Abstract
Free full text

Machine Learning Models Identify Inhibitors of SARS-CoV-2
Abstract
With the rapidly evolving SARS-CoV-2 variants of concern there is an urgent need for the discovery of further treatments for the coronavirus disease (COVID-19). Drug repurposing is one of the most rapid strategies for addressing this need and numerous compounds have already been selected for in vitro testing by several groups. These have led to a growing database of molecules with in vitro activity against the virus. Machine learning models can assist drug discovery through prediction of the best compounds based on previously published data. Herein we have implemented several machine learning methods to develop predictive models from recent SARS-CoV-2 in vitro inhibition data and used them to prioritize additional FDA approved compounds for in vitro testing selected from our in-house compound library. From the compounds predicted with a Bayesian machine learning model, lumefantrine, an antimalarial was selected for testing and showed limited antiviral activity in cell-based assays while demonstrating binding (Kd 259nM) to the spike protein using microscale thermophoresis. Several other compounds which we prioritized have since been tested by others and were also found to be active in vitro. This combined machine learning and in vitro testing approach can be expanded to virtually screen available molecules with predicted activity against SARS-CoV-2 reference WIV04 strain and circulating variants of concern. In the process of this work, we have created multiple iterations of machine learning models that can be used as a prioritization tool for SARS-CoV-2 antiviral drug discovery programs. The very latest model for SARS-CoV-2 with over 500 compounds is now freely available at www.assaycentral.org.
Introduction
In December 2019, several cases of pneumonia with unknown etiology started to arise in Wuhan, China. A new betacoronavirus was identified and named SARS-CoV-2 due to its high similarity with previous SARS-CoV.1,2 This virus causes the disease which has been called COVID-19.3 Since then, SARS-CoV-2 has rapidly spread worldwide prompting the World Health Organization to declare the outbreak a pandemic, with more than 1.5 million cases confirmed in less than 100 days.4 The high infection rate has caused considerable stress on global healthcare systems leading to more than 194 million people have been infected and more than 4.1 million deaths.5
The SARS-CoV-2 pandemic has started a worldwide effort to discover treatments that could prevent further COVID-19 deaths and decrease numbers hospitalized as well as the length of hospitalization for patients.6 Drug repurposing is one of the main strategies being used to accelerate this as most preclinical stages are removed and a promising drug could potentially move directly to Phase II clinical studies or beyond by using an approved, safe drug.7,8 When we started the current study (in early 2020) most SARS-CoV-2 in vitro inhibition studies relied on small to medium scale assays with high throughput screens (HTS) campaigns testing specific FDA-approved drugs and compounds that have previously shown inhibition against different betacoronaviruses or specific antiviral targets.9–17 Since then, large-scale screens have tested 1425 compounds in Huh7 cells, identifying 11 molecules with activity IC50 < 1 μM.18 Another large screen of 1528 compounds in Vero cells resulted in 19 hits with 4 possessing IC50’s of ~1 μM.19 A recent screen of the Prestwick library in hPSC lung organoids led to 3 hits.20 One of the largest screens to date in Vero cells used 12,000 clinical stage or FDA approved compounds in the ReFRAME library and resulted in 21 hits.21 This latter study represents an example of a dataset of molecules which has not been made available to the public as yet. With many HTS performed and data published, ChEMBL and PubChem rapidly started to gather and curate most of this data, making it easier for everyone to access and use it for different cheminformatics methods that can assist the COVID19 drug discovery process.22
Quantitative Structure Activity Relationship (QSAR) analyses from previous in vitro data has been widely used to assist drug discovery in both industry and academia.23 In the past few years, the rise of machine learning has also expanded to drug discovery, with different methods being implemented in a wide range of areas from predicting synthetic routes to biological activity.24,25 Many examples show that prioritizing compounds from machine learning and QSAR models can increase the success rate and save resources.23 Here we have implemented several machine learning methods to develop predictive models from recent public SARS-CoV-2 in vitro inhibition data and then used them to prioritize compounds from different compound libraries for in vitro testing. These efforts will add to the growing list of drugs under assessment for COVID-19.26
EXPERIMENTAL SECTION
Data Curation
Data from the first drug repurposing campaigns for SARS-CoV-2 were used to build a dataset from whole cell inhibition assays.9,10,13,15,16 In assays with several Multiplicity of Infection (MOI) the one closer to the whole dataset was chosen. In machine learning model generation, duplicate compounds with finite activities are averaged into a single entry. Due to the potential for diminished activity, when duplicate compounds were present, only the most active one was retained in the dataset. Additionally, compounds with ambiguous dose-response curves were discarded. Datasets were built with Molecular Notebook software (Molecular Materials Informatics, Inc). In order to evaluate the model performance on an external testing set, a total of 30 molecules was collated from different studies.11,12,21,27–30
Assay Central®
The Assay Central® software (AC) has been previously described.25,31–39 AC employs a series of rules for the detection of problem data for automated structure standardization to generate high-quality data sets and Bayesian machine learning models capable of predicting potential bioactivity for proposed compounds. AC was used to prepare and merge data sets, as well as generate Bayesian models using the ECFP6 descriptor and five-fold cross validation. During model generation, training compounds are standardized (i.e. salts were removed, corresponding acids neutralized), and thresholds for binary activity classification are applied to optimize internal five-fold cross validation metrics. For predictions, AC workflows assign a probability score and applicability score to prospective compounds according to a user-specified model, with prediction scores greater than 0.5 considered active.
Additional Machine Learning Methods
Additional Machine learning algorithms such as Bernoulli Naïve Bayes (bnb), AdaBoost Decision trees (ada), Random Forest (rf), support vector machine classifier (svc), k-Nearest Neighbors (knn) and Deep Learning (DL) were also implemented with ECFP6 fingerprints and five-fold cross validation. Details for the development of these models was previously described in our earlier articles.33,37,38 Bayesian models were also generated with Discovery Studio (Biovia, San Diego, CA) using ECFP6 descriptors where the top and bottom scoring fingerprints were selected for qualitative comparison.
