Abstract
Free full text

Cytomegalovirus infection in malignant pleural mesothelioma
Abstract
Human cytomegalovirus (HCMV) is a highly prevalent herpes virus which persists as a latent infection and has been detected in several different tumor types. HCMV disease is rare but may occur in high-risk settings, often manifesting as a pulmonary infection. To date HCMV has not been investigated in malignant pleural mesothelioma (MPM). In a consecutive case series of 144 MPM patients we evaluated two biomarkers of HCMV: IgG serostatus (defined as positive and negative) and DNAemia (>100 copies/mL of cell free HCMV DNA in serum). Approximately half of the MPM patient population was HCMV IgG seropositive (51%). HCMV DNAemia was highly prevalent (79%) in MPM and independent of IgG serostatus. DNAemia levels consistent with high level current infection (>1000 copies/mL serum) were present in 41% of patients. Neither IgG serostatus nor DNAemia were associated with patient survival. In tissues, we observed that HCMV DNA was present in 48% of tumors (n = 40) and only 29% of normal pleural tissue obtained from individuals without malignancy (n = 21). Our results suggest nearly half of MPM patients have a high level current HCMV infection at the time of treatment and that pleural tissue may be a reservoir for latent HCMV infection. These findings warrant further investigation to determine the full spectrum of pulmonary infections in MPM patients, and whether treatment for high level current HCMV infection may improve patient outcomes.
Introduction
Malignant Pleural Mesothelioma (MPM) is a rare cancer that develops in the protective lining around the lungs and is tightly linked to asbestos exposure [1]. Once diagnosed, the prognosis is very poor, with a 5-year survival rate of 9% [1, 2]. Factors that contribute to worse patient survival include advanced age, male gender, history of asbestos exposure, and non-epithelial histology subtype [3, 4]. Many patients with malignant mesothelioma ultimately die from pneumonia, respiratory failure or heart complications that are worsened by having mesothelioma [2, 5].
It has been suggested that the SV40 virus may contribute to MPM etiology, although the weight of evidence indicates it is not causal but may act as a co-factor [6]. This raises the question whether additional viruses might contribute to MPM. Human cytomegalovirus (HCMV) is a common pathogen with onco-modulatory properties [7–9]. HCMV is a member of the herpesvirus family and is a highly prevalent infection with approximately 50% of the population testing seropositive by age 50 [10]. HCMV infection is typically asymptomatic. However, in situations of immune suppression [11–13], including organ or stem cell transplant [14–16] and HIV [17, 18], infection can advance to HCMV disease, often manifesting as HCMV pneumonia [19]. Active HCMV infection has been described in approximately 30% of critically ill patients without a history of HIV or transplant [20]. In addition, HCMV DNAemia, indicative of an active viral infection, has been reported in patients with solid tumor malignancy [21–24].
The role of HCMV in cancer has primarily focused on the presence of virus in tumors [21, 22, 25–27]. Less well described is the epidemiology of active HCMV infection in solid tumor cancer patients. Whether HCMV, either at the tumor site or as an active DNAemia infection, is present in MPM and contributing to patient outcomes is an as-yet unexplored area of research. In this study we evaluated the prevalence of HCMV IgG seropositivity and HCMV serum DNAemia in MPM patients, as well as the prevalence of HCMV in tumor and normal pleural tissue. In addition, we evaluated whether these measures of HCMV were associated with patient survival.
Materials and methods
Study population
The study population has been previously described [28]. Briefly, study staff were embedded in the International Mesothelioma Program at the Brigham and Women’s Hospital between May 2000 and May 2005. Staff approached patients at their initial clinic visit to participate in the research study. Eligibility requirements included age greater than 18 years, diagnosis of MPM based on pathology and ability to complete a questionnaire. At the time of consent into the study, trained interviewers worked with participants to complete a detailed questionnaire covering lifestyle habits, occupational history with an emphasis on possible encounters with asbestiform material, personal medical history, and other potential carcinogen exposures (questionnaire available upon request). Consent was obtained to abstract medical records, and a blood sample was obtained for use in research. The current study included 144 participants from the original epidemiologic study with sufficient available sample for CMV testing. This retrospective analysis using a deidentified dataset and specimens was reviewed by the University of Minnesota IRB and considered an exempt study (IRB: 0809E47928).
Serum biomarkers of HCMV in MPM patients
HCMV IgG serology was performed on the COBAS e411 Analyzer (Roche Diagnostics, Indianapolis, IN) utilizing 150μL of serum, and an internal positive and negative CMV serological control. All IgG serology was performed at the Advanced Research and Diagnostic Laboratory (ARDL, Medical School, University of Minnesota, Twin-Cities). IgG status was categorized as negative or positive if the IgG value was over or below the threshold for reactivity (3 COI). To assess HCMV DNAemia, DNA was extracted from 200ul of serum using the Qiagen QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). Serum samples were previously kept at -80°C until DNA extraction. The standard DNA extraction protocol was amended to include a third column wash prior to the final elution of DNA into AE Buffer (Qiagen). 2μL of DNA were used in a dPCR reaction designed to quantify copies of the gBa). Primer sequences were 5’-TACCCCTATCGCGTGTGTTC-3’ and 3’-ATAGGAGGCGCCACGTATTC-5’, and FAM/TAM probe (5’-TTGCTGCCCAGCAGATAAGTGGTG). These primers amplify a 254 bp product (82,475 to 82,728 bp). DNA was partitioned into a Fluidigm Biomark 37k IFC (Fluidigm Co. San Francisco, CA), which segregates 48 samples into 36,960 individual PCR reactions. PCR amplification, signal capture and quantification then occurred on the Biomark HD instrument and Digital PCR Analysis tool. Positive control DNA (Reference Material 2366a from the National Institute of Standards and Technology) and negative template controls were included on each IFC. All samples were run on duplicate arrays on different days, and discordant results run a third time. HCMV DNA concentrations were reported in copies/mL of serum. Prevalent HCMV was defined as DNA levels above 100 copies/mL which correlates with the assay’s validated Limit of Detection (LoD). Patients with HCMV levels above 1000 copies/mL were categorized as having a high level current HCMV infection. Patients with levels below 1000 copies/mL were categorized as having a low level current HCMV infection.
