Abstract
Free full text

NCI Imaging Data Commons
Abstract
The National Cancer Institute (NCI) Cancer Research Data Commons (CRDC) aims to establish a national cloud-based data science infrastructure. Imaging Data Commons (IDC) is a new component of CRDC supported by the Cancer Moonshot. The goal of IDC is to enable a broad spectrum of cancer researchers, with and without imaging expertise, to easily access and explore the value of deidentified imaging data and to support integrated analyses with nonimaging data. We achieve this goal by colocating versatile imaging collections with cloud-based computing resources and data exploration, visualization, and analysis tools. The IDC pilot was released in October 2020 and is being continuously populated with radiology and histopathology collections. IDC provides access to curated imaging collections, accompanied by documentation, a user forum, and a growing number of analysis use cases that aim to demonstrate the value of a data commons framework applied to cancer imaging research.
Significance:
This study introduces NCI Imaging Data Commons, a new repository of the NCI Cancer Research Data Commons, which will support cancer imaging research on the cloud.
Introduction
Scalable on-demand access to managed configurable cloud resources offers unprecedented opportunities in supporting cancer research. The cloud-computing paradigm of colocating large multifaceted datasets with the compute resources, and bringing tools to the data instead of downloading the data for analysis, has the potential to address numerous challenges associated with big data research (e.g., storage and bandwidth constraints, and reproducibility of the analysis). The National Cancer Institute (NCI) Cancer Research Data Commons (CRDC), a component of a national cancer data ecosystem (1), is a cloud-based environment that aims to realize the promise of the cloud (2, 3). Primary components of CRDC include cloud-based domain-specific data repositories (4) and analysis-focused cloud resources (5–7). The NCI Imaging Data Commons (IDC) is a new data repository of CRDC that colocates imaging data with the compute resources and analysis tools within the CRDC cloud environment and provides researchers with access to (i) cancer image collections, (ii) infrastructure for exploration of metadata and imaging data, and (iii) interfaces to other components of CRDC enabling integrated analysis across various data types contained in CRDC (i.e., matching genomic and proteomic data).
Following the guiding principles of CRDC, IDC builds on the strengths of the established efforts to collect and share FAIR (Findable Accessible Interoperable Reusable; ref. 8) imaging data, and especially that of The Cancer Imaging Archive (TCIA; ref. 9). While TCIA has been successful in supporting researchers that utilize the traditional approach of downloading image data for analysis using local resources, IDC aims to make public TCIA collections available within, and tightly integrated with, the CRDC cloud environment, expanding the scope over time to include data from sources other than TCIA. To organize imaging data collected at multiple sites and by different modalities, IDC uses an extensible and documented standards-based approach to enable search operations and interoperability with analysis tools. IDC relies on the DICOM (Digital Imaging and Communications in Medicine) standard (10) for the definition of the data model and interfaces for accessing data, and for harmonizing the representation of data and metadata.
The role of IDC extends beyond establishing an infrastructure for cloud-based cancer imaging research. We are actively developing use cases demonstrating how this infrastructure can be utilized efficiently for research tasks that would be more difficult to achieve “on premises”. All of the code developed by the project is being shared under nonrestrictive open source licenses, and much of the code has been contributed back to established libraries and toolkits as a way to contribute further to the scientific community.
In this report we introduce IDC, describing its overall architecture and components as well as the current status and the priorities of the project.
Materials and Methods
Cloud platform
We chose to implement IDC using a combination of commercially available tools and capabilities provided by the Google Cloud Platform (GCP) and its Healthcare API, together with a range of open source components, as shown in Fig. 1. The choice of GCP was motivated by our desire to expediently deliver robust industry-grade infrastructure and ensure its integration with the existing components of CRDC. GCP implements a range of capabilities to support administration and security of the system, and provides a continuously evolving set of tools for scalable analysis of big data. Being one of the major cloud provider platforms, GCP is already used by the CRDC Cloud Resources: FireCloud (6), the Institute of Systems Biology Cancer Gateway in the Cloud (ISB-CGC; ref. 5), and Seven Bridges Genomics Cancer Genomics Cloud (SBG-CGC; ref. 7). Our prior experience building ISB-CGC (5) allowed us to leverage its components in establishing IDC. The GCP Healthcare API provides support for “DICOM stores,” which are accessible via the standard DICOMweb interface. The API includes tools for exporting DICOM metadata into BigQuery tables. BigQuery is a GCP scalable data warehouse solution based on Dremel (11), which enables high performance queries of very large tables using Structured Query Language (SQL) compliant with the SQL 2011 standard.