Model Performance
Machine learning model performance was evaluated with different metrics: accuracy, recall, precision, specificity, F1-score, area under receiver operating characteristic curve, Cohen’s kappa, and the Matthews correlation coefficient. The statistics were calculated for both training data with five-fold cross validation, to evaluate training performance, as well as in external testing set, to evaluate model performance in predicting data outside the training set.
Principal Component Analysis
Principal Component Analysis (PCA) was computed for both the SARS-CoV-2 data set as well as SARS-CoV-2 with different compound libraries to assess its chemical space. The scikit-learn40 (0.22.2) PCA algorithm was used to reduce feature dimensionality to three using different molecular descriptors (MW, MolLogP, NR, NArR, NRB, HBA, HBD) and also with EFCP6 fingerprints. Molecular descriptors and fingerprints were generated from the cheminformatics library RDkit (2020.03.1).
Applicability and Reliability Domain Assessment
In order to check if it is valid to apply the model for compounds being predicted and how reliable the predictions are, an applicability and reliability domain assessment was performed. First, the compound applicability within the model is assessed comparing its similarity with the model’s data using both molecular and fingerprint descriptors. If the molecule satisfies both criteria it is considered within the applicability domain and goes to the reliability domain assessment.
The first criterion for the applicability assessment is determined based on whether it fits within the range of the key molecular descriptors of the training set (MW, MolLogP, NRB, TPSA, HBA, HBD). If at least four properties lie within the maximum and minimum values of the model’s data, the molecule is considered similar and goes to the next criterion. The second criterion relies on structural fragment-based similarity measured with Tanimoto coefficient using MACCS fingerprints. The similarity of the MACCS fingerprints for the query compound and all training data is computed using the Tanimoto score. Only 5% of the training set compounds that are most similar to the query compound is used for evaluation (i.e. if the training set has 100 molecules only 5 molecules with more similarity to the query compound are used for the next evaluation). If the Tanimoto score exceeds 0.5 against the 5% of the training set compounds, the model is considered to have enough structural fragments overlap with the query compound and thus the compound goes onto the reliability assessment.
The reliability domain assessment implements k-means clustering methods based on ECFC6 fingerprints to classify the predictions from very high to low reliability. The reliability class depends on four criteria: distance from the major central point of the training data, distance from the closest cluster, closest cluster density and closest cluster distance within the chemical space. Each criterion has different weights and scores, with the second and third having higher priority. If the compound scores 1 in each criterion it is classified as very highly reliable, if that is not the case only the two higher priority criteria are considered for the next classes. The compound is classified as highly reliable if scores a total of 2, moderately reliable if it scores between −1 and 2 or low reliability if it scores less than or equal to −1 in the two higher priority criteria. The scores for each criterion as well as its definition are extensively described in the Supplemental Methods.
Docking in SARS-CoV-2 Spike protein
A region was selected for docking based on the crystal structure interface between the COVID-2 Spike receptor binding domain (RBD) and Angiotensin-converting enzyme 2 (ACE2) using Discovery Studio (Biovia, San Diego CA). CDOCKER was used to generate multiple poses of lumefantrine at this interface using rigid docking within the site of docking generated from the receptor cavities at this interface (9.7 Å radius). Docking parameters were set to default (top 10 hits retained). Ligand interaction energy calculated between the compound and receptor was done post in situ ligand minimization. Both the ligand and receptor within the sphere of docking were considered flexible during this minimization. The minimizing algorithm was “Smart Minimizer” with 1000 max steps and a minimization RMS gradient of 0.001 and an electrostatic spherical cutoff distance of 12 Å.
Expression and purification of Spike RBD of SARS-CoV-2
A codon-optimized gene encoding for SARS-CoV-2 (331 to 528 amino acids, QIS60558.1) was expressed in Expi293 cells (Thermo Fisher Scientific) with human serum albumin secretion signal sequence and fusion tags (6xHistidine tag, Halo tag, and TwinStrep tag) as described before.41 S1 RBD was purified from the culture supernatant by nickel–nitrilotriacetic acid agarose (Qiagen), and purity was confirmed to by >95% as judged by coomassie stained SDS-PAGE. The purified RBD protein was buffer exchanged to 1x PBS prior to analysis by Microscale Thermophoresis.
Microscale Thermophoresis
We used Microscale thermophoresis (MST) to detect binding of lumefantrine to the Spike RBD protein. The experiments were performed according to the manufacturer’s instructions (NanoTemper). Briefly, for protein labeling, 6 μM of protein was used with 3-fold excess NHS dye in MST Buffer (HEPES 10 mM pH 7.4, NaCl 150 mM), using Monolith Protein Labeling Kit RED-NHS 2nd Generation (Amine Reactive). Free dye was removed, and protein eluted in MST buffer, and centrifuged at 15 k rcf for 10 min. Binding affinity measurements were determined using NanoTemper’s Monolith NT.115 Pico (NanoTemper) and were performed using 5 nM protein a serial dilution of compounds, starting at 100 µM in MST buffer containing 5 % glycerol, 1 mM β-Mercaptoethanol and 0.1 % Triton X-100. Spike RBD was incubated at room temperature in presence of compounds for 20 min prior measurement. Samples were then loaded into sixteen standard capillaries (NanoTemper Technologies) and fluorescence was recorded for 20 s using 20 % laser power and 40 % MST power. The temperature of the instrument was set to 23°C for all measurements. After recording the MST time traces, data were analyzed. KD value was calculated from ligand concentration-dependent changes in the fraction bound (Fbound) of Dye-Spike RBD after 10 s of thermophoresis. The assay was performed in quadruplicate and the values reported were generated through the usage of MO Affinity Analysis software (NanoTemper Technologies).
Cell assays
Chemicals and reagents
Lumefantrine was purchased from MedChemExpress (MCE, Monmouth Junction, NJ).
