Abstract
Background
Pancreatic cancer is one of the most serious and lethal human cancers with a snowballing incidence around the world. The natural product celastrol has also been widely documented as a potent anti-inflammatory, anti-angiogenic, and anti-oxidant.Purpose
To elucidate the antitumor effect of celastrol on pancreatic cancer cells and its modulatory role on whole genome expression.Methods
The antitumor activity of celastrol on a panel of pancreatic cancer cells has been evaluated by Sulforhodamine B assay. Caspase 3/7 and histone-associated DNA fragments assays were done for apoptosis measurement. Additionally, prostaglandin (PGE2) inhibition was evaluated. Moreover, a microarray gene expression profiling was carried out to detect possible key players that modulate the antitumor effects of celastrol on cells of pancreatic cancer.Results
Our findings indicated that celastrol suppresses the cellular growth of pancreatic cancer cells, induces apoptosis, and inhibits PGE2 production. Celastrol modulated many signaling genes and its cytotoxic effect was mainly mediated via over-expression of ATF3 and DDIT3, and down-expression of RRM2 and MCM4.Conclusion
The current study aims to be a starting point to generate a hypothesis on the most significant regulatory genes and for a full dissection of the celastrol possible effects on each single gene to prevent the pancreatic cancer growth.Free full text
Celastrol Modulates Multiple Signaling Pathways to Inhibit Proliferation of Pancreatic Cancer via DDIT3 and ATF3 Up-Regulation and RRM2 and MCM4 Down-Regulation
Abstract
Background
Pancreatic cancer is one of the most serious and lethal human cancers with a snowballing incidence around the world. The natural product celastrol has also been widely documented as a potent anti-inflammatory, anti-angiogenic, and anti-oxidant.
Purpose
To elucidate the antitumor effect of celastrol on pancreatic cancer cells and its modulatory role on whole genome expression.
Methods
The antitumor activity of celastrol on a panel of pancreatic cancer cells has been evaluated by Sulforhodamine B assay. Caspase 3/7 and histone-associated DNA fragments assays were done for apoptosis measurement. Additionally, prostaglandin (PGE2) inhibition was evaluated. Moreover, a microarray gene expression profiling was carried out to detect possible key players that modulate the antitumor effects of celastrol on cells of pancreatic cancer.
Results
Our findings indicated that celastrol suppresses the cellular growth of pancreatic cancer cells, induces apoptosis, and inhibits PGE2 production. Celastrol modulated many signaling genes and its cytotoxic effect was mainly mediated via over-expression of ATF3 and DDIT3, and down-expression of RRM2 and MCM4.
Conclusion
The current study aims to be a starting point to generate a hypothesis on the most significant regulatory genes and for a full dissection of the celastrol possible effects on each single gene to prevent the pancreatic cancer growth.
Graphical Abstract

Introduction
Pancreatic cancer is ranked among the top five death-related-malignancies in the United States and accounts for over 43,000 deaths annually and an estimated 53,000 new diagnosed cases.1,2 The survival rate still remains one of the lowest among other types of malignant diseases, with 5% of patients living over 5 years.1−5 Early diagnosis of the tumor is considered the best tool to combat the disease. However, in practice, this is considered as an unrealistic option for several reasons. Surgical intervention, on the other hand, is the best treatment option so far for pancreatic cancer patients, despite its low survival rates (20%). Moreover, it has been estimated that nearly 80% of the patients do not undergo surgical resection due to the difficulties in early diagnosis. Alternatively, chemotherapy or chemoradiotherapy is still an available therapeutic option, in particular for patients who suffer from local unresectable or advanced metastatic tumors.6
The use of natural products and their derivatives has always rising attention to mitigate the seriousness of various diseases including cancers; that is gripping the attention of search groups worldwide.7–13 Recently, the immense bioactivities of natural products have been pinpointed; that they can modulate the signaling pathways in tumor cells via activation or suppression of multiple cellular pathways.3,8,14–18 Celastrol, a quinone methide triterpenoid, is a natural product extracted from Tripterygium wilfordii Hook F. (“Thunder of God Vine”) plant and is used widely in TCM (traditional Chinese medications). The biological activities of celastrol have been extensively explored; it has been documented that celastrol has anticancer activity against various tumors, including leukemia, melanoma glioma, and prostate cancer.19–28 The celastrol anticancer activity was owed to its capability to compromise different cellular mechanisms in cancer cells. Celastrol is found to suppress tumor necrotic factor (TNF) induced cancer cells metastasis, nuclear factor (NF)-kB signaling, heat shock protein (HSP) 90, extracellular signal-regulated kinase (ERK), proteasome, and modulates TRAIL-induced apoptosis.29–35
To our knowledge, the antitumor activity of celastrol on pancreatic cancer in different proliferation stages and its influence on the expression pattern of the whole genome is not yet identified. In the current work, we aimed to evaluate the inhibitory effects of celastrol on the growth of human pancreatic malignant cells at different stages. In this context, we investigated the antitumor activity of celastrol on numerous cell lines representing drug resistant patterns and diverse differentiation stages of pancreatic cancer in comparison to normal immortalized pancreatic cells.