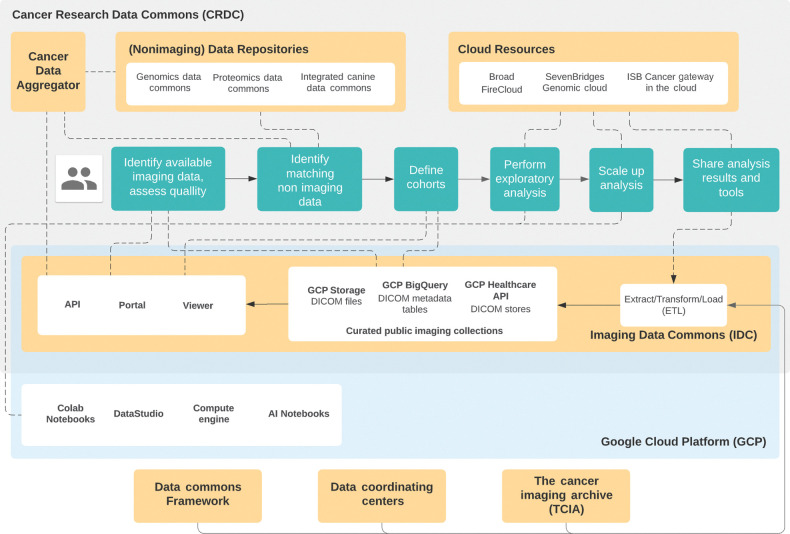
High-level diagram of relevant components of the Imaging Data Commons and related entities, and their relation to the steps of the envisioned CRDC user flow with the emphasis on imaging applications. Green boxes correspond to the envisioned user flow. IDC Extract Transform Load (ETL) process maintains the content of the data collected by external entities (e.g., TCIA) colocated with the various cloud-based tools, such as those maintained by Cloud Resources or by the Google Cloud Platform. The data can be accessed using both the interactive components (e.g., IDC Portal and Viewer) and programmatic APIs.
Portal
Similar to the already established nodes of CRDC, the IDC search portal provides an interface for exploring available data, defining cohorts of cases, and summarizing attributes of the cohort (see Supplementary Videos S1 and S2). The portal supports exploration of the metadata, imaging data, and image-derived data. The IDC portal shares the code base with the ISB-CGC (5) portal. The faceted search utilizes Apache Solr (12) populated from BigQuery content to reduce latency of certain types of queries (e.g., support of facet counting). In the current deployment of IDC, radiology images are displayed with the open source OHIF Viewer (13), which uses DICOMweb to access the IDC data. The OHIF Viewer is being actively developed, with the IDC project being one of many contributors. As IDC evolves to support new data types, alternative viewers specializing in viewing specific types of images may be integrated with the platform in the future. To address the need for display of brightfield and fluorescence microscopy images in DICOM format, IDC is working to leverage the Slim viewer (https://github.com/mghcomputationalpathology/slim). Like the OHIF Viewer, Slim viewer is a serverless single-page application that facilitates interactive visualization, in this case for digital slide microscopy images. Slim also supports image annotations in the web browser, relying on DICOMweb to query and dynamically retrieve image data from the DICOM store just as in the radiology case.
Data modeling
IDC will host a variety of cancer imaging data. While the initial focus is to support radiology data, IDC aims to provide similar capabilities for collections of brightfield microscopy, multi-channel immunofluorescence, and other imaging modalities. Equally important is the ability to support the results obtained by analysis of imaging data, such as annotations of image regions of interest or various descriptors of image findings. DICOM defines data models and standard information objects that cover a significant portion of the expected needs in communicating image analysis results (14–16). It can also be extended to support new types of data, wherever possible retaining compatibility with legacy systems (17). IDC relies on the data model defined by the DICOM standard and on the definitions of the DICOM objects to ensure their validity. DICOM is harmonized with several other healthcare standards [e.g., BRIDG (18) and HL7 (19)] and relies on standard vocabularies and ontologies (20), thus facilitating integration of IDC imaging data with other types of data within CRDC.