Vero 76 cells Reduction of virus-induced cytopathic effect (Primary CPE assay)
Confluent or near-confluent cell culture monolayers of Vero 76 cells were prepared in 96-well disposable microplates the day before testing. Cells were maintained in MEM supplemented with 5% FBS. For antiviral assays the same medium was used but with FBS reduced to 2% and supplemented with 50 µg/ml gentamicin. The test compound was prepared at four serial log10 concentrations. Five microwells were used per dilution: three for infected cultures and two for uninfected toxicity cultures. Controls for the experiment consist of six microwells that were infected and not treated (virus controls) and six that were untreated and uninfected (cell controls) on every plate. A known active drug was tested in parallel as a positive control drug using the same method as is applied for test compounds. The positive control was tested with every test run.
Growth media was removed from the cells and the test compound was applied in 0.1 ml volume to wells at 2X concentration. Virus, normally at ~60 CCID50 (50% cell culture infectious dose) in 0.1 ml volume was added to the wells designated for virus infection. Medium devoid of virus was placed in toxicity control wells and cell control wells. Plates were incubated at 37 °C with 5% CO2 until marked CPE (>80% CPE for most virus strains) was observed in virus control wells. The plates were then stained with 0.011% neutral red for approximately two hours at 37°C in a 5% CO2 incubator. The neutral red medium was removed, and the cells rinsed 1X with phosphate buffered solution (PBS) to remove residual dye. The PBS was completely removed, and the incorporated neutral red eluted with 50% Sorensen’s citrate buffer/50% ethanol for at least 30 minutes. Neutral red dye penetrates living cells, thus, the more intense the red color, the larger the number of viable cells present in the wells. The dye content in each well was quantified using a spectrophotometer at 540 nm wavelength. The dye content in each set of wells was converted to a percentage of dye present in untreated control wells. The 50% effective (EC50, virus-inhibitory) concentrations and 50% cytotoxic (CC50, cell-inhibitory) concentrations are then calculated by regression analysis. The quotient of CC50 divided by EC50 gives the selectivity index (SI) value. Compounds showing SI values ≥10 are considered active.
Vero 76 cells Reduction of virus yield (Secondary VYR assay)
Active compounds were further tested in a confirmatory assay. This assay was set up like the methodology described above only eight half-log10 concentrations of inhibitor were tested for antiviral activity and cytotoxicity. After sufficient virus replication occurs (3 days for SARS-CoV-2), a sample of supernatant was taken from each infected well (three replicate wells are pooled) and tested immediately or held frozen at −80 °C for later virus titer determination. After maximum CPE was observed, the viable plates were stained with neutral red dye. The incorporated dye content was quantified as described above to generate the EC50 and CC50 values. The VYR test is a direct determination of how much the test compound inhibits virus replication. Virus yielded in the presence of test compound was titrated and compared to virus titers from the untreated virus controls. Samples were collected 3 days after infection. Titration of the viral samples (collected as described in the paragraph above) was performed by endpoint dilution.42 Serial 1/10 dilutions of virus were made and plated into 4 replicate wells containing fresh cell monolayers of Vero 76 cells. Plates were then incubated, and cells scored for presence or absence of virus after distinct CPE was observed (3 days after infection), and the CCID50 calculated using the Reed-Muench method.42 The 90% (one log10) effective concentration (EC90) was calculated by regression analysis by plotting the log10 of the inhibitor concentration versus log10 of virus produced at each concentration. Dividing EC90 by the CC50 gives the SI value for this test.
Calu3 cells
Calu3 (ATCC, HTB-55) cells were pretreated with test compounds for 2 hours prior to continuous infection with SARS-CoV-2 (isolate USA WA1/2020) at a MOI=0.5. Forty-eight hours post-infection, cells were fixed, immunostained, and imaged by automated microscopy for infection (dsRNA+ cells/total cell number) and cell number. Sample well data was normalized to aggregated DMSO control wells and plotted versus drug concentration to determine the IC50 (infection: blue) and CC50 (toxicity: green).
Caco-2 cells Virus Yield Reduction
For the Caco-2 VYR assay, the methodology was identical to the Vero 76 cell assay other than the insufficient CPE is observed on Caco-2 cells to allow EC50 calculations. Supernatant from the Caco-2 cells were collected on day 3 post-infection and titrated on Vero 76 cells for virus titer as before.
Cytotoxicity
Vero CCL81 cells and A549 cells were cultivated at 5% CO2 and 37°C using Dulbecco’s Modified Eagle Medium supplemented with 10% heat-inactivated fetal bovine serum. For this experiment, Vero cells were seeded at a density of 10⁴ cells/ well in a 96 well plate prior incubation with a serial dilution of compounds of interest and controls for 72 h. After drug treatment, cells were next incubated with 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (Sigma- Aldrich M5655) for 4 h followed by formazan crystal solubilization with isopropanol and absorbance readings at OD570. Cellular viability was expressed as a percentage relative to vehicle treated control.
Results
Data Curation
In vitro SARS-CoV-2 data was initially collated from five drug repurposing studies leading to a data set of 63 molecules with mean activity of 15.94 ± 22.45 μM.9,10,13,15,16 The external testing set collated from different studies has 30 molecules and a mean activity of 34 ± 42 μM.11,12,21,29,30 Most assays were performed with different Vero cell lines and inhibition was measured with viral RNA quantification, cytopathogenic effects or immunofluorescence methods with MOI and incubation time varying from 0.01–0.05 and 24–72 h respectively (Figure S1). The threshold set for activity classification by the Bayesian model generated with AC was 6.65 μM, with a final ratio of 52% actives in the training set and 37% in the external test set. The molecules in both the training and test set are available in the Supplemental Data.
Machine Learning Models
Machine learning models were developed with AC as well as several other machine learning methods available to us. This five-fold cross validation comparison shows the different prediction statistics for all machine learning algorithms implemented with the training data only (Table 1). AC outperformed all of these at the same threshold of 6.65 μM with Rf coming the next closest. These machine learning models were used for external validation.
Table 1.
