacement, or indirect ones, such as transcriptional upregulation of the utrophin promoter, post‐transcriptional regulation and protein/mRNA stabilisation (see Table 1).
TABLE 1
Mechanisms of action of potential drugs that could modulate utrophin expression
| Direct mechanisms | Protein replacement | TAT‐μUtrn [46, 47] |
|---|---|---|
| Gene therapy | μUtrn [48] | |
| Indirect mechanisms | Acting at utrophin A promoter level |
Artificial zinc finger transcription factors (ZF‐ATFs): Jazz [49], Bagly [50], Utroup [51], JZif1 [52]. Aryl hydrocarbon receptors (AhR) antagonists [53]: Ezutromid or SMTC0011 [54] and SMT022357[42] Other small molecules: Nabumetone [55], Heregulin [56, 57], Okadaic acid [58], Adiponectin [59, 60] |
| Oxidative phenotype promoters |
Via peroxisome proliferator‐activated receptor (PPAR) agonists: GW501516 Via AMPK activators: AICAR Metformin [61] Adiponectin Obestatin [62] Quercetin Resveratrol [63] | |
| nNOS activation |
L‐arginine L‐citrulline | |
| mRNA stabilisation at 5’ UTR and 3’UTR level |
eEF1A2/IRES‐mediated translation: Betaxolol [64, 65], Pravastatina and 6α‐methylprednisolone‐21 sodium succinate (PDN) [66] via microRNA targeting: Let‐7c, miR‐150, miR‐196b, miR‐296‐5p, miR‐133b AntimiR 206 via p38 MAPK/KSRP: Heparin [67], Heparin/AICAR [68], Heparin/GALGT2 [69] Celecoxib[70] Anisomycin [71] Trichostatin A | |
| Protein stabilisation |
GalNAc2 [75] |
DIRECT UTROPHIN REPLACEMENT
Protein replacement
Direct protein replacement using recombinant full‐length or truncated utrophin is an attractive potential method to increase utrophin levels in vivo directly.
Systemic administration of a recombinant ‘micro‐utrophin’ (μUtrn) protein combined with the cell‐penetrating TAT protein (TAT‐μUtrn), the transduction domain of the HIV‐1, can functionally form a μUtrophin‐glycoprotein complex at the sarcolemma. This therapeutic strategy is able to mitigate the dystrophic phenotype of mdx mice, improving contractile strength [35]. TAT‐μUtrn also ameliorates the phenotype of dystrophin/utrophin double‐knockout (dko) mice, increasing skeletal muscle strength and improving activity and life span compared to placebo [36].
Although this therapeutic strategy looks promising, posology and administration limit its use; it would be necessary to give frequent, high‐dose injections that could eventually trigger a harmful immune response. Nevertheless, this approach might be combined with other therapies to increase utrophin expression.
Gene therapy
Developing gene therapy treatments for Duchene muscular dystrophy is challenging for three main reasons: first, both full‐length DMD or UTRN genes, and even their cDNAs, are too large and need to be engineered into truncated (‘mini’ or ‘micro’) constructs in order to be packaged into adeno‐associated virus (AAV), which are currently the most commonly used delivery vectors [80]; the second limitation is the possibility of inducing a cellular immune response to the new dystrophin generated and/or against the AAV vector [81]; the third and major challenge is the difficulty to achieve body‐wide transduction into human muscle fibres. Nevertheless, micro‐dystrophin (µDys) gene therapy using AAV vectors has recently been carefully optimised, leading to promising data in murine and canine DMD models [82, 83, 84] and phase I/II clinical trials in DMD patients that are currently ongoing.[85]
A similar pathway has been followed in the development of utrophin gene therapy alternatives. Several preclinical studies using ‘micro‐utrophin’ (µUtrn) gene delivery have been reported in the last years; studies conducted using AAV‐µUtrn in mdx mice reported restoration of the DGC, prevention of myofibre degeneration, normalisation of serum CK levels and improvement of muscle function [86]. Moreover, additional studies in double knockout (dko) mice and canine X‐linked muscular dystrophy dogs have shown that µUtrn improves their severe pathological dystrophic phenotype [87]. Modulation of utrophin expression could potentially treat many disease manifestations since AAV‐μUtrn transgene administration functionally replaces dystrophin in the heart and ameliorates the skeletal and cardiac muscle phenotype in the D2/mdx mouse model [88]. In addition, the ex vivo UTRN gene correction of mouse dystrophic iPS cells by µUtrn gene transfection and subsequent transplantation into dystrophic dko mice has also demonstrated DGC restoration and improvement of contractile strength [89].
