Abstract
Question
Cystic fibrosis (CF) is due to pathogenic variants in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Recent improvements have enabled pharmacological therapy aiming at restoring mutated CFTR expression and function. CFTR "modulators" have revolutionised the CF therapeutic landscape, particularly the last approved, Trikafta. This drug combination is indicated by the United States Food and Drug Administration and very recently by the European Medicines Agency for genotypes carrying at least one copy of CFTR with the F508del pathogenic variant. However, several genotypes are not yet eligible for Trikafta treatment.Materials/patients and methods
We exploited an innovative cellular approach allowing highly efficient in vitro expansion of airway epithelial stem cells (AESCs) through conditional reprogramming from nasal brushing of CF patients. This approach, coupled to the development of AESC-derived personalised disease models, as organoids and air-liquid interface (ALI) cultures, revealed highly suitable for CFTR pharmacological testing.Results and answer to the question
We fully validated the experimental models and implemented the CFTR functional assays and biochemical CFTR protein characterisation, which allowed the evaluation of the efficacy of clinically available modulators in restoring CFTR maturation and function of each patient-derived "avatar" (theratyping). F508del homozygous genotypes, used as controls, confirmed the higher clinical activity of Trikafta in comparison with older modulators. In addition, Trikafta showed its efficacy on three rare genotypes previously not eligible for treatment with modulators, opening the way to clinical translation. Finally, encouraging results for innovative drug combinations were obtained.Free full text

Theratyping cystic fibrosis in vitro in ALI culture and organoid models generated from patient-derived nasal epithelial conditionally reprogrammed stem cells
Abstract
Question
Cystic fibrosis (CF) is due to pathogenic variants in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Recent improvements have enabled pharmacological therapy aiming at restoring mutated CFTR expression and function. CFTR “modulators” have revolutionised the CF therapeutic landscape, particularly the last approved, Trikafta. This drug combination is indicated by the United States Food and Drug Administration and very recently by the European Medicines Agency for genotypes carrying at least one copy of CFTR with the F508del pathogenic variant. However, several genotypes are not yet eligible for Trikafta treatment.
Materials/patients and methods
We exploited an innovative cellular approach allowing highly efficient in vitro expansion of airway epithelial stem cells (AESCs) through conditional reprogramming from nasal brushing of CF patients. This approach, coupled to the development of AESC-derived personalised disease models, as organoids and air–liquid interface (ALI) cultures, revealed highly suitable for CFTR pharmacological testing.
Results and answer to the question
We fully validated the experimental models and implemented the CFTR functional assays and biochemical CFTR protein characterisation, which allowed the evaluation of the efficacy of clinically available modulators in restoring CFTR maturation and function of each patient-derived “avatar” (theratyping). F508del homozygous genotypes, used as controls, confirmed the higher clinical activity of Trikafta in comparison with older modulators. In addition, Trikafta showed its efficacy on three rare genotypes previously not eligible for treatment with modulators, opening the way to clinical translation. Finally, encouraging results for innovative drug combinations were obtained.
Short abstract
Despite approval of Trikafta, a fraction of cystic fibrosis patients with rare genotypes are still lacking modulator therapies. Conditionally reprogrammed nasal cell-based in vitro models may allow theratyping for each patient for personalised treatment. https://bit.ly/3ylOJ28
Introduction
Cystic fibrosis (CF) is caused by pathogenic variants of the cystic fibrosis transmembrane conductance regulator (CFTR) gene [1]. In the lungs of CF patients, defective CFTR protein results in a dehydrated surface liquid and compromised mucociliary clearance. The resulting thick and dense mucus makes the airway prone to chronic infection and inflammation, which consequently leads to airway structure damage and, eventually, respiratory failure. In past years, therapies have been mainly symptomatic [2]. More than 2100 different variants (CFTR1 database) of CFTR have been described and, initially, grouped into six classes based on their phenotypic effects [3]. Class II pathogenic variants cause protein trafficking defects, which may result in premature degradation of CFTR or accumulation of immature nonfunctional protein in the cytoplasm. The most common pathogenic variant, F508del, belongs to this class. Recently, advances in the understanding of the molecular genetics of CF have led to the development of novel mutation-specific therapies [4]. For example, drugs targeting class II pathogenic variants would correct protein folding and endoplasmic reticulum export defects, while those targeting class III pathogenic variants would restore correct gating. Current therapies approved for specific CFTR variants, all from Vertex Pharmaceuticals, consist of the CFTR potentiator ivacaftor/VX770 (commercial name Kalydeco) or combination of the potentiator ivacaftor with the correctors lumacaftor/VX809 (Orkambi) or tezacaftor/VX661 (Symdeko). Last year, after highly promising clinical trials, the triple combination Trikafta, consisting of ivacaftor-tezacaftor-elexacaftor/VX445 (an innovative corrector), has been approved by the United States Food and Drug Administration (FDA) for genotypes bearing at least one copy of F508del and is proving highly encouraging so far [5, 6]. In recent months this drug combination has been approved for commercialisation in the European Union (EU) by the European Medicines Agency (with the name Kaftrio) for the same genotypes. Nevertheless, a large number of pathogenic genotypes are not included in the list of genotypes eligible for Trikafta treatment and the drug is not yet provided by the national health systems of EU states.