Security
As a government-owned system, IDC is required to obtain and maintain data security at the Federal Information Security Modernization Act (FISMA) Low level. While FISMA Low is less demanding than higher levels, this requirement has major implications on allocation of the engineering effort for the implementation and upkeep of the security, logging, and reporting procedures, and for the users interacting with the system. IDC cannot host data that contains Protected Health Information (PHI). Deidentification is performed outside of IDC and is currently done through TCIA, and in the future via additional Data Coordinating Centers. Deidentification procedures implemented at additional future sources of data would need to be independently vetted before the data contributed by those sources can be hosted by IDC. While no PHI data can be included in the collections hosted by IDC, IDC users wishing to combine nonpublic data with the public collections can do this using CRDC Cloud Resources, which have FISMA Moderate designation, or using independent cloud projects with access to IDC public resources.
Development process and governance
The IDC development is supported through a contract between Leidos Biomedical Research and The Brigham and Women's Hospital with specific deliverables. Strategic guidance is provided by the National Cancer Institute, the Frederick National Laboratory for Cancer Research, advisory boards and stakeholders. IDC embraces the main principles of Agile development methodology, including incremental development and continuous customer involvement. While IDC is not required to use only open source components, all of the code developed by IDC is being released under permissive open source licenses. Our intent is to enable reuse of the individual open source components to support replication of the relevant capabilities of IDC.
Results
The pilot of IDC was released in October 2020, and its high-level organization, relationship to the other components of CRDC, and interaction with the user flow are summarized in Fig. 1. Included in the release were 28 collections of the TCIA: radiology images related to The Cancer Genome Atlas (TCGA) project and several collections prioritized to establish the capabilities of IDC in handling image-derived data (e.g., LIDC-IDRI and NSCLC-Radiomics collections). Access to the data is available from the GCP “requester pays” storage buckets (i.e., a user-provided Google billing project is required to read the data, although loading content onto a GCP VM is free). DICOM and collection-level metadata is available from the BigQuery tables and does not require a project configured with billing. The IDC portal (available at https://imaging.datacommons.cancer.gov, also see Fig. 2) allows users to define cohorts based on a subset of metadata, provides graphical summaries of the cohort attributes, and integrates a customized OHIF Viewer that supports visualization of both the images and image annotations (specifically, visualization of DICOM Segmentation and Radiotherapy Structure Set is supported, including multiplanar reformatting). All of the software components developed by the IDC team are available under the dedicated GitHub organization (https://github.com/ImagingDataCommons). Improvements and new features for the OHIF Viewer are developed in its main repository or the repositories of underlying libraries.
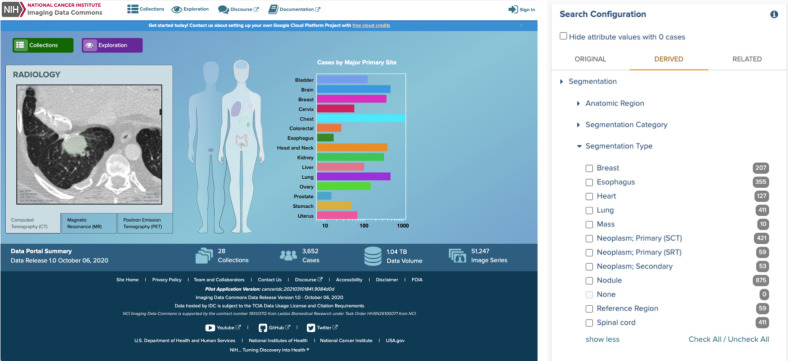
Elements of IDC Portal user interface. Left, front page of the IDC Portal for the pilot (preproduction) release of the platform, available at https://imaging.datacommons.cancer.gov. Right, example of filters available for defining cohort based on the attributes describing segmentation results available in IDC.
IDC enables the following user flow (also see Fig. 1). The portal's faceted search (21) user interface (UI) will typically serve as the entry point for the new users, allowing them to explore the data (both by viewing the images and searching the metadata) and build cohorts (see Supplementary Video S1). Alternatively, users will be able to utilize the IDC API, which we intend to be functionally equivalent to the IDC Portal, to form and interact with the cohorts. Metadata attributes that are not available via the IDC Portal can be explored using BigQuery or DataStudio (see Supplementary Video S2 and S3). Standard SQL and BigQuery APIs are available for interrogating the metadata and fine-tuning the definition of the cohort. Users can spot-check data quality by analyzing metadata and examining data in the IDC Viewer, which can be done either through the portal, or by configuring the viewer URL directly to show specific imaging studies. Data quality checks can utilize existing, continuously evolving general purpose cloud-based tools, such as Colab Notebooks [cloud-hosted GPU-enabled virtual machines (VM) with the Jupyter Notebook interface] or Google DataStudio (interactive platforms for building data dashboards; see Supplementary Video S3). At the next level, the user can initialize a cloud-based instance of a VM configured with the familiar desktop-based analysis tools to experiment with customized processing and visualizations on a subset of cases (see Supplementary Video S4). Once the analysis workflow is established, it can be applied at scale to the entire cohort utilizing either general-purpose pipelining tools (22), or the CRDC Cloud Resources (5–7). Ability to identify matching data in other repositories of CRDC is being provided by the Cancer Data Aggregator (CDA; ref. 23) APIs currently under development.