Apart from all these promising results, delivering µUtrn instead of µDys has a potential advantage: a lower risk to elicit an immune response since utrophin is naturally expressed at low levels in DMD patients. A recent study performed in the German shorthaired pointer deletional‐null canine model (GSHPMD), reported a strong systemic cell‐mediated immune response against µDys but not to µUtrn. This supports the use of non‐immunogenic utrophin‐based gene therapy approach for DMD [90]. Moreover, overexpression of utrophin rather than dystrophin could prevent the use of expensive and potentially toxic adjuvant immunosuppressive drug therapies [86].
INDIRECT MECHANISMS FOR UTROPHIN OVEREXPRESSION
Transcriptional upregulation
The utrophin A promoter contains several regulatory motifs that could activate utrophin overexpression (Figure 3). The E‐box and N‐box motifs are essential for myogenic differentiation, and synaptic expression of utrophin A [91]. The E‐box motif is a binding site to myogenic factors like MyoD, myogenin, Myf5 and MRF4 [91]. In contrast, the N‐box motif is targeted by the ETS‐related transcription factor complex GA‐binding protein (GABP) α/β, which is activated by nerve‐derived and transcription factors. Besides, Sp binding sites targeted by Sp1 and Sp3 zinc finger transcription factors may establish a cooperative interaction with GABP to stimulate the utrophin promoter [92] (Figure 3). Moreover, it has been recently shown that utrophin A promoter contains a PPRE site targeted by the peroxisome‐proliferator‐activated receptor beta/delta (PPAR‐β/δ). This PPRE site can also be stimulated through 5′ adenosine monophosphate‐activated protein kinase (AMPK) and sirtuin 1 (SIRT‐1) signalling pathways that activate the peroxisome proliferator‐activated receptor‐gamma coactivator 1α (PGC‐1α) which, in turn, activates either PPARβ/δ or GABPα/β.
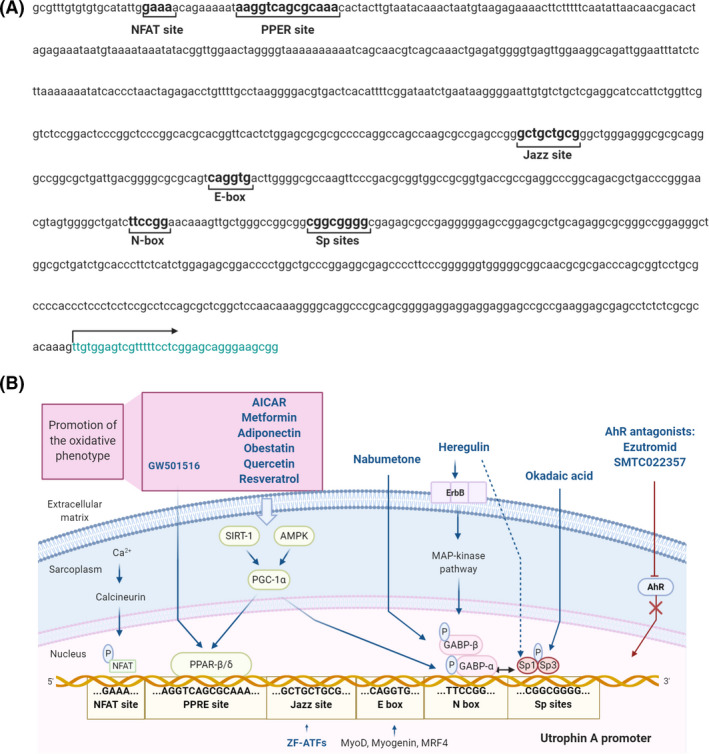
Utrophin A promoter transcriptional activation pathways. (A) Nucleotide sequence of the human UTRN gene promoter including the transcriptional regulatory elements: NFAT binding site, PPRE site, Jazz binding site, E‐box site, N‐box site and Sp binding sites. The arrow indicates the transcription starting site. (B) Representation of the utrophin A promoter regulatory binding sites, their transcriptional upregulation mechanisms and the compounds involved in utrophin upregulation through these signalling pathways (in blue). The compounds that act through promotion of the slow and oxidative phenotype are included in the purple box. Created with BioRender.com
The calcineurin‐nuclear factor of activated T‐cells (NFAT) calcium‐dependent signalling cascade is another pathway that positively regulates utrophin expression in the skeletal muscle. In this pathway, calcineurin dephosphorylates NFAT, enabling its entry into the nucleus and subsequent activation of the utrophin A promoter [93] (Figure 3).