Materials and Methods
Pancreatic Cell Lines
Eight pancreatic human cancer cell lines were obtained from the American Type Culture Collection (Rockville, USA) and were used in this study: Suit-007, Suit-2, BxPC-3, Capan-2, Capan-1, Miapaca-2, Colo-357, and Panc-1 as well as the normal immortalized pancreatic cell line HPDE. Cell lines were grown and treated with celastrol or control dimethyl sulfoxide (DMSO) as previously indicated.5,8,13,16,18,36–39
Effect of Celastrol on Cellular Proliferation
To evaluate the anti-proliferative effects of celastrol on cells of pancreatic cancer, the Sulforhodamine B (SRB) assay was accomplished to evaluate the cellular protein content.4,5,9,10,13,17 Briefly, cell lines were incubated and gradually treated for 48 hours with DMSO or increasing doses of celastrol. Then, cells were fixed with trichloroacetic acid (TCA) (10%) and stained with SRB fluorescent dye for 30 minutes. The excess dye was repeatedly washed with 1% acetic acid and the SRB bounded to cellular proteins was then dissolved in 10 mM Tris base. The absorbance was measured at 510 nm in a reader (Molecular Devices, Sunnyvale, CA, USA).
Evaluation of the Apoptotic Effects of Celastrol on Pancreatic Cancer Cell Lines
The apoptotic ability of celastrol was determined by measuring degradation of DNA and histone-associated DNA fragments. The apoptotic effects of celastrol on BxPC-3, Panc-1, and Miapaca-2 cancer cells were evaluated using the ELISA kit for detection of cell death (Roche Diagnostics GmbH, Germany). Briefly, cells were either dosed with or without celastrol (½ IC50, IC50, or 2 IC50) for 6 hours. The cytoplasm of untreated and treated cells was then transferred to coated plates with Biotin-binding protein streptavidin. The plates were incubated with biotinylated histone antibody and peroxidase-tagged mouse anti-human DNA, and the absorbance was measured at 405 nm (Molecular Devices, Sunnyvale, CA, USA).
Evaluation of Caspase-3/7 Activity
Caspases are protease enzymes that play crucial roles in apoptosis. In this context, the apoptotic effects of celastrol on pancreatic cell line Panc-1 were further confirmed through measuring caspase 3/7 quantities using assay kit (Caspase-Glo 3/7) (Promega, Fitchburg, MA, USA) as previously performed.8,9,18,37,40–42 Briefly, cells were dosed with or without celastrol (½ IC50, IC50, or 2 IC50) for 6 hours. Reagent Caspase-Glo 3/7 was prepared, added in equal volumes to cells, mixed gently, and incubated for 1 hour at 25°C. The luminescence was detected, and the activity of caspase was presented as a percentage of the untreated control.