HCMV DNA in pleural tissue
DNA derived from frozen resected tumor was available for 40 MPM patients [29]. Normal pleural tissue samples, also frozen following resection, were obtained from the Brigham and Women’s Hospital tissue bank from patients without mesothelioma, with no known asbestos exposure and no evidence of pleural disease (n = 21). These samples were from patients in the lung transplant program (both donors and recipients) or patients undergoing lung surgery for other disease processes that did not involve the pleura. In all control pleural samples, there was no gross or microscopic evidence of pleural pathology. DNA was extracted using the QIAamp DNA mini kit (Qiagen, Valencia, CA). Digital PCR for detection of HCMV DNA was as described above. The DNA extractions were not done with a standardized mass input, therefore results for tissue are reported as HCMV positive or HCMV negative.
Lung cancer biobank samples
As a comparison group we evaluated serum from participants in the Masonic Cancer Center lung cancer biorepository; all with metastatic lung cancer. Beginning in March 2017 study staff monitored clinic schedules for patients that met eligibility criteria and approached them for participation in the biobank initiative. Eligibility requirements included adult patients with newly diagnosed, untreated metastatic lung cancer. A smoking history questionnaire was administered. Medical history, medication use, occupational and environmental exposure history, and demographic information were collected. All patients provided written, informed consent. A serum sample obtained at the time of participant consent (baseline visit) was tested for the presence of HCMV DNAemia (n = 22); all available samples in the biobank were tested. No a priori sample size calculations were performed, and all available samples were tested. The average age of the participants was 64 years (SD 9.8), 50% were women.
Statistical analysis
Histology for MPM was assessed through pathology reports and classified as epithelioid or non-epithelioid (sarcomatoid or biphasic). The distribution of age (years), gender, history of asbestos exposure, and histology were compared across HCMV groups using t-tests and chi-square tests. Patient survival information was ascertained using the National Death Index and last known clinic visit. The relationship between HCMV and patient survival was evaluated with the log-rank test comparing Kaplan-Meier survival probability plot strata. In addition, Cox proportional hazards models were utilized to adjust for potential confounding variables, and likelihood ratio tests were used to examine statistical significance. All statistical analyses were performed in R 3.6.3 and data was visualized using ggplot2 and ggfortify.
Results
The clinical characteristics of MPM patients included in this study are summarized in Table 1. Among the 144 patients, 113 were HCMV DNAemia positive (79%), and 59 (41%) had DNAemia levels indicative of an high level current HCMV infection (>1000 copies/mL serum). HCMV IgG seroprevalence was 52%, and serostatus was not associated with the presence nor level of HCMV DNAemia. In addition, age, sex, histology, and asbestos exposure were similar across HCMV DNAemia groups. The prevalence of HCMV DNAemia was similar in a comparison group of 22 metastatic lung cancer patients (80%), however, none of the metastatic lung cancer patients had HCMV viral loads greater than 1000 copies/mL.
Table 1
| HCMV DNAemia (copies/mL) | ||||
|---|---|---|---|---|
| Characteristic | <100 | 100–1000 | >1000 | p-value |
| Below LOQ | Low level infection | High level infection | ||
| n (%) | 31 (21%) | 53 (37%) | 60 (42%) | |
| Age | 0.67 | |||
| mean (SD) | 62.7 (±11.7) | 62.3 (±11.4) | 62.5 (±10.2) | |
| Sex | 0.56 | |||
| Female | 7 (5%) | 9 (6%) | 9 (5%) | |
| Male | 22 (15%) | 44 (31%) | 51 (35%) | |
| Asbestos Exposure | 0.22 | |||
| No | 6 (4%) | 9 (6%) | 17 (12%) | |
| Yes | 25 (17%) | 44 (31%) | 43 (30%) | |
| Histology | 0.92 | |||
| Epithelioid | 22 (15%) | 36 (25%) | 39 (27%) | |
| Biphasic | 6 (5%) | 13 (9%) | 18 (13%) | |
| Sarcomatoid | 3 (2%) | 4 (3%) | 3 (2%) | |
| IgG | 0.84 | |||
| Negative | 16 (11%) | 24 (17%) | 30 (20%) | |
| Positive | 15 (10%) | 29 (20%) | 31 (22%) | |
Next, we evaluated the presence of HCMV DNA in both mesothelioma tumor and normal pleura tissues. DNA from tumor specimens collected at surgical resection was available for a subset of patients; 19/40 (48%) were HCMV DNA positive (Table 2). There was no association between tumor HCMV status and serum HCMV DNAemia, nor between tumor status and IgG seropositivity (Table 2). To evaluate whether HCMV infection was specific to malignant tissue, we assessed HCMV DNA in 21 normal pleura specimens from individuals without cancer; 6/21 (29%) were HCMV DNA positive (p = 0.47). Presence of CMV in the tumor was not associated with patient survival (S1 Fig).
Table 2
| Tumor HCMV DNA status | |||
|---|---|---|---|
| Negative | Positive | p-value | |
| (n = 21) | (n = 19) | ||
| Serum DNAemia | 0.47 | ||
| < LOQ | 6 (60%) | 4 (40%) | |
| Low level infection | 10 (56%) | 8 (44%) | |
| High level infection | 5 (42%) | 7 (58%) | |
| IgG | 0.13 | ||
| Negative | 16 (48%) | 17 (52%) | |
| Positive | 5 (71%) | 2 (29%) | |
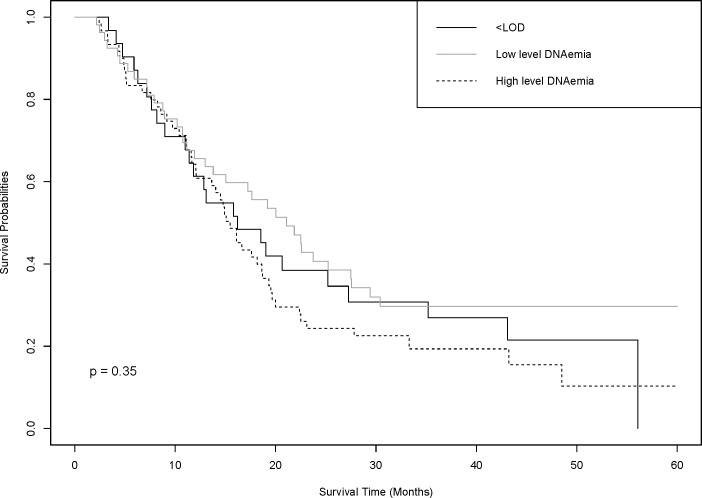
Finally, we evaluated HCMV and patient survival. The median patient survival was 17.2 months. Median survival time was similar for those with no DNAemia (16.2 months) and those with high DNAemia >1000 copies/mL (15.6 months), while those with low level DNAemia had the longest survival time (21.1 months) (Fig 1). After adjusting for age, gender and histology there was no evidence for differences in survival for either low-level DNAemia (HR 1.01, 95% CI 0.59–1.71) nor high-level DNAemia (HR 1.26, 95% CI 0.77–2.08) compared to those with no DNAemia. Similarly, IgG status was not associated with patient survival (HR 1.03, 95% CI 0.65–1.44) (S2 Fig).