Utrophin upregulation by stimulating utrophin A promoter activity is a promising pharmacological approach that has been extensively investigated using different strategies. One laboratory's proposal was the engineered artificial zinc finger transcription factors (ZF‐ATFs) called ‘Jazz’, capable of binding the utrophin A promoter both in humans and mice. Systemic delivery of ZF‐ATFs with AAVs can induce a significant rescue of muscle function in dystrophic mdx mice through utrophin upregulation [94]. Indeed, several ‘Jazz’ factors have shown remarkable efficacy in ameliorating the pathological phenotype of mdx mice and improving the morphology and plasticity of neuromuscular junctions [52]. Among them, ‘JZif1’, the most recently upgraded version, was developed using the backbone of the well‐characterised Zif268/EGR1 human transcription factor to minimise immunogenicity and facilitate its clinical application.
Thousands of candidates from drug libraries have been tested by high‐throughput screening (HTS) assays in order to find small molecules acting at the utrophin A promotor level. In these cell‐based assays, a reporter gene (usually luciferase) is linked to the utrophin promoter [55, 95]. Small molecules offer several advantages, such as improved delivery and bioavailability compared to gene therapy or protein replacement and the possibility of testing compounds already approved for clinical use. Indeed, drug repurposing to other indications may accelerate their transfer to the clinic and improve their chances of success. In different studies, both repurposing and newly synthesised compounds have shown promising results at preclinical level, with dose‐dependent activation of the utrophin promoter such as nabumetone, heregulin and okadaic acid. Some of them, like ezutromid, have already reached clinical trials.
Nabumetone is a long‐acting nonsteroidal anti‐inflammatory drug, specifically a COX‐1/COX‐2 inhibitor that shows a preference for COX‐2 inhibition in vitro. It is used for pain and inflammation management in osteoarthritis and rheumatoid arthritis, and it is an example of pharmacological repurposing for DMD. HTS assays in C2C12 muscle cells demonstrated that nabumetone could activate utrophin A promoter and upregulate endogenous utrophin at mRNA and protein level [55].
Heregulin is a small nerve‐derived grow factor capable of transactivating utrophin A promoter via the N‐box motif. Utrophin transcription induced by heregulin‐mediated activation of GABPα/β occurs through the extracellular‐signal related kinase (ERK) signalling pathway via the interaction of heregulin with the ErbB tyrosine kinase receptor [56, 57]. Intraperitoneal injections of a small heregulin peptide in mdx mice resulted in upregulation of utrophin, together with a marked functional improvement of the muscle pathology [96].
Recently, it has been shown that okadaic acid, a selective inhibitor of PP1 and PP2A phosphatases, can induce utrophin A promoter activation during myogenesis through Sp1 phosphorylation. There is evidence that okadaic acid increases utrophin A mRNA levels increased by around two‐fold in C2C12 myoblasts, but not in myotubes [58].
Ezutromid (SMTC1100) was the first orally bioavailable utrophin regulator that showed increased UTRN transcription. It was identified following a HTS strategy with a luciferase reporter‐linked assay in murine H2K cells. Later, in vitro assays in human myoblasts demonstrated an increase in utrophin expression at mRNA and protein levels after ezutromid treatment, and further in vivo assays demonstrated that once‐a‐day daily‐dosing of ezutromid in mdx mice increased utrophin levels, as well as muscle strength and resistance to exercise.