Inhibition Assay of Prostaglandin
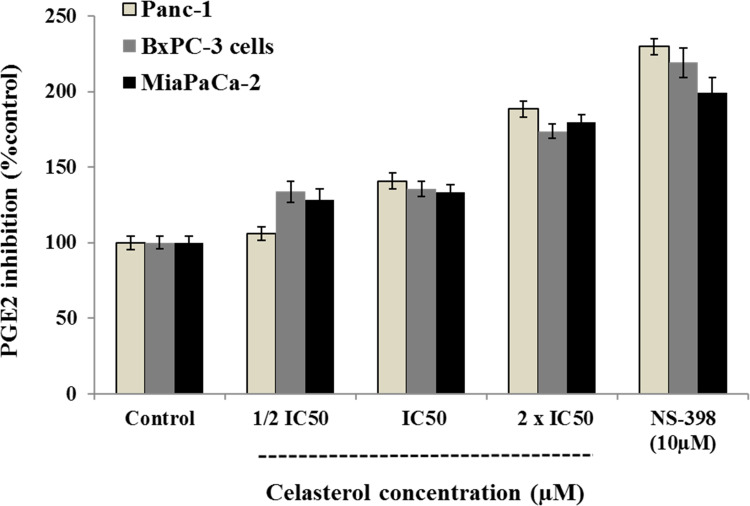
To evaluate the celastrol’s effect on the production of prostaglandin E2 (PGE2), the prostaglandin E2 assay Kit (Cayman Chemicals, MI, USA) was employed as indicated formerly.3,8,38,43 The assay is designed to detect and quantify the level of human PGE2 in biological samples. It is based on the competition between PGE2 and a PGE2 alkaline phosphatase tracer for a restricted amount of specific PGE2 monoclonal antibody. Furthermore, NS-398, non-steroidal anti-inflammatory, was employed as a positive control. Briefly, after treatment, cells were dosed with or without celastrol (½ IC50, IC50, or 2 IC50) for 24 hours and stimulated with arachidonic acid. The PGE2 production was assessed by a competitive enzyme immunoassay in accordance with the kit manufacturer’s procedure. The inhibitory effects of celastrol and positive control were presented as a percentage of the untreated control.
Microarray
In order to perform a whole genome expression analysis, the high COX-2 expressing BxPC-3 cells were either dosed with 10 μM celastrol or DMSO for 2 days. RNA was extracted, purified, and processed as we described previously.37–39,41 The Illumine human sentrix-8 v3. The Human sentrix Ref-8 Expression BeadChip contains eight arrays of more than 24,000 probes derived from the RefSeq database. These new Expression BeadChips provide high-quality performance with low rates of false differential expression. Here, the differential expression values are defined as genes showing a more than or equal to 1.5-fold difference in raw hybridization intensity (up or down) between two samples.
Validation of Microarray by RT-PCR
The significance of array results has been checked and validated by RT-PCR analysis for selected significantly regulated genes throughout the array study. The RT-PCR was performed with the same samples used for microarray according to QuantiTect SYBR Green Kit (Qiagen, Germany).37,41,44,45
Determination of the Tangled Effects of Celastrol on Signaling Pathways in Cells of Pancreatic Cancer
The Ingenuity Pathway Analysis (IPA) (www.ingenuity.com) (Mountain View, USA) was employed in our study to identify the expected molecular biomarkers list, networks and signaling pathways that have been modified after treatment of celastrol, as previously shown.3,5,8,16,36
Data Analysis and Statistical Analysis
Microarray data analysis, cluster analysis, normalization, and quality assessment were carried out with the MIAME compatible analysis where differences with a P-value <0.05 were only considered. Moreover, correspondence analysis was employed for cluster analysis.5,37,39,41 The assays were conducted in triplicate and the results are presented as the means±standard errors and P-values <0.05 were assumed to be statistically significant using two-way ANOVA test (Graphpad Prism 8 software).
Results
Celastrol Suppresses Cellular Proliferation of Cells of Pancreatic Cancer
The celastrol effects on cellular proliferation of eight different pancreatic cancer cell lines (Suit-2, Suit-007, BxPC-3, Capan-2, Colo-357, Miapaca-2, Panc-1, and Capan-1) and the normal immortalized pancreatic cells (HPDE) have been evaluated. SRB colorimetric assay has been used to evaluate the cellular proliferation with steady rising in celastrol dosing for 48 hours. Results showed that Miapaca-2 and BxPc-3 were the most affected cells (IC50=7.31±0.33 and 7.79±0.81, respectively), while Capan-2 and Panc-1 cells were the least affected cancer ones (IC50=11.46±0.83 and 16.15±0.97, respectively). Meanwhile, the normal HPDE cells were the least affected after treatment (IC50>20 µM) (Table 1).