Five-fold cross validation statistics for all SARS-CoV-2 machine learning models implemented using ECFP6 fingerprints.
| ACC | AUC | CK | MCC | Pr | Recall | Sp | F1 | |
|---|---|---|---|---|---|---|---|---|
| AC | 0.81 | 0.78 | 0.62 | 0.64 | 0.78 | 0.88 | 0.73 | 0.83 |
| rf | 0.75 | 0.74 | 0.49 | 0.5 | 0.73 | 0.82 | 0.67 | 0.77 |
| knn | 0.71 | 0.71 | 0.43 | 0.42 | 0.71 | 0.76 | 0.67 | 0.74 |
| svc | 0.7 | 0.69 | 0.39 | 0.4 | 0.68 | 0.79 | 0.6 | 0.73 |
| bnb | 0.68 | 0.68 | 0.36 | 0.36 | 0.7 | 0.7 | 0.67 | 0.7 |
| ada | 0.64 | 0.63 | 0.27 | 0.26 | 0.65 | 0.67 | 0.6 | 0.66 |
| DL | 0.65 | 0.65 | 0.3 | 0.3 | 0.66 | 0.67 | 0.63 | 0.66 |
ACC: Accuracy, AUC: Area under curve, CK: Cohen’s Kappa, MCC: Matthews correlation coefficient, Pr: Precision, Sp: Specificity, F1: F1 Score. bnb: Bernoulli Naïve Bayes, ada: AdaBoost Decision trees, rf: Random Forest, svc: support vector machine classifier, knn: k-Nearest Neighbors and DL: Deep Learning (DL).
External Validation
The performance of the machine learning models on the external testing data is shown in Table 2. The external validation was used to measure model performance using data from different studies outside of the training set. svc and knn had slightly better overall statistics with the best balance between recall and specificity when compared to all other machine learning models.
Table 2.
Prediction statistics with the external data for all SARS-CoV-2 machine learning models implemented.
| ACC | AUC | CK | MCC | Pr | Recall | Sp | F1 | |
|---|---|---|---|---|---|---|---|---|
| AC | 0.62 | 0.58 | 0.17 | 0.17 | 0.50 | 0.40 | 0.76 | 0.44 |
| rf | 0.63 | 0.57 | 0.10 | 0.11 | 0.42 | 0.30 | 0.80 | 0.35 |
| knn | 0.67 | 0.6 | 0.21 | 0.21 | 0.50 | 0.40 | 0.80 | 0.44 |
| svc | 0.70 | 0.57 | 0.34 | 0.34 | 0.54 | 0.60 | 0.75 | 0.57 |
| bnb | 0.50 | 0.49 | −0.09 | −0.09 | 0.27 | 0.30 | 0.60 | 0.28 |
| ada | 0.53 | 0.49 | 0.00 | 0.00 | 0.33 | 0.40 | 0.60 | 0.36 |
| DL | 0.63 | 0.56 | 0.15 | 0.15 | 0.44 | 0.40 | 0.75 | 0.42 |
ACC: Accuracy, AUC: Area under curve, CK: Cohen’s Kappa, MCC: Matthews correlation coefficient, Pr: Precision, Sp: Specificity, F1: F1 Score. bnb: Bernoulli Naïve Bayes, ada: AdaBoost Decision trees, rf: Random Forest, svc: support vector machine classifier, knn: k-Nearest Neighbors and DL: Deep Learning (DL).
Chemical Space
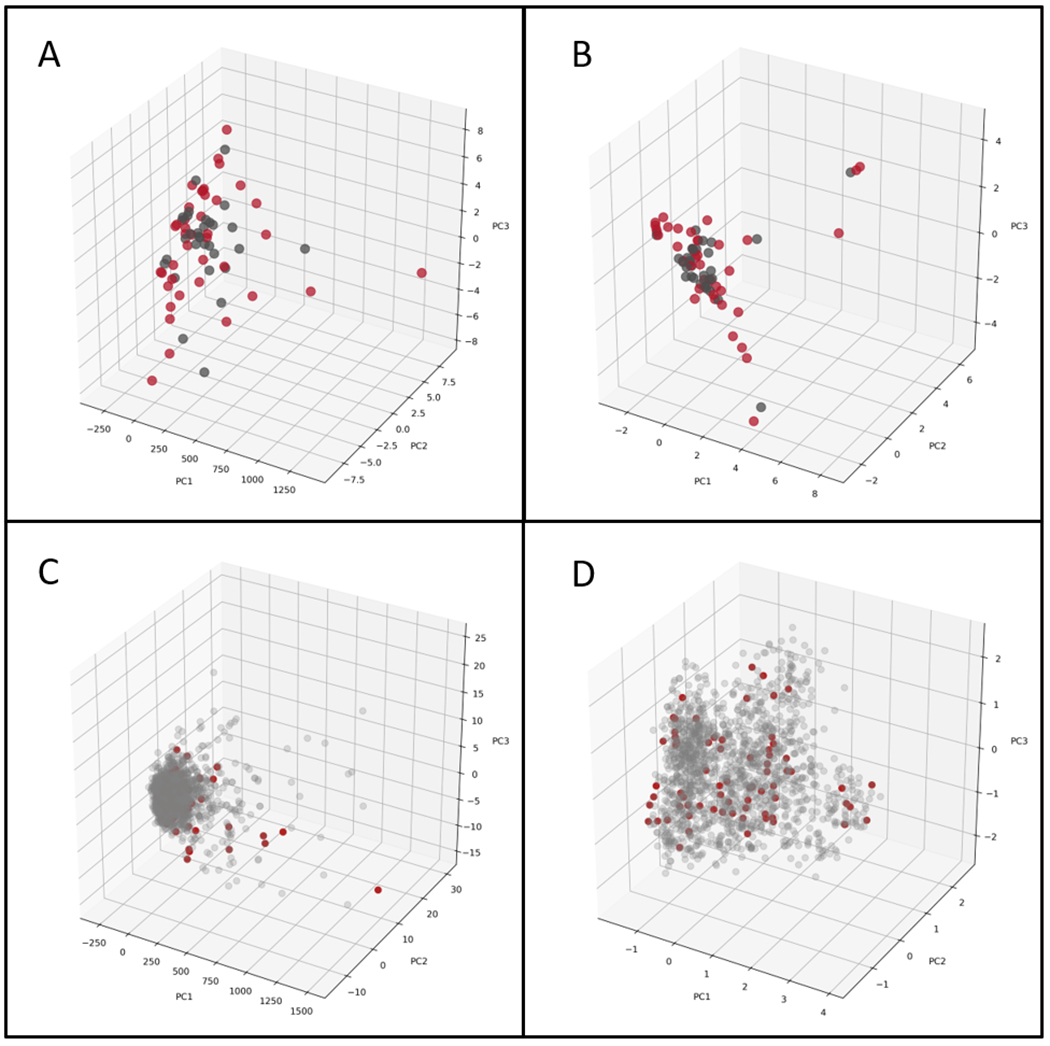
The PCA of the model training set alone shows that the SARS-CoV-2 chemical space is well distributed with active and inactive molecules well mixed when analyzed using molecular either fingerprint descriptors (Figure 1). When compared with the Prestwick Chemical Library (PwCL), a library of predominantly FDA approved drugs, the SARS-CoV-2 data lie within a big cluster with molecular descriptors and is more widely distributed when using the fingerprint descriptors (Figure 1C and andDD).