After these initial results, ezutromid was developed by Summit Therapeutics as a potential treatment for DMD and BMD. A Phase 1 placebo‐controlled randomised clinical trial in healthy male volunteers and a Phase 1b placebo‐controlled, randomised, double‐blind study in boys with DMD showed that it was safe and well‐tolerated. However, a Phase 2 clinical study (NCT02858362) failed to achieve both the primary (changes in leg muscle magnetic resonance parameters) and secondary endpoints (increased utrophin levels and decreased muscle damage). Based on these results, Summit Therapeutics abandoned the development program of ezutromid [97, 98]. Recent studies have elucidated the ezutromid mechanism of action as an aryl hydrocarbon receptor (AhR) antagonist [53, 99]. Similarly, other molecules that ameliorate mdx pathology like SMT022357 [53] or resveratrol [100] have also shown activity as AhR antagonists [100]. While the pathway between AhR antagonism and utrophin upregulation remains unknown, it seems to involve the stabilisation of active peroxisome proliferator‐activated receptor gamma coactivator (PGC1α) [101]. Indeed, moderately elevated levels of PGC1α ameliorate the dystrophic phenotype of mdx mice at the biochemical, histological and functional levels [102].
SMT022357, is a second‐generation compound structurally related to ezutromid, sharing the same mechanism of action but with improved physicochemical properties and a more robust metabolic profile. SMT022357 administration has been associated with an increase in utrophin expression in skeletal, respiratory and cardiac muscles and prevention of the dystrophic pathology in mdx mice [42].
Oxidative phenotype promotion
An alternative therapeutic strategy to increase utrophin expression in the skeletal muscle focuses on the upregulation of the slow oxidative myogenic program. Promotion of the slow oxidative phenotype has been achieved by different transcriptional and post‐transcriptional pathways showing utrophin overexpression (Figure 3). This strategy has demonstrated attenuation of the dystrophic pathology in mdx animals [103].
One mechanism reported is PPAR‐β/δ stimulation using the synthetic agonist GW501516. This molecule has also been found to stimulate utrophin A promoter in C2C12 muscle cells and improve sarcolemmal integrity in mdx mice, conferring protection against eccentric contraction‐induced damage to muscle [104].
Chronic activation of AMPK also promotes the slow oxidative phenotype. Treatment of mdx mice with 5‐aminoimidazole‐4‐carboxamide‐1‐β‐D‐ribofuranoside (AICAR) and other AMPK/PGC‐1α activators significantly enhanced utrophin expression and have proved to be beneficial for the dystrophic phenotype and rescue muscle function [103].
One of the best known pharmacologically AMPK activators is metformin, a widely prescribed oral antidiabetic drug that has reached clinical trials for DMD in combination with the NOS modulators L‐arginine and L‐citrulline. Metformin increases skeletal muscle utrophin content via AMPK activation and parallel or reciprocal increments in PGC‐1α and PPAR‐δ expression [61]. Skeletal muscle nNOS activation is also AMPK dependent [105]. However, the partial response to metformin treatment in mdx muscles combined with the reduced quantity of NO in some studies supports the notion of combined therapy for DMD patients [61, 106]. In combination with L‐arginine, metformin showed evident amelioration of muscular metabolism in the first proof‐of‐concept pilot study (NCT02516085) carried out in DMD patients. Results from another study, a randomised, double‐blind placebo‐controlled clinical trial with 47 ambulant DMD patients, combining L‐citrulline (an L‐arginine precursor) and metformin (NCT01995032), showed a clinically relevant but not statistically significant reduction in motor function decline in a specific subgroup of patients with no apparent side effects. Therefore, additional clinical trials are needed to validate this approach [107]. Interestingly, NOS‐based therapy by itself has also proved to increase utrophin expression. In this context, L‐arginine administration in mdx mice resulted in a nearly 2‐fold increase in utrophin in skeletal muscle, heart and brain, accompanied by an improvement of the dystrophic phenotype [108]. This study demonstrates that NOS expression has beneficial effects on skeletal muscle metabolism both in vitro and in vivo.