Table 1
Celastrol Inhibits Proliferation of Pancreatic Cancer Cell Lines. Eight Different Pancreatic Cancer Cell Lines Were Treated with Increasing Concentrations of Celastrol and IC50 Values Have Been Accordingly Calculated
| Cell Line | IC50 (µM) |
|---|---|
| Miapaca-2 cells | 7.31±0.33 |
| Capan-1 cells | 8.24±0.54 |
| Capan-2 cells | 11.46±0.83 |
| Suit-007 cells | 8.64±0.71 |
| BxPC-3 cells | 7.79±0.81 |
| Colo-357 cells | 8.76±0.56 |
| Panc-1 cells | 16.15±0.97 |
| Suit-2 cells | 10.38±0.92 |
| HPDE cells | ˃20 |
Celastrol Induces Apoptotic Cell Death
Induction of apoptotic cascade is a main chemotherapy-induced cell death mechanism.46 The apoptotic effects of celastrol on pancreatic resistant cell lines Miapaca-2, BxPC-3, and Panc-1 were evaluated by measurement of histone linked DNA fragments in cytosol and caspase 3/7 levels. Results revealed that celastrol provoked apoptosis via increasing the number of fragmented DNA released in cytoplasm (Figure 1) and, additionally, by increasing the amount of activated caspases 3/7 (Figure 2) in Panc-1, BxPC-3, and MiaPaCa-2 cell lines compared to untreated controls. The apoptotic effect was dose-dependent in all tested cell lines.
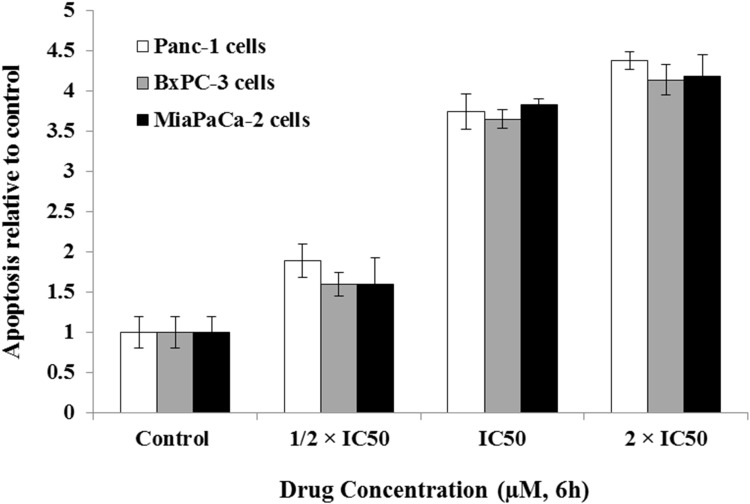
Celastrol induced apoptosis of cancer cell. Cell lines were treated with different concentrations of celastrol and histone-associated DNA fragments were quantified by SRB assay. The assay was repeated in triplicate and the results are shown as means±SEM.
Celastrol Prevents PGE2 Production
To appraise celastrol anti-inflammatory effect via PGE2 inhibition, we hired the high COX expression cell line (BxPC-3). Our findings indicated that celastrol considerably decreased PGE2 production openly with dose increasing confirming the anti-inflammatory effect of celastrol on pancreatic cancer cells and the possible beneficial effect for cancer treatment (Figure 3).
Target Genes and Signaling Pathways After BxPc-3 Cells Treatment with Celastrol
Microarray expression testing has been conceded as formerly designated37,39,41 using Illumine human sentrix-8 v3 (Chip 5388892088). For significance a threshold of typically 0.05 is chosen: Reject all as not significant, where p≥0.05. In our study, BxPC-3 cells were dosed with DMSO as control or 10 μM celastrol for 48 hours. Gene regulation outcomes indicated that celastrol drastically regulated 300 genes in BxPc-3 cells, with a minimum of 2-folds fluctuation in their expression. Two hundred and eight and 92 genes were significantly up- or down-regulated, respectively. The top 80 regulated genes are shown (Figure 4). The color-coded scale shows fold changes for upregulated and down-regulated genes. Regulated genes were primarily implicated in cyclin and cell cycle regulation. Importantly, array results agreed with RT-PCR results (Table 2) where both the DNA-damage-inducible transcript 4 (DDIT4), the Growth Arrest and DNA Damage Inducible Alpha (GADD45B) and Fos Proto-Oncogene (Fos) were commonly upregulated and the Ribonucleotide Reductase Regulatory Subunit M2 (RRM2) was commonly downregulated in both array and RT-PCR results after treatment.