Discussion
We evaluated HCMV among patients with MPM and observed HCMV DNAemia viral loads indicative of an active HCMV infection (>1000 copies/mL) in 41% of patients. However, we did not observe evidence that this HCMV viremia was driven by infection at the tumor site. Our data suggest that active HCMV infection may be an unrecognized complication in MPM patients, putting them at risk for HCMV pneumonia as is observed in bone marrow and solid organ transplant recipients [14, 30–33].
HCMV, as assessed by IgG serostatus, is highly prevalent in the global population [34]. In healthy persons, HCMV infection is often asymptomatic. However, HCMV reactivation is of high concern in immune suppressed populations where it can progress to end-organ disease [20, 35], often presenting as a pulmonary HCMV infection (pneumonia) [19, 33], an observation confirmed in a MCMV mouse model [11]. Immune compromised populations, such as those with HIV, are also at risk for HCMV disease [18, 31, 36]. We suspect that inflammation, a known trigger for HCMV reactivation from latency, in combination with immune compromise, is contributing to HCMV viral activity in a subset of MPM patients. An important future direction of this research is to determine whether MPM patients are experiencing HCMV viral complications which could be treated.
HCMV DNAemia is intermittently prevalent in healthy populations (6–24%) [37, 38] and may occur due to primary HCMV infection, a second infection with a new HCMV strain, or reactivation of HCMV from latency. The limited reports on HCMV DNAemia prevalence in solid tumor patients suggest that prevalence may vary depending on the timing of blood collection, ranging from 11–93% [23, 24]. A significant limitation of the current work is a lack of standardization of blood draw in relation to course of treatment, making our estimate of DNAemia prevalence unstable. A 2010 study reported that among solid tumor cancer patients with CMV DNAemia, lung cancer was the most common diagnosis (30/75), however data on DNAemia levels were not available [39]. In the current study we compared HCMV DNAemia in MPM and metastatic lung cancer patients and found DNAemia common in both patient groups, but that high DNAemia levels (>1000 copies/mL serum) were specific to MPM. A limitation of our data is the inability to distinguish between a new infection or reactivation from latency. In fact, there was a striking lack of concordance between IgG status and DNAemia, which would suggest many patients were CMV naive and experiencing a primary CMV infection. Alternatively, CMV status may be misclassified by either the DNAemia or serology measurements. Studies from blood bank donation have mixed findings on this issue. Larsson et al reported discordance between IgG and DNAemia measurements, and suggested that a substantial proportion of the population may harbor CMV without developing a CMV IgG response [40]. In contrast, Roback et al. [41] did not observe discordance between DNAemia and IgG and claim observations of discordance are likely due to technical artifact. A comprehensive evaluation of CMV in a prospective study design is warranted.
We have established that in the absence of malignancy HCMV DNA is present in pleural tissue, indicating this may be a natural reservoir for latent infection. A higher proportion of tumor tissue was DNA positive, and this is consistent with the growing body of literature demonstrating the presence of HCMV DNA or viral proteins in the tumor environment [27], as well as multiple tissues serving as reservoirs susceptible to reactivation during times of immune suppression [40, 42, 43]. Across cancer types HCMV is present at low levels in the tumor, and it is not present as a clonal infection. Here, we tested a small amount of tissue, and given the non-clonal nature of tumor HCMV infections our reported prevalence of 29% is likely an underestimate. We did not observe an association between tumor HCMV status and serum HCMV status. This too could be attributed to misclassification of tumor status given the small proportion of tumor material tested.
HCMV infection may be a biomarker that identifies patients who are the most immune-compromised or have the greatest tumor-associated inflammation. In addition, HCMV may be associated with immune benefit; there is a growing body of literature describing the emergence of adaptive Natural Killer (NK) cells with enhanced cytotoxic activity following HCMV reactivation [44, 45], and it is plausible that those with low level reactivation may have this immune benefit. Consistent with this hypothesis, those with high DNAemia had a shorter median survival than those with low level DNAemia. However, none of the observations regarding patient survival were statistically significant and further study is warranted. Specifically, capturing immune phenotypes and inflammation would provide clarity.
HCMV has been implicated multiple times in enhancing the malignancy of cancer cells and tumor associated cells. HCMV infection has been shown to modulate multiple molecular pathways involved in signal transduction of cellular activation [46, 47]. These onco-modulatory effects of HCMV on cellular metabolism are like those mediated by small DNA tumor viruses such as simian virus 40 (SV40) [6] and human adenovirus [48, 49]. In tumor cells HCMV-encoded regulatory proteins interfere with a variety of cellular signal transduction pathways leading to accelerated cell proliferation, enhanced survival, angiogenesis, cell motility and adhesion, thus enhancing the malignant behavior of tumor cells For this reason, potential treatment strategies such as immunotherapies using viral vectors such as HCMV could be employed and are currently being studied. The confirmation of the presence of HCMV is interesting as it provides a unique perspective that can guide future immunotherapies. There have already been promising results using CMV as a viral vector for many cancer immunotherapies [50].
A significant limitation of the present study is lack of treatment information. Patients were approached for study participation at a consultation visit. However, many did not seek treatment at the consenting hospital and treatment information is unavailable. An unexpected finding was the lack of association between HCMV DNAemia and IgG. This could occur if a subset of patients is HCMV naïve and experiencing a primary infection. Future work should include both replication in a second population of patients, and inclusion of IgM which would reflect new infections. Additionally, it would be useful to have bronchial swabs, or bronchial washing fluid (BAL) from each patient to confirm an active HCMV pneumonic infection. Lastly, no patient symptoms or co-infections were assessed at the time of blood draw. Osawa et al. 2009 reported that bacterial pneumonia was often present as a coinfection with HCMV viremia in solid tumor patients [20]. These shortcomings, and evaluation for HCMV pneumonia and bacterial infections, should be addressed in future work.