The hormone adiponectin protects the skeletal muscle against inflammation and injury via the AMPK‐SIRT1‐PGC‐1α signalling pathway. Treatment of myotubes from DMD patients with adiponectin leads to downregulation of the nuclear factor kappa B (NF‐κB) and inflammatory genes, together with an upregulation of utrophin [59]. Transgenic upregulation of adiponectin has demonstrated significant beneficial properties in dystrophic mdx muscles [109]. Recently, an orally administrable active adiponectin receptor agonist, called AdipoRon, has been identified. This small synthetic molecule has also proved to attenuate the dystrophic phenotype in mdx mice offering a promising therapeutic prospect for DMD patients [60].
In the same line, obestatin, an autocrine factor that controls the myogenic differentiation program, induces a skeletal muscle shift towards a more oxidative metabolic profile through mechanisms involving PGC1α and class II histone deacetylases (HDAC)/myocyte enhancer factor‐2 (Mef2). It has been reported that obestatin has shown activity in stabilising the sarcolemma of mdx skeletal muscle through the expression of utrophin, α‐syntrophin, β‐dystroglycan and α7β1‐integrin proteins, ameliorating the DMD phenotype [62].
Another molecule studied in preclinical assays that seems to upregulate utrophin through activation of the PGC‐1α pathway is quercetin [110]. Diet enriched with this flavonol seems to rescue dystrophic muscle in mdx mice and provide physiological cardioprotection [111, 112].
Finally, administration of the natural phenol resveratrol to mdx mice has also demonstrated stimulation of the SIRT1‐PGC‐1α pathway, a significant upregulation of utrophin expression, and activation of the slow, oxidative myogenic program in mdx mouse muscle [63].
Post‐transcriptional and translational events regulating utrophin isoform A
While utrophin upregulation at the transcriptional level has been widely investigated over the years, an increasing number of new studies support the importance of post‐transcriptional and translational regulator factors of utrophin in order to find new therapeutic targets (Figure 4).
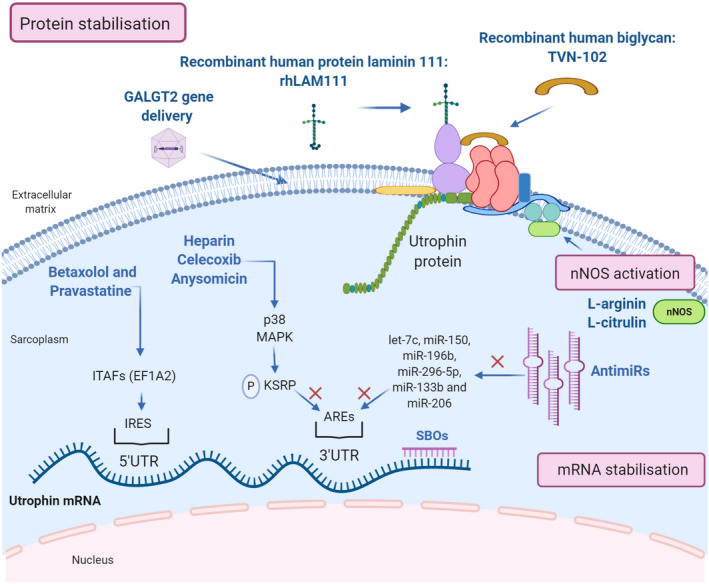
Therapeutic strategies for post‐transcriptional utrophin upregulation. Representation of the post‐transcriptional pathways to enhance utrophin expression: mRNA stabilisation, nNOS activation and protein stabilisation and the compounds acting through these mechanisms (in blue). Created with BioRender.com
Utrophin full‐length isoforms, A and B, have different 5′‐untranslated regions (5′‐UTRs). The skeletal muscle isoform, utrophin A, presents an internal ribosome entry site (IRES) at its 5′UTR that promotes expression through IRES‐dependent translational mechanisms [113]. IRES elements are thought to associate with the translational machinery, including some IRES trans‐acting factors (ITAFs). EF1A2 has been reported as a suitable ITAF able to modulate the activity of the utrophin A IRES. For clarity, we refer to utrophin isoform A as ‘utrophin’ in the manuscript.