Table 2
RT-PCR Verification of Microarrays Results for Some Selected Genes
| Official Gene Name | Gene Symbol | Fold Change (Microarray) | Fold Change (qRT-PCR) |
|---|---|---|---|
| Ribonucleotide Reductase Regulatory Subunit M2 | RRM2 | −2.27 | −3.81 |
| DNA-damage-inducible transcript 4 | DDIT4 | +14.06 | +8.22 |
| Growth Arrest and DNA Damage Inducible Alpha | GADD45B | +3.26 | +2.17 |
| Fos Proto-Oncogene | Fos | +21.9 | +11.52 |
Analysis of Canonical and Signaling Pathways
To recognize the canonical pathways and networks facilitating the celastrol inhibitory effects on the BxPC-3 cancer cell line, the IPA was employed.37 The IPA Graphical Summary gives an outline of the key biological themes in our IPA core analysis and explains how these thoughts communicate with each other, creating a coherent and comprehensible synopsis of the analysis. The topmost five regulated canonical pathways post-celastrol treatment were cyclins and cell cycle regulation (Figure 5), senescence pathway, cell cycle control and chromosomal regulation, cell cycle: G1/S checkpoint regulation (Figure 6), apoptosis of embryonic cell lines, estrogen-mediated S-phase entry, and prostatic cancer signaling. Figure 6 shows the number of upregulated genes (red) involved in cell cycle: G1/S checkpoint regulation like P27K1P1, and P18INK4C and additionally the number of downregulated genes (green) involved in cell cycle: G1/S checkpoint regulation like Cyclin B, Cyclin A, and CDK1.

Graphical summary for pathways regulated after celastrol treatment. The topmost five regulated canonical pathways post-celastrol treatment were cyclins and cell cycle regulation, senescence pathway, cell cycle control and chromosomal regulation, cell cycle: G1/S checkpoint regulation, apoptosis of embryonic cell lines, estrogen-mediated S-phase entry and prostatic cancer signaling. An asterisk (*) indicates that a given gene is represented in the microarray set with multiple probes.

Cyclin and cell cycle regulation pathway after treatment of pancreatic cancer cells with celastrol. Example of upregulated genes (red) involved in cell cycle: G1/S checkpoint regulation were P27K1P1, and P18INK4C while among downregulated genes (green) involved in cell cycle: G1/S checkpoint regulation were Cyclin B, Cyclin A, and CDK1. An asterisk (*) indicates that a given gene is represented in the microarray set with multiple probes.
Interestingly, IPA has recognized a massive number of regulated genes and were mainly involved in cell movement of cancer cell lines like CTNNB1, EGFR, ESR2, HIF1A, RELA, and WNT3A while some other genes were involved in cell death of various breast cancer cell lines like ECSIT, EWSR1, GPER1, KDM3A, MRTFB, and NCOA3. Moreover, DNA recombination, replication, and repair, cell cycle, survival, and death, and cellular development were among the highest regulated molecular and cellular functions after celastrol treatment.
Discussion
Natural products are an invaluable source of novel drug discovery because of their safety, potency, and lower cost. Due to deleterious side-effects of chemically synthesized therapeutics, especially anticancer; natural products are plausible alternatives and it is urgently needed to find safer natural remedies.35,47–49 Celastrol is a natural product obtained from Tripterygium wilfordii Hook F and was stated for its anticancer activities. Significantly, celastrol showed anticancer activity against various tumors as breast, prostate, and liver cancers, multiple myeloma, and glioma.21–28,35 Moreover, celastrol was used in treatments of chronic inflammation, autoimmune diseases, and neurodegenerative diseases.50 Despite there being some proposed mechanisms of celastrol’s action in suppression of cancer,35,50 its antitumor activity, especially in pancreatic cancer, has not been explored yet.