Conclusions
The findings from the present investigation add to a limited but increasing evidence base supporting the use of identifying HCMV infection or reactivation in cancer patient care. Our data support the hypothesis that active HCMV infection is a common unrecognized clinical event among mesothelioma patients that may be associated with poor patient survival.
Supporting information
S1 Fig
HCMV DNA status in tumor tissue and mesothelioma patient survival.MPM patients with HCMV DNA negative tumor tissue had a shorter survival period then MPM patients with HCMV DNA positive tumor tissue.
(TIF)
S2 Fig
HCMV IgG status and mesothelioma patient survival.Positive HCMV IgG status, indicating the presence of HCMV antibodies, was associated with lower survival when compared to those with negative HCMV IgG status. This association was not found to be statistically significant (p = 0.35).
(TIF)
Acknowledgments
We would like to acknowledge and thank the Advanced Research and Diagnostic Laboratory (ARDL) located at the University of Minnesota for completing all serology services for this project.
Abbreviations
| HCMV | Human Cytomegalovirus |
| MPM | Malignant Pleural Mesothelioma |
| qdPCR | Quantitative Digital Polymerase Chain Reaction |
Funding Statement
This work was supported by the National Institute of Health through grants awarded to NF (R35 CA197292 and P30 CA77598) and KK (R01 CA126939). The Masonic Cancer Center also supported the study through a grant given to HN (R35 00052643). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
References
Decision Letter 0
23 Dec 2020
PONE-D-20-34162
Cytomegalovirus infection in malignant pleural mesothelioma
PLOS ONE
Dear Dr. Hunter-Schlichting,
Thank you for submitting your manuscript to PLOS ONE. After careful consideration, we feel that it has merit but does not fully meet PLOS ONE’s publication criteria as it currently stands. Therefore, we invite you to submit a revised version of the manuscript that addresses the points raised during the review process.
Two expert reviewers have commented on your article, and their feedback is appended below. Overall the reviewers felt that results were publication-worthy and of interest to the scientific community, but additional information is needed before it can be published. Three major issues need to be addressed: 1. greater detail on experimental methods overall, 2. improved presentation and discussion of DNAemia results, and 3. expanded discussion with respect to HCMV biology. Specifically, more details on the methods is required, as noted by reviewer 1, particularly clarification on the sample preparation and analysis of HCMV serostatus. In addition, both reviewers ask for more information and clarification on the DNAemia status, both in the results (Fig 1) and discussion sections, particularly in comparison to the general population. Finally, the reviewers include several comments about typos, awkward working, or areas where enhanced or clarified discussion of the results is necessary. Your revised manuscript should effectively address each of the the reviewer's comments below.
Please submit your revised manuscript by Feb 06 2021 11:59PM. If you will need more time than this to complete your revisions, please reply to this message or contact the journal office at gro.solp@enosolp. When you're ready to submit your revision, log on to https://www.editorialmanager.com/pone/ and select the 'Submissions Needing Revision' folder to locate your manuscript file.
Please include the following items when submitting your revised manuscript:
A rebuttal letter that responds to each point raised by the academic editor and reviewer(s). You should upload this letter as a separate file labeled 'Response to Reviewers'.
A marked-up copy of your manuscript that highlights changes made to the original version. You should upload this as a separate file labeled 'Revised Manuscript with Track Changes'.
An unmarked version of your revised paper without tracked changes. You should upload this as a separate file labeled 'Manuscript'.
If you would like to make changes to your financial disclosure, please include your updated statement in your cover letter. Guidelines for resubmitting your figure files are available below the reviewer comments at the end of this letter.
If applicable, we recommend that you deposit your laboratory protocols in protocols.io to enhance the reproducibility of your results. Protocols.io assigns your protocol its own identifier (DOI) so that it can be cited independently in the future. For instructions see: http://journals.plos.org/plosone/s/submission-guidelines#loc-laboratory-protocols
We look forward to receiving your revised manuscript.
Kind regards,
Juliet V Spencer, Ph.D.
Academic Editor
PLOS ONE
Journal Requirements:
When submitting your revision, we need you to address these additional requirements.
1. Please ensure that your manuscript meets PLOS ONE's style requirements, including those for file naming. The PLOS ONE style templates can be found at
https://journals.plos.org/plosone/s/file?id=wjVg/PLOSOne_formatting_sample_main_body.pdf and
2. In your Methods section, please provide additional information about the participant recruitment method and the demographic details of your participants in both cohorts. Please ensure you have provided sufficient details to replicate the analyses such as: a) the recruitment date range (month and year), b) a description of how participants were recruited.
3. Please provide a sample size and power calculation in the Methods, or discuss the reasons for not performing one before study initiation.
4. Please include additional information regarding the survey or questionnaire used in the study and ensure that you have provided sufficient details that others could replicate the analyses. For instance, if you developed a questionnaire as part of this study and it is not under a copyright more restrictive than CC-BY, please include a copy, in both the original language and English, as Supporting Information.
5. Please note that PLOS does not permit references to “data not shown.” Authors should provide the relevant data within the manuscript, the Supporting Information files, or in a public repository. If the data are not a core part of the research study being presented, we ask that authors remove any references to these data.
6.Thank you for stating the following in the Acknowledgments Section of your manuscript:
"This work was supported in part by NIH P30 CA77598 utilizing the Masonic Cancer Center,
University of Minnesota shared resources and the sponsored by the R35 grant (00052643)"
We note that you have provided funding information that is not currently declared in your Funding Statement. However, funding information should not appear in the Acknowledgments section or other areas of your manuscript. We will only publish funding information present in the Funding Statement section of the online submission form.
Please remove any funding-related text from the manuscript and let us know how you would like to update your Funding Statement. Currently, your Funding Statement reads as follows:
"The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript."
Please include your amended statements within your cover letter; we will change the online submission form on your behalf.
7. Please include a caption for figure 1.
[Note: HTML markup is below. Please do not edit.]
Reviewers' comments:
Reviewer's Responses to Questions
Comments to the Author
1. Is the manuscript technically sound, and do the data support the conclusions?
The manuscript must describe a technically sound piece of scientific research with data that supports the conclusions. Experiments must have been conducted rigorously, with appropriate controls, replication, and sample sizes. The conclusions must be drawn appropriately based on the data presented.