A recent ELISA‐ based HTS assay has identified at least four FDA‐approved drugs that target eEF1A2 and cause at least a two‐fold increase in utrophin in C2C12 muscle cells. Among them, betaxolol and pravastatine, seem to improve the dystrophic phenotype of mdx mice via utrophin upregulation through IRES activation [64]. Moreover, in another study, utrophin protein levels are increased after 6α‐methylprednisolone‐21 sodium succinate (PDN) treatment of C2C12 myotubes, suggesting that glucocorticoid's mechanism in muscle cells could be at least partially explained by enhancement of utrophin translation due to IRES activation [66]. These studies highlight the increasing interest in using repurposed drugs to activate this specific pathway where endogenous utrophin levels in muscle are upregulated by promoting protein synthesis from already synthesised transcripts.
Expression of utrophin is also regulated at its UTR 3’ end, where a series of cis‐elements, including conserved AU‐rich elements (AREs), modulate the stability of utrophin mRNA transcripts. Different proteins can bind the AU‐rich elements at the 3'‐UTR and regulate mRNA stability either negatively or positively. For example, 3'‐UTR repression has been attributed to miRNAs and K‐homology splicing regulator protein (KSRP) binding to these sites.
Several miRNAs, including let‐7c, miR‐150, miR‐196b, miR‐296‐5p, miR‐133b and miR‐206 have been shown to repress utrophin expression [114, 115], and this has led to two therapeutic approaches: targeting the microRNAs directly by using antimiRs or blocking their binding site with site‐blocking oligonucleotides (SBOs). Both mechanisms have shown to upregulate utrophin expression and improve the dystrophic phenotype in vivo. Intraperitoneal injections of specific SBOs targeted to prevent let‐7c miRNA binding to the utrophin 3’UTR resulted in higher utrophin protein expression in skeletal muscles and improvement in the dystrophic phenotype in mdx mice [116, 117]. On the other hand, a 3‐month treatment with antiMiR‐206 increases utrophin in mdx mouse muscles compared to the untreated group [118].
Activating p38 mitogen‐activated protein kinase (MAPK) reduces KSRP availability to bind utrophin's 3’UTR AREs, resulting in increased stability of existing mRNAs, increased utrophin protein production and reduction of muscle damage [67]. At least three approved drugs and activators of p38 MAPK, heparin, celecoxib and anysomicin, have demonstrated a significant utrophin upregulation efficacy in different preclinical studies.
Heparin, which is an anticoagulant commonly used in the clinic, significantly increases utrophin levels both in C2C12 [67] myoblasts and mdx mouse dystrophic fibres, leading to substantial morphological and functional improvements [68]. In addition, combinatory treatment with heparin plus AICAR (an oxidative phenotype promoter compound mentioned previously) has an additive effect, increasing utrophin protein levels nearly 3‐fold in C2C12 myoblasts and mdx mouse muscle [68].
Celecoxib is a nonsteroidal anti‐inflammatory drug (NSAID) and a specific cyclo‐oxygenase (COX)‐2 inhibitor used for osteoarthritis and rheumatoid arthritis. This drug can activate p38 MAPK signalling in skeletal muscle cells. Treated mdx mice revealed a 1.5‐ to 2‐fold increase in utrophin expression in tibialis anterior, diaphragm and heart muscles, and ameliorated the dystrophic phenotype, improving muscle strength [70].
Anisomycin is an antibiotic identified by HTS assays. In C2C12 muscle cells, it induces a 2.5‐fold increase in utrophin levels in vitro. It is also reported to significantly increase utrophin protein in the diaphragm of mdx mice treated daily with a low dose [71].
Another recent HTS screening study, targeting the 5′ and 3′ untranslated regions (UTRs), identified 27 hit compounds capable of upregulating utrophin expression [119]. In this study, trichostatin A was identified as one of these hit compounds. Previous studies had demonstrated that trichostatin A could activate the utrophin promoter [55]. It also increases utrophin levels post‐transcriptionally by interacting with the 5′ and/or 3′UTR of the utrophin mRNA, resulting in a functional improvement of the mdx mouse. The remaining hits are yet to be further studied, but this is a good starting point for additional in vitro or in vivo assays.