In the current analysis, the growth inhibitory effects of celastrol have been assessed on diverse pancreatic cancer cell lines with diverse proliferation stages and different resistance pattern. It was obvious that celastrol inhibited cellular growth of all tested cell lines compared to normal cells and the effect was cell line dependent where results showed that Miapaca-2 and BxPC-3 were the most affected cells while Capan-2 and Panc-1 cells were the least affected cancer ones. Additionally, celastrol enhanced apoptosis in cancerous cells via activation of caspases 3/7 and an increasing amount of histone-linked DNA fragments released in cytoplasm. Moreover, the effect of celastrol on PGE2 (prostaglandin production) was also monitored where celastrol significantly inhibited PGE2 production in the BxPC-3 cell line (the highly COX-2 expressing cells).4,51 Furthermore, a comprehensive genome expression analysis was accomplished to identify the responsible genes for the growth inhibitory effects of celastrol, and its molecular targets in addition to the expected pathways that mediate its cellular effects in pancreatic cancer.
In the next few lines we will explain some genes up- and down-regulated significantly in array results and how they helped us to explore the possible anticancer effect of celastrol on pancreatic cancer. These modulated genes after treatment have been previously reported for their correlations with biologically important pathways, cancer development, apoptosis, cell migration, metastasis, angiogenesis, and cell cycle and cyclin dependent pathways.
DNA-damage-inducible transcript 3 (DDIT3/GADD153) is an endoplasmic reticulum stress (ERS) related protein which was previously reported to have a noteworthy role in cancer.52 Moreover, DDIT3 is known to play a crucial role in arresting apoptotic growth after DNA damage and after numerous stress circumstances, such as deprivation of nutrients and treatment with chemotherapeutic agents.5,53 Our array results showed that DDIT3 encoding gene was among significantly up-regulated genes after treating of BxPC-3 cells with celastrol, and that could explain in part celastrol growth inhibitory effects.
Activating transcription factor 3 (ATF3) shows a significant role in modulation of stress and inflammatory responses. ATF3 is an ATF/cyclic AMP response element-binding (ATF/CREB) transcription factors family member. Moreover, the role of ATF3 has been shown to be critical for cancer progression by promoting or suppressing apoptosis and cell proliferation.54,55 However, the fluctuating role of ATF3 on tumor progression, ATF3 notably exerts anticancer activity by regulation of CYR61.56 Recently, ATF3 was reported in hepatocellular carcinoma as a novel cancer suppressor gene.57 Our findings showed that ATF3 was one of the significantly up-regulated genes after treating cancer cells with celastrol, which could participate in the celastrol’s inhibitory effect on the growth of pancreatic cancer cells.
The rate-determining step in the synthesis of DNA is mediated by Ribonucleotide reductase (RRM). It was shown that RRM2 up-regulation is associated with gemcitabine anticancer resistance in pancreatic cancer patients. Furthermore, it was demonstrated that the efficacy of gemcitabine was enhanced by siRNA-based therapy of RRM2.5,58 Here, we report that RRM2 is down-regulated significantly in BxPC-3 cells after celastrol treatment. As far as we know, it is the first study that recognizes RRM2 as a candidate gene that can modulate the inhibitory effects of celastrol on pancreatic cancer cells growth.
Minichromosome maintenance (MCMs) family proteins play key roles in cellular replication and progression cycle. It was reported that the mRNA over-expression of MCM2, MCM4, and MCM10 genes were markedly enhanced lung adenocarcinoma, and over-expression of MCM2–5, MCM8, and MCM10 mRNA was linked to poor survival.59−62 Our results indicated that MCM4 was significantly down-regulated after cancer cells treatment with celastrol. Accordingly, MCM4 down-regulation could be a proposed molecular mechanism that describes celastrol’s inhibitory effect on pancreatic cancer cells.
In this study, we aimed to explore the exact celastrol’s mechanisms of action on pancreatic cancer cells based on the most pointedly up- or down-regulated key player genes and for a comprehensive functional analysis of each single gene in pancreatic cancer. In this context and after confirmation of the anti-inflammatory, apoptotic, and anti-proliferative effects of celastrol on cells of pancreatic cancer; cDNA microarray was chosen as the best choice technique to screen its effect on thousands of genes alongside. The results of microarray pointed out the celastrol effects are mediated via multiple signaling pathways, and many genes were significantly involved in its inhibitory effect on cancer cells.