Support and engagement of IDC users is a major priority for the project. To support user training and outreach, IDC is accompanied by online documentation, examples of Colab Notebooks (including those contributed by IDC users) and DataStudio dashboards interacting with the IDC-hosted data, as well as video tutorials (see Supplementary Videos S1–S4 included with this article). Further use cases demonstrating implementation of radiomics and pathomics analysis pipelines integrated with the IDC data are currently under development. Users can participate in the IDC online forum based on the Discourse platform. Complete analysis use cases that demonstrate the capabilities of IDC to support imaging research needs are being developed, with the first such use case replicating an earlier study by Hosny and colleagues (24) already available (see Supplementary Video S4).
Prospective IDC users can apply for free GCP credits to experiment with the resource and develop confidence with the cloud-based analysis. Experienced investigators can participate in the NIH Science and Technology Research Infrastructure for Discovery, Experimentation, and Sustainability (STRIDES) Initiative, which provides cloud training resources and discounted credits to all NIH-funded investigators and is intended to support production use of CRDC.
Discussion
We described the design and implementation of IDC and summarized capabilities of the IDC pilot available to the cancer research community. Early examples already show promise for the utility of cloud-hosted public imaging collections colocated with the compute resources and a growing number of tools to support data analysis. The IDC Portal supports exploration and cohort building from cloud-based data. The IDC Viewer provides unique and growing capabilities in supporting visualization of image annotations. Combined with BigQuery, IDC offers the unprecedented ability to access and explore DICOM metadata for public imaging collections from the IDC-maintained tables. The analysis use cases that accompany IDC illustrate the ease of access to the data from cloud-based tools and the potential to enable sharing of fully reproducible analysis pipelines to accompany academic manuscripts.
IDC is under active development to further enhance both the capabilities of IDC itself and its integration with the other components of CRDC. Immediate priorities for the development of IDC are the data versioning strategy (motivated by the updates to the released collections due to addition of new data, correction of errors, or mitigation of PHI leaks) and subsequent ingestion and periodic updates towards inclusion of all the public TCIA radiology collections. Support for digital pathology is also planned for the production release currently scheduled for Fall 2021. Existing public collections of digital pathology images, which are typically shared using vendor-specific formats, will be converted into a DICOM representation to better support metadata search and visualization. The datasets hosted by IDC will not be limited to human data, nor to the modalities currently available from TCIA. We expect IDC to host preclinical (mouse) and canine imaging data, as well as various types of images that will be shared by the NCI Human Tumor Atlas Network (HTAN; ref. 25). IDC will also include relevant non-cancer imaging collections, as prioritized by the NCI stakeholders, such as the recently announced COVID-19 collections released by TCIA.
Alongside replication of the imaging collections, IDC supports inclusion of image-derived data (e.g., annotations, measurements and regions of interest) and accompanying clinical data. Harmonization of clinical data is being done in coordination with the CRDC Center for Cancer Data Harmonization (CCDH; ref. 26) and the CDA teams. Harmonization of image-derived data is a major undertaking in the IDC data intake process. Common coded and structured data representation in standard formats (DICOM SR, SEG and RTSS) using standard coded concepts for fields and value sets (SNOMED, NCIt) is critical to enable metadata search across collections, to provide a consistent interface to the data for visualization and analysis tools, and for semantic interoperability between CRDC nodes. We are actively working on these harmonization tasks both for the retrospective collections and for prospective submission of analysis results to TCIA.
IDC will be relying on global unique identifiers (GUID) to support persistent referencing of the data. The CRDC Data Commons Framework is in the process of implementing the relevant parts of the Global Alliance for Genomics in Health (GA4GH; ref. 27) Data Repository Service (DRS) API (28) to support GUIDs for the data bundles at the selected levels of the DICOM hierarchical data model.