Reviewer #1: Yes
Reviewer #2: Partly
**********
2. Has the statistical analysis been performed appropriately and rigorously?
Reviewer #1: Yes
Reviewer #2: Yes
**********
3. Have the authors made all data underlying the findings in their manuscript fully available?
The PLOS Data policy requires authors to make all data underlying the findings described in their manuscript fully available without restriction, with rare exception (please refer to the Data Availability Statement in the manuscript PDF file). The data should be provided as part of the manuscript or its supporting information, or deposited to a public repository. For example, in addition to summary statistics, the data points behind means, medians and variance measures should be available. If there are restrictions on publicly sharing data—e.g. participant privacy or use of data from a third party—those must be specified.
Reviewer #1: Yes
Reviewer #2: Yes
**********
4. Is the manuscript presented in an intelligible fashion and written in standard English?
PLOS ONE does not copyedit accepted manuscripts, so the language in submitted articles must be clear, correct, and unambiguous. Any typographical or grammatical errors should be corrected at revision, so please note any specific errors here.
Reviewer #1: Yes
Reviewer #2: Yes
**********
5. Review Comments to the Author
Please use the space provided to explain your answers to the questions above. You may also include additional comments for the author, including concerns about dual publication, research ethics, or publication ethics. (Please upload your review as an attachment if it exceeds 20,000 characters)
Reviewer #1: Hunter-Schlichting and co-authors present an interesting preliminary study assessing the incident of HCMV infection in malignant pleural mesothelioma patients. While the study is small, the authors are reasonable in their assessment of the data and their conclusions, and fully outline the limitations to the patient cohort and reasonable analysis thereof. This is an interesting preliminary study, and the authors properly address all aspects of the patient cohort, analyses, and conclusions, with an emphasis on future study. This paper provides a useful addition to the literature and is scientifically sound. To that end, I have only a few minor comments:
1) The finding that more patients have positive DNAemia than are seropositive is surprising and quite strong even using a smaller study sample size. This finding could be highlighted more as this would be a very interesting follow up.
2) On the same note as point 1 above, the conclusions/discussion regarding a latent HCMV infection contributing to disease are not supported by the data presented here. The data suggests - both by higher DNAemia patients (although any detectable DNA typically suggests a non-latent viral state in patients) trending towards increased mortality, and the by the lack of detectable IgG in DNA+ patients; that an initial or reactivated viral infection is more likely associated. The authors do address this in the Discussion, but the emphasis on latent infection is overstated.
3) Why was the DNAemia reporting level changed between patient cohorts (page 7)? Serum was analyzed for DNA copies and grouped into three categories (undetected, DNA+, active infection (greater than 1000 copies/mL) - so negative, positive, active. Yet tissue samples, although run in the same manner were categorized as only negative or positive. Active infection could be defined using similar criteria here as in serum (i.e. greater than 1000 copies per ug tissue for example)? Or discuss limitations as to why not? This is relevant to the Discussion as well (third paragraph, last sentence), which states that "viremic levels (>100 copies/mL serum) were specific to MPM" which is not supported by the data in this cohort, although I agree that it does warrant further study in a larger cohort.
4) Additional information should be included as to whether patients were assessed for treatment status / immunosuppression at the time of sampling (table 1).
5) The prevalence of HCMV in these samples is a key piece of data. The data presented on page 8 (Table 1) however suggests that prevalence of HCMV is comparable to the general population and DNAemia is similar in similar sampled tissues (metastatic lung cancer, data not shown). This is a very key point to discuss since this could suggest that the findings of HCMV prevalence are no different than background population levels. This could be clarified in the Discussion where a couple of statements (i.e. third sentence of paragraph one - "suggest HCMV infection may be an unrecognized co-morbidity") are overstated.
6) There are some minor typos/formatting/missing punctuation, including in Table 2.
Reviewer #2: In the article entitled, “Cytomegalovirus infection in malignant pleural mesothelioma”, by Hunter-Schlichting, et al, the authors describe the prevalence of CMV in patients with malignant pleural mesothelioma (MPM). To the authors’ and this reviewer’s knowledge, this is the first investigation into the potential connection between CMV and MPM. Overall, the manuscript could be clarified in some places, which should not prove overly cumbersome but will undoubtedly improve readability. Also, some of the data should be more clearly presented (e.g. Fig 1), so the reader can appropriately interpret the data herein. Suggestions are provided below for the authors’ consideration:
Major:
1. Clarity to the methods is warranted.
a. Study population: how long between the time of consent and blood sampling?
b. Serum biomarkers: Stating “…serology was performed on the Roche e411 Analyzer…”, with no additional information as to how the samples were prepared, etc. is definitely not enough information. Similarly, how were the tissue samples prepared? In the same paragraph, what does “gBa region” refer to? Is it not just “gB”? Was an uninfected control used to determine baseline for the CMV DNA qPCR? This would allow one to determine if the primers detect background (in other words, it allows one to determine the baseline). This could also be why there was no association between IgG and DNAemia (which the authors describe as ‘unexpected’ in their Discussion, p13).
c. HCMV DNA in tissue: Were the normal tissue samples also freshly frozen? What were the methods used to extract the DNA from tissue?
d. Lung Cancer BioBank samples: “Blood was collected at specified time points during a patient’s treatment course” - Did all patients receive the same treatment course, and did treatment regimens differ between the two study sites? What were these specified time points?
2. Figure 1 is hard to evaluate. The lines are not labeled, making it difficult to interpret.
3. p11 – “HCMV…is highly prevalent in the global population and nearly ubiquitous in immunocompromised populations” – I would argue this is misleading, especially since the authors use the term ‘immunocompromised’ to describe transplant patients as well (who are technically immunosuppressed). Likewise, “HCMV infection may be a biomarker that identifies patients who are the most immune-compromised…” is misleading.
4. p12 – “…those with the longest survival had low level DNAemia” – Isn’t this not completely true when adjusted for age? Similarly, p14 (Conclusion section) – “…MPM patients with a negative or low level HCMV infection were greater than those with a high level HCM infection” – isn’t this not the case when adjusted for age? (Also, minor, but this last sentence should read “…HCMV infection”).