Applicability and Reliability Domain Assessment of External Test Set
The applicability and reliability domain assessment of the external test set was determined for each molecule as described in methods to see how the test set compares with the training data. Molecules in the applicability domain are considered suitable for the model predictions due to similarity based on structural and molecular properties with the training data, whereas the reliability value is a measurement of how reliable the predictions are and uses different clustering metrics to determine its value.
From 30 molecules in the external test set, 22 were within the training data applicability domain and had their reliability value calculated. Most molecules that fell within the applicability domain had high or very high reliability values, with only 36% showing moderate reliability, so, most molecules obey the similarity criteria and are not far away from dense clusters. In comparison, with the Assay Central applicability score, which accounts only for structural similarity of the query compound with the training data, only 10 molecules were considered within the domain with a higher reliability, suggesting it is likely more conservative. Indeed, with the external test and training set PCA we can see that most molecules superimpose with few of them distant from each other (Figure S2). Therefore, similarity together with clustering methods are more suitable for applicability and reliability assessment compared with only structural similarity, as seen by the PCA.
Prospective Prediction
A selection of FDA approved drugs available to us in our relatively small in-house compound collection was scored with the AC Bayesian model. A selection of some of the best scoring molecules (Table 3) was used to identify and prioritize compounds for in vitro testing. Not surprisingly, several of the top-ranked molecules are antimalarials like lumefantrine and artesunate or kinase inhibitors like nilotinib. AC Applicability score is the similarity of the compound with the training data, compounds are ranked by reliability which may provide some degree of confidence in these predictions.
Table 3.
Prospective prediction compounds predicted.
| Name | Prediction Score | AC Applicability Score | Reliability |
|---|---|---|---|
| Lumefantrine | 0.67 | 0.5 | High |
| Artesunate | 0.62 | 0.38 | High |
| Naloxone | 0.62 | 0.39 | High |
| Nilotinib | 0.70 | 0.70 | Moderate |
| Tiamulin | 0.70 | 0.40 | Moderate |
| Budesonide | 0.65 | 0.41 | Moderate |
| Tetrabenazine | 0.7 | 0.7 | Low |
Antiviral Activity Assays of Predicted Compounds
Lumefantrine was initially selected as it is a widely available antimalarial and was subsequently tested in Vero 76, Calu-3 and Caco-2 cells. The IC50 or EC90 data for each cell line were not indicative of useful in vitro activity (Table 4) when compared with the cytotoxicity (Figure S3). However, the Vero 76 neutral red assay data demonstrated an EC50 far lower than the CC50. Budesonide, tiamulin fumarate and tetrabenazine were also tested in Caco-2 cells and demonstrated inhibition comparable to cytotoxicity. Tiamulin had an EC90 that was lower than the CC50 (Table 4).
Table 4.
IC50, EC90 and CC50 values for lumefantrine Vero 76, Calu-3, and Caco-2.
| Compound | Cell line | Assay detail | IC50 or EC90 | CC50 | SI |
|---|---|---|---|---|---|
| Lumefantrine | Vero 76 | Visual (Cytopathic effect/Toxicity) | EC50 > 60 µM | >60 µM | 0 |
| Lumefantrine | Vero 76 | Neutral Red (Cytopathic effect/Toxicity) | EC50 54 µM | 177 µM | 3.2 |
| Lumefantrine | Calu-3 | - | IC50 >20 µM | >20 µM | 0 |
| Lumefantrine | Caco-2 | - | EC9010 µM | 10 µM | 0 |
| Budesonide | Caco-2 | Visual (Virus yield reduction)/Neutral Red (Toxicity) | EC90 >10 µM | 10 µM | 0 |
| Tiamulin fumarate | Caco-2 | Visual (Virus yield reduction)/Neutral Red (Toxicity) | EC90 65 µM | >100 µM | > 1.5 |
| Tetrabenazine | Caco-2 | Visual (Virus yield reduction)/Neutral Red (Toxicity) | EC90 >100 | >100 µM | 0 |
Microscale Thermophoresis
Microscale Thermophoresis (MST) was employed to measure lumefantrine’s binding affinity to the SARS-Cov2 Spike RBD protein. MST is a sensitive method that can be used to assess biomolecular interactions in solution for a variety of binding partners of various molecular sizes.43,44 Change in its thermophoretic movement45 allows quantifying the affinity of the interaction between the binding partners. Figure 2 shows that Lumefantrine binds to the Spike RBD with an estimated Kd of ~250 nM.