Utrophin‐glycoprotein complex stabilisation
Utrophin complex stabilisation is an alternative mechanism that has gained strength in the last years with promising results. One example of this approach is the extracellular matrix biglycan, a proteoglycan that plays an essential role in muscle development. Biglycan is a component of the DGC/UGC, where it regulates the expression of sarcoglycans, dystrobrevins, syntrophins and nNOS, by recruiting utrophin to the plasma membrane. In humans and mice, biglycan is most highly expressed in immature and regenerating muscle [22]. Several studies in mdx mice have shown that systemically administered recombinant human biglycan upregulates utrophin and other DGC components at the sarcolemma, while ameliorating muscle pathology and improving muscle structure and function with no obvious toxicity.[72, 73] Tivorsan Pharmaceuticals is currently developing a potential treatment for DMD and BMD called TVN‐102, a recombinant human biglycan that can be systemically administrated [120]. The FDA granted TVN‐102 orphan drug status in 2016. Meanwhile, Tivorsan Pharmaceuticals has completed pharmacological studies in rats and non‐human primates in order to determine the safe starting dose in clinical trials, planned to be initiated soon.
Similarly, the recombinant human protein laminin‐111 (rhLAM111), another extracellular matrix protein, has shown to upregulate other proteins such as utrophin and α7β1 integrin, both capable of restoring muscle cell adhesion and stimulating muscle regeneration in DMD patients. Research in mdx mouse has demonstrated that rhLAM111 can strengthen muscles and improve muscle function. The underlying mechanisms of action reported involved elevated levels of different compensatory proteins and utrophin increases of 1.3‐fold. However, it is not completely clear if this increase in utrophin is sufficient to induce a phenotypical improvement [76, 77]. Indeed, some studies claim that higher utrophin concentrations (1.5/2‐fold increase) are necessary to achieve a therapeutic effect [38]. In any case, recent results show that laminin prevents muscle disease progression in the golden retriever muscular dystrophy (GRMD) dog model of DMD and, thus, it could be a novel protein therapy for DMD patients [120].
Overexpression of CT‐GalNAc 2 (cytotoxic T‐cell N‐acetylgalactosamine transferase), or Galgt2 protein, has been shown to increase synapse‐associated proteins, including utrophin, and enhances its transportation to the sarcolemma [75]. AAV‐mediated GALGT2 gene delivery has shown protection in both wild‐type and dystrophin‐mdx skeletal myofibres from eccentric contraction‐induced injury. It also prevents muscular dystrophy and ameliorates the phenotype in different animal models [121, 122]. Following these studies, the first clinical trial of AAVrh74‐mediated GALGT2 gene delivery in DMD boys began recruiting in 2018.
CONCLUDING REMARKS
Utrophin upregulation is a promising therapeutic approach, applicable for all DMD and BMD patients, that has demonstrated functionally compensation for the lack of dystrophin, improving the pathological phenotype in different dystrophic models.
Many pathways involved in utrophin expression are currently being explored, and some of them have only started to be elucidated. There are high expectations in many compounds that have demonstrated efficacy in activating utrophin expression in preclinical assays. However, the amount of utrophin required by dystrophic patients to achieve a relevant clinical benefit remains to be determined. Hopefully, soon some of these molecules will reach clinical studies and become therapeutic options for the Duchenne community.
CONFLICT OF INTERESTS
A. V.‐I. and V. A.‐G. are shareholders of Miramoon Pharma SL, a company developing DMD treatments, but not related to the focus of this manuscript. All authors report no known competing financial interests or personal relationships that could have appeared to influence the work reported in this manuscript.
AUTHOR CONTRIBUTIONS
P. S.‐M., investigation, writing of original draft, review and editing, visualisation. L. d.‐l.‐P.‐O., investigation, writing ‐ review and editing. A. L.‐M., investigation, writing ‐ review and editing. A. V.‐I. writing ‐ review and editing. V. A.‐G.: conceptualisation, writing or original draft, review and editing, visualisation, supervision, project administration and funding acquisition.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1111/nan.12735.