Minor:
1. For ease in reviewing, it would help a ton to have line numbers.
2. Various points in the manuscript should be clarified:
a. What is meant by “HCMV…has been observed as viremia in various cancer patients and the tumor environment in different cancers” (p4)?
b. p4 – “>50% of the population infected by ages 75-80.” This should be double-checked. It’s more like >50% by 40 years of age.
c. What is meant by “Less is known about the experience of HCMV viremia in cancer patients…” (p4-5)?
d. The data re: 21 normal pleura samples – this is not included in the tables is it?
e. p10, p11, p12 – “HCMV biomarkers” should actually be defined.
f. p12 – what is meant by a “robust NK cell phenotype”?
3. Figures should be presented in order of appearance. As is, the supplemental figures need to be reversed. Also, I would suggest the authors put the supplemental figures in the main body of the manuscript as opposed to including them as supplemental figures.
4. The authors should include an overall “take-home” message for their findings at the end of the results section. As is, this section just drops off, leaving the reader wondering what the overall take is.
5. p11 – The authors state that HCMV infection could put MPM patients at risk for HCMV-associated pneumonia. Do any of these patients the authors evaluated present with this symptom?
6. p12 – I appreciate the authors’ idea that pleural tissue may serve as a site of latency. But it may also be worth noting that infiltrating, latently infected monocytes that are recruited to the ‘injured’ pleura could be a source of reactivating virus in this tissue. Just a thought.
**********
6. PLOS authors have the option to publish the peer review history of their article (what does this mean?). If published, this will include your full peer review and any attached files.
If you choose “no”, your identity will remain anonymous but your review may still be made public.
Do you want your identity to be public for this peer review? For information about this choice, including consent withdrawal, please see our Privacy Policy.
Reviewer #1: No
Reviewer #2: No
[NOTE: If reviewer comments were submitted as an attachment file, they will be attached to this email and accessible via the submission site. Please log into your account, locate the manuscript record, and check for the action link "View Attachments". If this link does not appear, there are no attachment files.]
While revising your submission, please upload your figure files to the Preflight Analysis and Conversion Engine (PACE) digital diagnostic tool, https://pacev2.apexcovantage.com/. PACE helps ensure that figures meet PLOS requirements. To use PACE, you must first register as a user. Registration is free. Then, login and navigate to the UPLOAD tab, where you will find detailed instructions on how to use the tool. If you encounter any issues or have any questions when using PACE, please email PLOS at gro.solp@serugif. Please note that Supporting Information files do not need this step.
Author response to Decision Letter 0
14 Jun 2021
Seven issues were raised by the academic editor. A response to each of these issues is provided below.
1. Please ensure that your manuscript meets PLOS ONE’s style requirements, including those for file naming.
This has been done.
2. In your Methods section, please provide additional information about the participant recruitment method and the demographic details of your participants in both cohorts.
We have added additional text to the methods section describing participant recruitment for both the mesothelioma study and the lung cancer biobank. The demographic characteristics of the lung cancer biobank participants are now included in the text of the methods.
Please ensure you have provided sufficient details to replicate the analyses such as: a) the recruitment date range (month and year) and b) a description of how participants were recruited.
These details have been added to the methods section.
3. Please provide a sample size and power calculation in the Methods or discuss the reasons for not performing one before study initiation.
No a priori sample size calculations were performed. We tested all available samples from the mesothelioma study and lung cancer biobank. A statement to that effect was present in the original description of the mesothelioma study population, and a similar statement has now been added to the lung cancer biobank description.
4. Please include additional information regarding the survey or questionnaire used in the study and ensure that you have provided sufficient details that others could replicate the analyses. For instance, if you developed a questionnaire as part of this study and it is not under a copyright more restrictive than CC-BY, please include a copy, in both the original language and English, as Supporting Information.
The epidemiologic study was conducted over 20 years ago and the questionnaire is not available. The data used in this study are not specific to the questionnaire, with the exception of asbestos exposure. In our analysis we utilized self-report of asbestos exposure and it was not associated with CMV status. The other variables investigated (age, sex, and histology) are easily replicated in a clinical study using data from the health record.
5. Please note that PLOS does not permit references to “data not shown.” Authors should provide the relevant data within the manuscript, the Supporting Information files, or in a public repository. If the data are not a core part of the research study being presented, we ask that authors remove any references to these data.
The data are now more fully described in the text of the manuscript, and “data not shown” has been removed.
6. Thank you for stating the following in the Acknowledgments Section of your manuscript: "This work was supported in part by NIH P30 CA77598 utilizing the Masonic Cancer Center, University of Minnesota shared resources and the sponsored by the R35 grant (00052643)" We note that you have provided funding information that is not currently declared in your Funding Statement. However, funding information should not appear in the Acknowledgments section or other areas of your manuscript. We will only publish funding information present in the Funding Statement section of the online submission form. Please remove any funding-related text from the manuscript and let us know how you would like to update your Funding Statement. Currently, your Funding Statement reads as follows:"The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript." Please include your amended statements within your cover letter; we will change the online submission form on your behalf.
We have removed from the funding information from the Acknowledgments Section of the manuscript and have an updated Funding Statement in the cover letter. It is repeated here: “This work was supported by the following NIH grants: R35 CA197292, R01 CA126939 and P30 CA77598. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript".
7. Please include a caption for figure 1.
A caption has now been added to Figure 1.
Reviewer #1
1) The finding that more patients have positive DNAemia than are seropositive is surprising and quite strong even using a smaller study sample size. This finding could be highlighted more as this would be a very interesting follow up.
We agree that the lack of concordance between DNAemia and serostatus is surprising. Among our patient population it is possible that patients were CMV naive and are now experiencing a primary CMV infection. Alternatively, CMV status may be misclassified, either due to technical artifact or lack of antibody response. We have broadened our discussion of DNA-IgG discordance to describe these possibilities and present our finding in context of what has been observed in the literature. Specifically, there are two large observational studies from blood bank donation centers. One observed discordance similar to what we observe in our study, and these authors suggest that a substantial proportion of the population may be “CMV carriers” who have CMV but did not develop an IgG response. The second observed no discordance and suggested that if discordance is observed it is likely a technical artifact or arises due to new primary infection.