MicroScale Thermophoresis binding analysis for the interaction between Spike RBD and lumefantrine. The concentration of labeled Spike RBD was maintained at 5 nM, and the ligand concentration varied from 125 μM to 3.8 nM. The serial titrations result in measurable changes in the fluorescence signal within a temperature gradient that was used to calculate the dissociation constant (Kd = 259 ± 78 nM). The curve is shown as Fraction Bound [-] against lumefantrine concentration on a log scale.
Docking in the Spike Protein
The energy of interaction of each of the docked poses of lumefantrine was calculated following a ligand minimization step and the most energetically favorable pose is displayed (−145.35 kcal/mol) (Figure S4).
Discussion
One of the challenges for addressing novel viral outbreaks like SARS-CoV-2 is the selection of drugs to test. Testing capacity, even for in vitro antiviral activities is likely to be very low at the onset of an outbreak, making compound selection even more critical. In the case of SARS-CoV-2, much of the initial focus early on was on molecules that had previously shown activity against the related viruses SARS or MERS.10,46 The training set for the current model is therefore not a random sampling of drug property space as in many cases it is biased towards molecules with some history against these viruses. When compared with the PwCL, a library of mostly FDA approved drugs, all molecules superimpose in the same property space highlighting the suitability of the model for drug repurposing. Even with a relatively small training dataset the first SARS-CoV-2 machine learning models evaluated have shown acceptable five-fold cross validation statistics, with almost all metrics greater than random and AUC >0.75 for AC (Table 1). When compared with various machine learning methods AC outperforms all of them with the SARS-CoV-2 training set, but this may be due to the threshold being set as optimal for AC. However, choosing different values could imbalance the training set as well as remove important compounds from the active group. More important than performance of the training set is that of the external test set, since prospective predictions are the goal of such models. For external validation all machine learning models had generally poor performance, with AUC of 0.6 (Table 2) when compared to the training set 5 fold cross validation (Table 1). Taking into account the small number of molecules and that some test set molecules lie outside the applicability domain, the performance is however acceptable. svc had the highest overall score for the external test set, predicting 60% of the active molecules which is in contrast to this models modest performance in five-fold cross validation. The performance of svc in predicting this biological activity is in accordance with several studies in different datasets.33,37,38,47 Therefore, the machine learning models described here appear suitable for prospective predictions.
The applicability and reliability assessment shows that 73% of the test set molecules lie within the model applicability domain with high to moderate reliability, so poor performance in external validation occurs because there is not a clear boundary in the model’s feature space that can correctly classify external data. Increasing the number of molecules might incorporate new features in both actives and inactive molecules which can also increase model performance in both training and external data.
The SARS-CoV-2 training and test set can also be merged to increase data set size and applicability domain. When this is done the AC model with merged training and test data has slightly worse statistics (ACC: 0.76, AUC:0.79, CK: 0.53, MCC: 0.75, Pr: 0.76, Recall: 0.76, Sp: 0.77, F1: 0.76), but a higher applicability domain. The PCA of the training and test data confirms this wide chemical property space (Figure S2), the PCA of the updated model (training+test set) is much more balanced and broader than the earlier one (Figure S2) versus Figure 1B. Without external validation, we cannot assess how predictions of compounds outside the applicability domain perform. As model statistics were comparable it is expected that compounds outside of this applicability domain would obviously have unreliable predictions, however this may be offset by a higher domain which can increase reliability of some compounds.
The molecules in the dataset do not have a common scaffold, but there are several common structural features that occur in active/inactive molecules that can be highlighted, such as tertiary amines and aliphatic chains in active molecules and phenyl rings and peptide molecule features in inactive molecules (Figure S5). These most common active features appear in chloroquine, tripanarol and tilorone, while the inactive features appear in darunavir, amprenavir and ritonavir (Figure 3). The lack of common scaffolds and features that appear in more than 30% of the active or inactive molecules shows how different and diverse the active molecules are, which also makes classification models a relatively difficult task.
The performance of a predictive model is highly dependent on the curation and the quality of the data used. One of the main problems that comes from building models with biological data from different laboratories is data reproducibility and assay standardization.48 Cell based assays of viral infections have many parameters that can affect the compound potency, e.g., cell lines, MOI and assay readout as well as other factors.49 From all inhibition assays for SARS-CoV-2 collated to date, most studies use MOI of 0.01–0.05 (73% of data), different Vero cell lines (77% of data) and qRT-PCR (60% of data), however there is no clear definition of compound addition time post infection (Figure S1).
Besides this, even assays with the same or similar conditions have differences in ‘control’ compounds such as the use of chloroquine or remdesivir (e.g. Vero cells (EC50 1.65 μM), human epithelial cultures (EC50 0.01 μM) and Calu-3 (EC50 0.28 μM))50 which can impact machine learning model building. If we keep only studies with the most in common, there is likely not enough data to build a model, while merging all studies will have problems caused by the retention of data with different assay parameters. It was previously shown that for Ebola infections in VeroE6 cells the change in the compound potency at different time post infection is lower when using MOI of 0.01–0.1 therefore, merging different assays with the same cell line and low MOI is likely a good choice to avoid data inconsistency.49
It should be noted that most of the in vitro data collated to date uses Vero or Vero E6 cells for inhibition assays. Although these cells lines have high ACE2 expression levels, they lack a TMPRSS2 gene. Priming of viral S proteins can occur with the host cell protease TMPRSS2 and Cathepsin L and is essential for SARS-CoV-2 entry.51,52 Therefore, inhibition assays with cells that do not express TMPRSS2 should be avoided as they might miss compounds that could inhibit the protein and instead find compounds that prevent virus entry by inhibiting only Cathepsin L. In order to avoid these problems with the TMPRSS2 and Cathepsin L gene, cell lines like Calu-3 or modified Vero cell lines should be used instead.53 SARS-CoV-2 Spike contains a furin cleavage site, which may reduce the dependence of SARS-CoV-2 on target cell proteases (TMPRSS2/ cathepsin L) for entry.51,54 Furin is also abundantly expressed in human bronchial epithelial cells, thus potentially extending its cellular tropism.55
From the 7 compounds prioritized for testing in our laboratory using the machine learning model, lumefantrine was prioritized for in vitro testing due to limited testing capabilities available to us at the time. We tested this molecule in Vero 76, Caco-2 and Calu-3 but these did not indicate significant activity. Interestingly, while this study was in progress, we also became aware of recently published work describing an EC50 23.17 µM and CC50 >100 µM SI > 4 in Vero E6 cells 56,57 approximately 2 fold more potent than in this study. We also performed experiments to clarify the potential mechanism of action of lumefantrine. We measured binding of lumefantrine to the glycosylated Spike RBD protein from SARS-Cov-2 using microscale thermophoresis. The dissociation constant KD determined using this technique is 259 nM. This Kd is ~ 17 times weaker when compared to ACE2, which has been reported to be ~ 15 nM by different techniques.58,59 Binding affinity experiments using MST were performed with the RBD, which binds to ACE2. Despite lumefantrine binding to the Spike RBD, this affinity might not be sufficiently high enough to compete with ACE2 or lumefantrine may instead bind to the RBD in a location without affecting binding to the ACE2 receptor.