IDC is in its early days. There are numerous questions relating to costs of conducting imaging research in the cloud and limiting the risk of runaway processes. Repositories of reusable image analysis tools that are easily accessible from cloud workflows [with the relevant existing platforms including Dockstore (29) and ModelHub.AI (30) ]need to be established. Integrative analysis of data across CRDC nodes needs to be enabled. We hope to engage the future users of IDC, as well as contributors and maintainers of emerging repositories of cancer research tools, through venues such as the IDC online forum (https://discourse.canceridc.dev/). Working together, we can answer these questions and develop new components of the CRDC ecosystem to support a broad range of cancer imaging research use cases. With the pilot release, we introduce an early example of the capabilities and the potential for applying the data commons concepts to the imaging space.
Authors' Disclosures
A. Fedorov reports other support from Leidos Biomedical Research during the conduct of the study. W.J. Longabaugh reports other support from Leidos Biomedical Research during the conduct of the study. D.A. Clunie reports grants from NIH during the conduct of the study; personal fees from Flywheel.io, Health Care Technology Services, Imago Medical Systems, Kela Health, Lunit Inc., maiData, Medigate, BioClinica, Inc., and personal fees from Koninklijke Philips NV outside the submitted work; and Editor of DICOM Standard [contracted by Medical Imaging & Technology Alliance (MITA)]. S. Pieper reports personal fees from US NIH (NCI) during the conduct of the study and grants from US NIH outside the submitted work. H.J. Aerts reports grants from NIH during the conduct of the study and personal fees from Onc.AI outside the submitted work. A. Homeyer reports grants from Leidos Biomedical Research, Inc. during the conduct of the study. R. Lewis reports personal fees from Leidos Biomedical Research during the conduct of the study. A. Akbarzadeh reports other support from Leidos Biomedical Research during the conduct of the study. W. Clifford reports other support from Leidos Biomedical Research during the conduct of the study. H. Höfener reports grants from Leidos Biomedical Research, Inc. during the conduct of the study. S. Paquette reports other support from Leidos Biomedical Research during the conduct of the study. J. Petts reports grants from NIH during the conduct of the study. D.P. Schacherer reports grants from Leidos Biomedical Research, Inc. during the conduct of the study. M. Tian reports other support from Leidos Biomedical Research during the conduct of the study. G. White reports other support from Leidos Biomedical Research during the conduct of the study. E. Ziegler reports personal fees from Radical Imaging LLC during the conduct of the study and personal fees from Radical Imaging LLC outside the submitted work. I. Shmulevich reports other support from Leidos during the conduct of the study. U. Wagner reports other support from National Cancer Institute during the conduct of the study. R. Kikinis reports other support from Leidos Biomedical Research during the conduct of the study. No disclosures were reported by the other authors.
Supplementary Material
Supplemental video 1
Supplemental video 2
Supplemental video 3
Supplemental video 4
Acknowledgments
The authors acknowledge the support of NCI Communications in refining the video materials accompanying this submission. This project has been funded in whole or in part with Federal funds from the NCI, NIH, under task order no. HHSN26110071 under contract no. HHSN261201500003l.
This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Task Order No. HHSN26110071 under Contract No. HHSN261201500003l.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclaimer
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Authors' Contributions
A. Fedorov: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. W.J.R. Longabaugh: Conceptualization, resources, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. D. Pot: Conceptualization, resources, supervision, funding acquisition, validation, investigation, methodology, writing–review and editing, information security. D.A. Clunie: Conceptualization, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. S. Pieper: Conceptualization, software, supervision, validation, investigation, visualization, methodology, writing–review and editing. H.J.W.L. Aerts: Conceptualization, resources, data curation, software, supervision, validation, investigation, visualization, methodology, writing–review and editing. A. Homeyer: Conceptualization, resources, software, formal analysis, supervision, validation, investigation, visualization, methodology, writing–review and editing. R. Lewis: Resources, software, supervision, visualization, writing–review and editing. A. Akbarzadeh: Data curation, software, investigation, visualization, writing–review and editing. D. Bontempi: Data curation, software, validation, investigation, visualization, writing–review and editing. W. Clifford: Data curation, software, validation, investigation, writing–review and editing. M.D. Herrmann: Conceptualization, resources, data curation, software, supervision, validation, investigation, visualization, methodology, writing–review and editing. H. Höfener: Conceptualization, data curation, software, supervision, validation, investigation, visualization, methodology, writing–review and editing. I. Octaviano: Software, visualization, writing–review and editing. C. Osborne: Information security. S. Paquette: Conceptualization, software, investigation, methodology, writing–review and editing. J. Petts: Software, investigation, visualization. D. Punzo: Software, investigation, visualization. M. Reyes: Software, validation, investigation. D.P. Schacherer: Data curation, software, validation, investigation, visualization, methodology. M. Tian: Software, validation. G. White: Software, validation, investigation, methodology. E. Ziegler: Conceptualization, software, supervision, validation, investigation, visualization, methodology. I. Shmulevich: Conceptualization, formal analysis, investigation, writing–review and editing. T. Pihl: Resources, supervision, project administration. U. Wagner: Resources, supervision, project administration, writing–review and editing. K. Farahani: Resources, supervision, project administration, writing–review and editing. R. Kikinis: Conceptualization, resources, supervision, funding acquisition, validation, investigation, methodology, project administration, writing–review and editing.