2) On the same note as point 1 above, the conclusions/discussion regarding a latent HCMV infection contributing to disease are not supported by the data presented here. The data suggests - both by higher DNAemia patients (although any detectable DNA typically suggests a non-latent viral state in patients) trending towards increased mortality, and the by the lack of detectable IgG in DNA+ patients; that an initial or reactivated viral infection is more likely associated. The authors do address this in the Discussion, but the emphasis on latent infection is overstated.
We agree with the reviewer that we need to be more careful with our language regarding CMV status throughout the manuscript and recognize that we cannot distinguish between primary and reactivated infection. Based on the reviewers comments we now refer to “low level current infection” and “high level current infection” with regard to the serum DNAemia data. The manuscript has been edited accordingly.
3) Why was the DNAemia reporting level changed between patient cohorts (page 7)? Serum was analyzed for DNA copies and grouped into three categories (undetected, DNA+, active infection (greater than 1000 copies/mL) - so negative, positive, active. Yet tissue samples, although run in the same manner were categorized as only negative or positive. Active infection could be defined using similar criteria here as in serum (i.e. greater than 1000 copies per ug tissue for example)? Or discuss limitations as to why not? This is relevant to the Discussion as well (third paragraph, last sentence), which states that "viremic levels (>100 copies/mL serum) were specific to MPM" which is not supported by the data in this cohort, although I agree that it does warrant further study in a larger cohort.
For the serum data it is possible to back-calculate the dPCR results as copies/mL serum. However, for the tumor data we were not confident in making a similar calculation, largely because the input tumor mass for the extractions was not constant across samples. There is a large clinical body of literature that equates serum/plasma DNAemia with the presence of a current replicating CMV infection. The same is not true with the tumor tissue. For these reasons we felt most comfortable classifying the tumor as positive or negative. We have now included this information in the methods section.
4) Additional information should be included as to whether patients were assessed for treatment status / immunosuppression at the time of sampling (table 1).
We acknowledge that this is an important issue. Unfortunately, treatment status information is not available on this study. We have now directly addressed this limitation in our discussion section.
5) The prevalence of HCMV in these samples is a key piece of data. The data presented on page 8 (Table 1) however suggests that prevalence of HCMV is comparable to the general population and DNAemia is similar in similar sampled tissues (metastatic lung cancer, data not shown). This is a very key point to discuss since this could suggest that the findings of HCMV prevalence are no different than background population levels. This could be clarified in the Discussion where a couple of statements (i.e. third sentence of paragraph one - "suggest HCMV infection may be an unrecognized co-morbidity") are overstated.
We agree with the reviewer that the prevalence of HCMV IgG seropositivity is comparable to the general population, and that DNAemia is much higher in these cancer patients than what would be expected in the general population. We have updated the manuscript to include information from the literature on the prevalence of DNAemia in the general population, which is dramatically lower than we observe in these two patient populations. We have now added emphasis in the discussion that IgG prevalence is similar to the general population, and that active HCMV infection is much higher than expected. We have carefully reviewed the language in the discussion and revised the sentence highlighted by the reviewer to indicate that it is active HCMV infection that may be an unrecognized co-morbidity,
6) There are some minor typos/formatting/missing punctuations, including in Table 2
We thank the reviewer for this feedback and have edited the manuscript accordingly.
Reviewer #2
Major:
1. Clarity to the methods is warranted.
a. Study population: how long between the time of consent and blood sampling?
This information was not collected as part of the original epidemiology study; this limitation is now addressed in the discussion section.
b. Serum biomarkers: Stating “…serology was performed on the Roche e411 Analyzer…”, with no additional information as to how the samples were prepared, etc. is definitely not enough information. Similarly, how were the tissue samples prepared? In the same paragraph, what does “gBa region” refer to? Is it not just “gB”? Was an uninfected control used to determine baseline for the CMV DNA qPCR? This would allow one to determine if the primers detect background (in other words, it allows one to determine the baseline). This could also be why there was no association between IgG and DNAemia (which the authors describe as ‘unexpected’ in their Discussion, p13).
We have added the requested information to the methods section. Controls, both positive and negative, were used to ensure accuracy of the assay.
c. HCMV DNA in tissue: Were the normal tissue samples also freshly frozen? What were the methods used to extract the DNA from tissue?
Yes, the normal tissue samples were also freshly frozen. DNA was extracted using a Qiagen kit. These details are now included in the methods section.
d. Lung Cancer BioBank samples: “Blood was collected at specified time points during a patient’s treatment course” - Did all patients receive the same treatment course, and did treatment regimens differ between the two study sites? What were these specified time points?
We have clarified the methods to indicate that we tested a blood sample taken at the time of study enrollment, prior to treatment. There was only one site for the enrollment of participants in the Lung Cancer BioBank.
2. Figure 1 is hard to evaluate. The lines are not labeled, making it difficult to interpret.
Figure 1 has been updated; the lines are labeled.
3. p11 – “HCMV…is highly prevalent in the global population and nearly ubiquitous in immunocompromised populations” – I would argue this is misleading, especially since the authors use the term ‘immunocompromised’ to describe transplant patients as well (who are technically immunosuppressed). Likewise, “HCMV infection may be a biomarker that identifies patients who are the most immune-compromised…” is misleading.
Thank you for these suggestions, we have edited the manuscript accordingly.
4. p12 – “…those with the longest survival had low level DNAemia” – Isn’t this not completely true when adjusted for age? Similarly, p14 (Conclusion section) – “…MPM patients with a negative or low level HCMV infection were greater than those with a high level HCM infection” – isn’t this not the case when adjusted for age? (Also, minor, but this last sentence should read “…HCMV infection”).
We have revised the text as suggested.
Minor:
1. For ease in reviewing, it would help a ton to have line numbers.
Line numbers have been added.
2. Various points in the manuscript should be clarified:
a. What is meant by “HCMV…has been observed as viremia in various cancer patients and the tumor environment in different cancers” (p4)?
We have edited the text to read: “The role of HCMV in cancer has primarily focused on the presence of virus in tumors (21,22,25–27). Less well described is the epidemiology of active HCMV infection in solid tumor cancer patients.”
b. p4 – “>50% of the population infected by ages 75-80.” This should be double-checked. It’s more like >50% by 40 years of age.
We have revised the manuscript to say “... is a highly prevalent infection with approximately 50% of the population testing seropositive by age 50”, and reference NHANES.
c. What is meant by “Less is known about the experience of HCMV viremia in cancer patients…” (p4-5)?