Notes
Soblechero‐Martín P, López‐Martinez A, de la Puente‐Ovejero L, Vallejo‐Illarramendi A, Arechavala‐Gomeza V. Utrophin modulator drugs as potential therapies for Duchenne and Becker muscular dystrophies. Neuropathol Appl Neurobiol. 2021;47:711–723. 10.1111/nan.12735 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
Funding information
This work was supported by funding from Health Institute Carlos III (ISCIII, Spain) and the European Regional Development Fund, (ERDF/FEDER), ‘A way of making Europe’: Grant PI15/00333; Basque Government (grants 2016111029, 2018222035 and 2020333012) and Duchenne Parent Project Spain (grant 05/2016). P. S‐M holds a Rio Hortega Fellowship from ISCIII (CM19/00104). V.A‐G holds a Miguel Servet Fellowship from the ISCIII (CPII17/00004), part‐funded by ERDF/FEDER. A. L‐M acknowledges funding by Biocruces Bizkaia Health Research Institute (BC/I/DIV/19/001). V.A‐G also acknowledges funding from Ikerbasque (Basque Foundation for Science). None of this funding represents a conflict of interest with the content of this review.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analysed in this study.
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1111/nan.12735
Read article for free, from open access legal sources, via Unpaywall:
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1111/nan.12735
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/107104536
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1111/nan.12735
Article citations
The Role of MicroRNA in the Pathogenesis of Duchenne Muscular Dystrophy.
Int J Mol Sci, 25(11):6108, 01 Jun 2024
Cited by: 2 articles | PMID: 38892293 | PMCID: PMC11172814
Review Free full text in Europe PMC
Molecular and Biochemical Therapeutic Strategies for Duchenne Muscular Dystrophy.
Neurol Int, 16(4):731-760, 05 Jul 2024
Cited by: 0 articles | PMID: 39051216 | PMCID: PMC11270304
Review Free full text in Europe PMC
3D-QSAR Modeling on 2-Pyrimidine Carbohydrazides as Utrophin Modulators for the Treatment of Duchenne Muscular Dystrophy by Combining CoMFA, CoMSIA, and Molecular Docking Studies.
ACS Omega, 9(23):24707-24720, 31 May 2024
Cited by: 0 articles | PMID: 38882130 | PMCID: PMC11171099
How Can Proteomics Help to Elucidate the Pathophysiological Crosstalk in Muscular Dystrophy and Associated Multi-System Dysfunction?
Proteomes, 12(1):4, 16 Jan 2024
Cited by: 1 article | PMID: 38250815 | PMCID: PMC10801633
Dystrophin- and Utrophin-Based Therapeutic Approaches for Treatment of Duchenne Muscular Dystrophy: A Comparative Review.
BioDrugs, 38(1):95-119, 02 Nov 2023
Cited by: 0 articles | PMID: 37917377 | PMCID: PMC10789850
Review Free full text in Europe PMC
Go to all (18) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Clinical Trials
- (1 citation) ClinicalTrials.gov - NCT01995032
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Correlation of Utrophin Levels with the Dystrophin Protein Complex and Muscle Fibre Regeneration in Duchenne and Becker Muscular Dystrophy Muscle Biopsies.
PLoS One, 11(3):e0150818, 14 Mar 2016
Cited by: 28 articles | PMID: 26974331 | PMCID: PMC4790853
Embryonic myosin is a regeneration marker to monitor utrophin-based therapies for DMD.
Hum Mol Genet, 28(2):307-319, 01 Jan 2019
Cited by: 23 articles | PMID: 30304405 | PMCID: PMC6322073
Therapeutic strategies for Duchenne and Becker dystrophies.
Int Rev Cytol, 240:1-30, 01 Jan 2004
Cited by: 14 articles | PMID: 15548414
Review
Alternative utrophin mRNAs contribute to phenotypic differences between dystrophin-deficient mice and Duchenne muscular dystrophy.
FEBS Lett, 592(11):1856-1869, 30 May 2018
Cited by: 11 articles | PMID: 29772070 | PMCID: PMC6032923
Funding
Funders who supported this work.

 1
,
4
1
,
4