Finally, the outcome from the presented microarray results is considered a starting point to identify the exact molecular mechanisms of celastrol anticancer activities. This study gives precise insights about the prime targets of celastrol which may inspire us and others to further investigate celastrol and similar compounds for their pharmacological actions.
Conclusion
The current analysis obviously demonstrated that celastrol is a promising antitumor agent. Celastrol inhibited pancreatic cancer proliferation through affecting several signaling pathways. The celastrol’s inhibitory effects on pancreatic cancer was basically attributed to DDIT3 and ATF3 activation and suppression of RRM2 and MCM4 genes. The microarray results and the expression analysis information that have been described here could be a starting point to recognize target genes and for a more thorough investigation of the functional role of each gene that describes the celastrol activity in prevention of cancer.
Acknowledgment
We are very thankful of Dr. Jörg Hoheisel, German Cancer Research Center (DKFZ), Heidelberg, Germany, for his continous support, and help. Additionally, the authors appreciate the efforts of Dr. Oliver Heil, Genomics and Proteomics Core Facility, DKFZ, Heidelberg, Germany, for his help in microarray data preparation and analysis.
Disclosure
The authors declare no conflicts of interest in this work.
References
Articles from OncoTargets and Therapy are provided here courtesy of Dove Press
Full text links
Read article at publisher's site: https://doi.org/10.2147/ott.s313933
Read article for free, from open access legal sources, via Unpaywall:
https://www.dovepress.com/getfile.php?fileID=70884
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/108441157
Article citations
Unraveling the Impact of Six Pentacyclic Triterpenes Regulating Metabolic Pathways on Lung Carcinoma Cells.
Pharmaceuticals (Basel), 17(6):694, 28 May 2024
Cited by: 1 article | PMID: 38931361
Multi-omics pan-cancer analyses identify MCM4 as a promising prognostic and diagnostic biomarker.
Sci Rep, 14(1):6517, 18 Mar 2024
Cited by: 0 articles | PMID: 38499612 | PMCID: PMC10948783
Elucidating the role of phytocompounds from Brassica oleracea var. italic (Broccoli) on hyperthyroidism: an in-silico approach.
In Silico Pharmacol, 12(1):6, 04 Jan 2024
Cited by: 0 articles | PMID: 38187876
Immune modulation and prognostic significance of MCM10 in pan-cancer: a comprehensive analysis.
Am J Transl Res, 15(11):6451-6463, 15 Nov 2023
Cited by: 1 article | PMID: 38074804 | PMCID: PMC10703649
Design, synthesis and antitumor evaluation of novel pyrazolo[3,4-d]pyrimidines incorporating different amino acid conjugates as potential DHFR inhibitors.
J Enzyme Inhib Med Chem, 38(1):203-215, 01 Dec 2023
Cited by: 5 articles | PMID: 36382444 | PMCID: PMC9673804
Go to all (29) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Celastrol suppresses human pancreatic cancer via m6A-YTHDF3-mediated downregulation of Claspin and Bcl-2.
Discov Oncol, 14(1):233, 18 Dec 2023
Cited by: 0 articles | PMID: 38110764 | PMCID: PMC10728403
The growth inhibitory effect of gambogic acid on pancreatic cancer cells.
Naunyn Schmiedebergs Arch Pharmacol, 391(5):551-560, 15 Mar 2018
Cited by: 11 articles | PMID: 29546614
Celastrol inhibits tumor cell proliferation and promotes apoptosis through the activation of c-Jun N-terminal kinase and suppression of PI3 K/Akt signaling pathways.
Apoptosis, 16(10):1028-1041, 01 Oct 2011
Cited by: 96 articles | PMID: 21786165
Celastrol induces apoptosis in non-small-cell lung cancer A549 cells through activation of mitochondria- and Fas/FasL-mediated pathways.
Toxicol In Vitro, 25(5):1027-1032, 03 Apr 2011
Cited by: 51 articles | PMID: 21466843