We have clarified and expanded this text. Our intent is to convey that active HCMV infection (viremia) has to date been understudied in cancer patients, outside the context of bone marrow transplant. Primarily the literature regarding HCMV in non-hematologic malignancy has been focused on detecting HCMV in the tumor environment.
d. The data re: 21 normal pleura samples – this is not included in the tables is it?
The reviewer is correct, the normal pleura data is only present in the text of the manuscript.
e. p10, p11, p12 – “HCMV biomarkers” should actually be defined.
This comment has been addressed.
f. p12 – what is meant by a “robust NK cell phenotype”?
This text has been clarified
3. Figures should be presented in order of appearance. As is, the supplemental figures need to be reversed. Also, I would suggest the authors put the supplemental figures in the main body of the manuscript as opposed to including them as supplemental figures.
We have corrected the order of appearance of the supplemental figures. We defer to the editor whether these should be supplemental or included in the main body of the manuscript.
4. The authors should include an overall “take-home” message for their findings at the end of the results section. As is, this section just drops off, leaving the reader wondering what the overall take is.
A brief summary of the results is now included at the end of the results section.
5. p11 – The authors state that HCMV infection could put MPM patients at risk for HCMV-associated pneumonia. Do any of these patients the authors evaluated present with this symptom?
Unfortunately, we do not have that information available.
6. p12 – I appreciate the authors’ idea that pleural tissue may serve as a site of latency. But it may also be worth noting that infiltrating, latently infected monocytes that are recruited to the ‘injured’ pleura could be a source of reactivating virus in this tissue. Just a thought.
We really like this idea and thank the reviewer for the suggestion.
Attachment
Submitted filename: Master response to reviewers submit.docx
Decision Letter 1
21 Jun 2021
Cytomegalovirus infection in malignant pleural mesothelioma
PONE-D-20-34162R1
Dear Dr. Hunter-Schlichting,
We’re pleased to inform you that your manuscript has been judged scientifically suitable for publication and will be formally accepted for publication once it meets all outstanding technical requirements.
Within one week, you’ll receive an e-mail detailing the required amendments. When these have been addressed, you’ll receive a formal acceptance letter and your manuscript will be scheduled for publication.
An invoice for payment will follow shortly after the formal acceptance. To ensure an efficient process, please log into Editorial Manager at http://www.editorialmanager.com/pone/, click the 'Update My Information' link at the top of the page, and double check that your user information is up-to-date. If you have any billing related questions, please contact our Author Billing department directly at gro.solp@gnillibrohtua.
If your institution or institutions have a press office, please notify them about your upcoming paper to help maximize its impact. If they’ll be preparing press materials, please inform our press team as soon as possible -- no later than 48 hours after receiving the formal acceptance. Your manuscript will remain under strict press embargo until 2 pm Eastern Time on the date of publication. For more information, please contact gro.solp@sserpeno.
Kind regards,
Juliet V Spencer, Ph.D.
Academic Editor
PLOS ONE
Additional Editor Comments (optional):
Reviewers' comments:
Acceptance letter
29 Jul 2021
PONE-D-20-34162R1
Cytomegalovirus infection in malignant pleural mesothelioma
Dear Dr. Hunter-Schlichting:
I'm pleased to inform you that your manuscript has been deemed suitable for publication in PLOS ONE. Congratulations! Your manuscript is now with our production department.
If your institution or institutions have a press office, please let them know about your upcoming paper now to help maximize its impact. If they'll be preparing press materials, please inform our press team within the next 48 hours. Your manuscript will remain under strict press embargo until 2 pm Eastern Time on the date of publication. For more information please contact gro.solp@sserpeno.
If we can help with anything else, please email us at gro.solp@enosolp.
Thank you for submitting your work to PLOS ONE and supporting open access.
Kind regards,
PLOS ONE Editorial Office Staff
on behalf of
Dr. Juliet V Spencer
Academic Editor
PLOS ONE
Articles from PLOS ONE are provided here courtesy of PLOS
Full text links
Read article at publisher's site: https://doi.org/10.1371/journal.pone.0254136
Read article for free, from open access legal sources, via Unpaywall:
https://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0254136&type=printable
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/111725930
Article citations
The power of mumps virus: Matrix protein activates apoptotic pathways in human colorectal cell lines.
PLoS One, 18(12):e0295819, 13 Dec 2023
Cited by: 1 article | PMID: 38091318 | PMCID: PMC10718445
Prevalence of active cytomegalovirus infection at diagnosis of ovarian cancer and during chemotherapy and subsequent changes in cognitive functioning.
BMC Cancer, 23(1):1057, 03 Nov 2023
Cited by: 1 article | PMID: 37923995 | PMCID: PMC10623703
Current management of CMV infection in cancer patients (solid tumors). Epidemiology and therapeutic strategies.
Rev Esp Quimioter, 35 Suppl 3:74-79, 24 Oct 2022
Cited by: 6 articles | PMID: 36285863 | PMCID: PMC9717468
Review Free full text in Europe PMC
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Assessment of human cytomegalovirus co-infection in Egyptian chronic HCV patients.
Virol J, 8:343, 10 Jul 2011
Cited by: 6 articles | PMID: 21740595 | PMCID: PMC3145597
Correlation between systemic lupus erythematosus and cytomegalovirus infection detected by different methods.
Clin Rheumatol, 34(4):691-698, 10 Mar 2015
Cited by: 24 articles | PMID: 25750182
Correspondence Between Cytomegalovirus Immunoglobulin-G Levels Measured in Saliva and Serum.
Front Immunol, 11:2095, 28 Aug 2020
Cited by: 1 article | PMID: 32983163 | PMCID: PMC7484902
Detection of Cytomegalovirus (CMV) Infection in Wheezing Infants by Urine DNA and Serum IgG Testing.
Med Sci Monit, 23:1242-1246, 11 Mar 2017
Cited by: 2 articles | PMID: 28283676 | PMCID: PMC5358860
Funding
Funders who supported this work.
NCI NIH HHS (4)
Grant ID: P30 CA077598
Grant ID: R01 CA120528
Grant ID: R01 CA126939
Grant ID: R35 CA197292
National Institutes of Health (1)
Grant ID: P30 CA77598
masonic cancer center, university of minnesota (1)
Grant ID: R35 00052643
national institutes of health (2)
Grant ID: R35 CA197292
Grant ID: R01 CA126939