References
Full text links
Read article at publisher's site: https://doi.org/10.1158/0008-5472.can-21-0950
Read article for free, from open access legal sources, via Unpaywall:
https://cancerres.aacrjournals.org/content/canres/81/16/4188.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/108552860
Article citations
Ten challenges and opportunities in computational immuno-oncology.
J Immunother Cancer, 12(10):e009721, 26 Oct 2024
Cited by: 0 articles | PMID: 39461879 | PMCID: PMC11529678
Review Free full text in Europe PMC
Recommendations for the creation of benchmark datasets for reproducible artificial intelligence in radiology.
Insights Imaging, 15(1):248, 14 Oct 2024
Cited by: 0 articles | PMID: 39400639 | PMCID: PMC11473745
NCI's Proteomic Data Commons: A Cloud-Based Proteomics Repository Empowering Comprehensive Cancer Analysis through Cross-Referencing with Genomic and Imaging Data.
Cancer Res Commun, 4(9):2480-2488, 01 Sep 2024
Cited by: 0 articles | PMID: 39225545 | PMCID: PMC11413857
Summary of the National Cancer Institute 2023 Virtual Workshop on Medical Image De-identification-Part 1: Report of the MIDI Task Group - Best Practices and Recommendations, Tools for Conventional Approaches to De-identification, International Approaches to De-identification, and Industry Panel on Image De-identification.
J Imaging Inform Med, 12 Jul 2024
Cited by: 0 articles | PMID: 38997571
Summary of the National Cancer Institute 2023 Virtual Workshop on Medical Image De-identification-Part 2: Pathology Whole Slide Image De-identification, De-facing, the Role of AI in Image De-identification, and the NCI MIDI Datasets and Pipeline.
J Imaging Inform Med, 09 Jul 2024
Cited by: 0 articles | PMID: 38980626
Go to all (30) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
NCI Cancer Research Data Commons: Cloud-Based Analytic Resources.
Cancer Res, 84(9):1396-1403, 01 May 2024
Cited by: 5 articles | PMID: 38488504 | PMCID: PMC11063685
Review Free full text in Europe PMC
National Cancer Institute Imaging Data Commons: Toward Transparency, Reproducibility, and Scalability in Imaging Artificial Intelligence.
Radiographics, 43(12):e230180, 01 Dec 2023
Cited by: 7 articles | PMID: 37999984
AI-Generated Annotations Dataset for Diverse Cancer Radiology Collections in NCI Image Data Commons.
Sci Data, 11(1):1165, 23 Oct 2024
Cited by: 0 articles | PMID: 39443503 | PMCID: PMC11500357
NCI Cancer Research Data Commons: Lessons Learned and Future State.
Cancer Res, 84(9):1404-1409, 01 May 2024
Cited by: 4 articles | PMID: 38488510 | PMCID: PMC11063686
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCI NIH HHS (6)
Grant ID: HHSN261201000031C
Grant ID: HHSN261201500001C
Grant ID: HHSN261201500001W
Grant ID: HHSN261201500003I
Grant ID: HHSN261201500003C
Grant ID: HHSN261201500001G
NIBIB NIH HHS (2)
Grant ID: P41 EB015898
Grant ID: P41 EB028741
National Cancer Institute (1)
Grant ID: Task Order No. HHSN26110071 under Contract No. HHSN2612015000031

 1,*
1,* 