Lumefantrine is a first-line antimalarial used only in combination with artemether to treat uncomplicated P. falciparum malaria.60 The artemisinin-based combination therapy artemether-lumefantrine (AL; Coartem) has been approved by the FDA since 2009 as a treatment for uncomplicated malaria. The exact mechanism of action is unknown with some studies suggesting the inhibition of nucleic acid and protein synthesis through inhibition of β-haematin formation.61 Although several antimalarials have been evaluated as antivirals against different viruses62–65, lumefantrine has only been tested in combination with artemether in a few observational studies, without supporting in vitro data.66 The drug combination reduced viral load in the urine of children infected with HCMV and malaria, and showed less efficacy in reducing the risk of death by Ebola infection when compared with artesunate-amodiaquine.67,68 Future studies could be performed to compare the activity of lumefantrine and artemether-lumefantrine in SARS-CoV-2 infections since it is the most widely used antimalarial combination and could also represent an accessible treatment in some countries. Lumefantrine is metabolized in the liver to desbutyl-lumefantrine, which has a longer half-life and shows higher potency against P.falciparum infections, therefore the metabolite could also be tested to see if it also has antiviral activity.69 It should be noted that lumefantrine is not as potent against SARS-CoV-2 as other antimalarials such as pyronaridine, and this may be due to numerous factors such as cell penetration and differences in structure which could be important for the targeting or mechanism.70
In the process of this work, new data was continually being published and the machine learning models were regularly updated to increase performance in terms of both training and external test set validation. The latest model for SARS-CoV-2 is available at www.assaycentral.org and consists of over 500 molecules (Figure S6). The higher number of molecules compared to the first model shows how fast data was published on SARS-CoV-2, keeping track and curating this are of great importance for future machine learning applications. Despite the ChEMBL effort to gather and curate all assays related to the virus further detailed literature review is still important, since some pre-prints and papers are not included in these databases.
The addition of new compounds improved the AUC score and expanded the chemical space, in accordance with our previous discussions. As more high-throughput screening campaigns for SARS-CoV-2 were performed, the number of inactive molecules available increased, allowing one to test the best active/inactive ratio with the addition of inactive molecules through random selection. The inactive molecules not used for model building can be further used to filter out false positives after compound predictions, increasing the prediction outcome. Beyond that, the higher model specificity due to more inactive molecules, reduces the number of positive predictions by prioritizing the major class classification, with a smaller list of positive predictions it becomes easier to choose which molecules are going to be tested in-vitro thus highlighting the importance of HTS data for model building.
Artesunate and nilotinib were predicted with our model, however they were active in different cell lines as published by others. Artesunate had an IC50 of 1.76 µM in Calu-3 cells65, while nilotinib had sub-micromolar potency in Vero cells71–73, showing that machine learning models built with different sources of SARS-CoV-2 data can be useful to assist COVID-19 drug discovery. Budesonide was also predicted with our model and it was previously shown to have inhibitory activity for HCoV-229E replication and cytokine production in human respiratory epithelial cells in vitro, in combination with glycopyrronium and formoterol.74 The activity against SARS-CoV-2 in Caco-2 cells was comparable to the CC50, indicative of no activity. Currently, budesonide is in several clinical trials with patients with COVID19, including investigation as a treatment for COVID-19 patients who are not in hospital, to verify if daily high dose inhaled corticosteroids for 28 days will reduce the chances of severe respiratory illness needing hospitalization (NCT04416399). The literature was silent on the in vitro activity of tiamulin, naloxone and tetrabenzine against SARS-CoV-2. When tested in Caco-2 cells in this study tiamulin demonstrated weak inhibition (EC90 65 µM, CC50 >100 µM) while tetrabenazine was inactive with an EC90 identical to the CC50 (Table 4). As we have frequently observed, activity testing in multiple different cell lines is important before ruling compounds out from further testing.
Conclusion
In conclusion, we have shown machine learning models perform well with internal cross validation, external validation, however prospective prediction is much more difficult due to the limited availability of in vitro testing. Importantly, machine learning enabled us to find additional active molecules for SARS-CoV-2 as validated either by ourselves or others. These machine learning models could also be used to prioritize further compounds in future which have both a high prediction score and reliability. This will be expected to return more reliable predictions that when combined with drug discovery expertise can help prioritize compounds in future for in vitro testing. These efforts also complement the increasing number of examples of applying machine learning methods to SARS-CoV-2 drug discovery in order to find new molecules for clinical testing.75–79
Supplementary Material
Supplemental data
SARS-CoV-2 external test set
SARS-CoV-2 training set
SARS-CoV-2 latest dataset
Acknowledgements
We would like to kindly acknowledge Dr. Nancy Baker and Natasha Baker for their help in collating SARS-CoV-2 published data. We graciously thank Dr. Sara Cherry and Dr. David Schultz for the Calu-3 high-content SARS-CoV-2 studies performed by the University of Pennsylvania High-throughput Screening Core and the Cherry laboratory.
Dr. Mindy Davis and colleagues are gratefully acknowledged for assistance with the NIAID virus screening capabilities. We also kindly acknowledge Dr’s Sungjun Beck, Nathan Beutler, Thomas Rogers, Frank Scholle, Ethan James Fritch, Nathaniel John Moorman, Ralph S. Baric and Kenneth H. Pearce for many discussions and collaborations. Dr. Alex M. Clark is acknowledged for assistance with Assay Central.
Grant information
Per Subcontract: “This material is based upon work supported by the Defense Advanced Research Projects Agency (DARPA) under Contract No. HR001119C0108.” Per DISTAR Form: “The views, opinions, and/or findings expressed are those of the author(s) and should not be interpreted as representing the official views or policies of the Department of Defense or the U.S. Government.”
We kindly acknowledge NIH funding: R44GM122196–02A1 from NIGMS (PI – Sean Ekins) and support from DARPA (HR0011–19-C-0108; PI: P. Madrid) is gratefully acknowledged. Distribution Statement “A” (Approved for Public Release, Distribution Unlimited). The views, opinions, and/or findings expressed are those of the author and should not be interpreted as representing the official views or policies of the Department of Defense or the U.S. Government. FAPESP funding: 2019/25407–2 (PI – Glaucius Oliva), 2019/27626–3 (PI- Tatyana Tavella). This project was also supported by the North Carolina Policy Collaboratory at the University of North Carolina at Chapel Hill with funding from the North Carolina Coronavirus Relief Fund established and appropriated by the North Carolina General Assembly. FAPESP grant # 2020/05369–6 (PI Fabio Costa).
Collaborations Pharmaceuticals, Inc. has utilized the non-clinical and pre-clinical services program offered by the National Institute of Allergy and Infectious Diseases.
Footnotes
Conflicts of interest
SE is CEO and owner of Collaborations Pharmaceuticals, Inc. DHF, KMZ, TRL, AP are employees of Collaborations Pharmaceuticals, Inc. Other authors have no conflicts.
Supporting Information Available
Supporting information available includes supplemental figures describing cell-based parameters, PCA of test and training set, cytotoxicity of lumefantrine in different cell lines, docking of lumefantrine in the spike protein, good and bad molecular features for the Bayesian machine learning model and an ROC plot for 5-fold cross validation. We also include supplemental methods describing the reliability domain criteria definition and scores and tables describing the training set, external test data and supplemental references. All sdf files for datasets described are available on FigShare (see below).
Data and software availability
We have made the datasets available on FigShare (https://figshare.com/account/home#/projects/118959). The models generated with commercial software are available upon request. The latest iterations of the COVID-19 models are available at www.assaycentral.org.
References
Full text links
Read article at publisher's site: https://doi.org/10.1021/acs.jcim.1c00683
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8574161
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/111774814
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1021/acs.jcim.1c00683
Article citations
Significance of Artificial Intelligence in the Study of Virus-Host Cell Interactions.
Biomolecules, 14(8):911, 26 Jul 2024
Cited by: 0 articles | PMID: 39199298 | PMCID: PMC11352483
Review Free full text in Europe PMC
Identification of SARS-CoV-2 Main Protease Inhibitors Using Chemical Similarity Analysis Combined with Machine Learning.
Pharmaceuticals (Basel), 17(2):240, 12 Feb 2024
Cited by: 1 article | PMID: 38399455 | PMCID: PMC10892746
How Deep Learning in Antiviral Molecular Profiling Identified Anti-SARS-CoV-2 Inhibitors.
Biomedicines, 11(12):3134, 24 Nov 2023
Cited by: 1 article | PMID: 38137356 | PMCID: PMC10740425
Practical guidelines for the use of gradient boosting for molecular property prediction.
J Cheminform, 15(1):73, 28 Aug 2023
Cited by: 6 articles | PMID: 37641120 | PMCID: PMC10464382
A review of SARS-CoV-2 drug repurposing: databases and machine learning models.
Front Pharmacol, 14:1182465, 04 Aug 2023
Cited by: 5 articles | PMID: 37601065 | PMCID: PMC10436567
Review Free full text in Europe PMC
Go to all (19) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
Clinical Trials
- (1 citation) ClinicalTrials.gov - NCT04416399
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Combined deep learning and molecular docking simulations approach identifies potentially effective FDA approved drugs for repurposing against SARS-CoV-2.
Comput Biol Med, 141:105049, 20 Nov 2021
Cited by: 17 articles | PMID: 34823857 | PMCID: PMC8604796
Identification of novel compounds against three targets of SARS CoV-2 coronavirus by combined virtual screening and supervised machine learning.
Comput Biol Med, 133:104359, 30 Mar 2021
Cited by: 84 articles | PMID: 33845270 | PMCID: PMC8008812
In silico prediction of potential inhibitors for the main protease of SARS-CoV-2 using molecular docking and dynamics simulation based drug-repurposing.
J Infect Public Health, 13(9):1210-1223, 16 Jun 2020
Cited by: 106 articles | PMID: 32561274 | PMCID: PMC7297718
Staying Ahead of the Game: How SARS-CoV-2 has Accelerated the Application of Machine Learning in Pandemic Management.
BioDrugs, 37(5):649-674, 18 Jul 2023
Cited by: 1 article | PMID: 37464099
Review
Funding
Funders who supported this work.
Funda????o de Amparo ?? Pesquisa do Estado de S??o Paulo (3)
Grant ID: 2020/05369-6
0 publications
Grant ID: 2019/27626-3
0 publications
Grant ID: 2019/25407-2
0 publications
NCCIH NIH HHS (1)
Grant ID: R43 AT010585
NIGMS NIH HHS (1)
Grant ID: R44 GM122196
National Center for Complementary and Integrative Health (1)
Grant ID: 1R43AT010585-01
North Carolina General Assembly
U.S. Department of Defense (1)
Grant ID: HR0011-19-C-0108
U.S. Department of Health and Human Services (1)
Grant ID: R44GM122196-02A1