Drugs allowing fully improved restoration of CFTR function are still needed, and development of more effective ones for all patients with CF is challenging. This is especially true for those with rare pathogenic variants, whose clinical trials are hampered by the scarcity of patients. The success of personalised therapy of CF has been hindered by poor functional characterisation of CFTR variants and from the limited number of mutational classes studied so far. Moreover, the great majority of CFTR pathogenic variants show multiple biochemical defects and seem to belong to multiple mutational classes. For this reason, a reclassification of CFTR pathogenic variants in a wider number of mutational types is underway, each with a more specific multiclass functional description [7]. This enhanced classification is aimed at identifying specific mutational types responding to specific therapy, an approach called “theratyping” [8] (a methodology approved in the United States by the FDA).
Various cellular in vitro CF models have been used to validate personalised experimental strategies, including immortalised human bronchial epithelial cell lines and primary airway epithelial cells; the former with the limitations of immortalised cells and the latter with the limitation of low efficiency of culture establishment and limited supply of experimental material. Based on their intrinsic properties, stem cells would allow the generation of large amounts of functionally differentiated cells, representing the best cellular candidates for experimental CF models. The possibility to obtain with high efficiency large amounts of primary airway epithelial stem cells (AESCs) from CF patients with various gene defects would represent a pivotal advancement for CF research, particularly for the investigation of treatment response. Of note, CF patients with rare genetic variants, their low frequency being a major obstacle for the accomplishment of valuable clinical trials, would particularly benefit from improved and personalised in vitro disease models [8–10].
The so-called “culture reprogramming condition” (CRC), consisting of co-culture of primary epithelial cells with irradiated mouse fibroblasts as a feeder layer, in the presence of the rho-kinase (Rock) inhibitor Y-27632, induces reprogramming of differentiated epithelial cells associated to active cell proliferation, generating long-term cultures of stem-like epithelial cells from several tissues, including respiratory epithelium (pulmonary, bronchial, tracheal) [11, 12]. We and others have demonstrated that CRC cultures can be obtained with very high efficiency from patient respiratory tissue, generating large amounts of AESCs endowed with the ability to differentiate into mature mucociliary cells [13–16]. The possibility to expand CRC stem-like cells from lung tissue of CF patients and their ability to generate differentiated cells in vitro have been demonstrated [15]. The availability of large sources of AESC cells, from CF patients with different genotypes, would enable the improvement of both pharmacological and genetic therapeutic approaches for CF. However, this approach has been limited so far by the low availability of patient lung tissue or the invasiveness of procedures for bronchial brush biopsies. Viable nasal epithelial cells can be obtained through a much less invasive procedure, such as superficial nasal brushing. The ease with which AESCs can be obtained allows the availability of experimental material from each patient, supporting the investigation of virtually any genotype [17]. Nasal brushing is feasible in both adults and infants and allows harvesting of respiratory cells suitable for cell culture, although the yield of primary nasal cells has been very low and establishment of long-term cultures highly inefficient, so far [17, 18]. Here, we generated large amounts of AESCs using the CRC method from nasal brushing samples obtained from CF patients. We used these stem cells (CF-CRC) to model CF variants in vitro, by generating both respiratory tissue in air–liquid interface (ALI) conditions and three-dimensional organoids, mimicking the diseased respiratory tissue. These patient-specific models were highly suitable for testing basal activity and drug response of various genotypes not previously eligible for approved drugs, with the concrete possibility of a clinical translation.
Materials and methods
A detailed description of materials and methods can be found in the supplementary material.
Results
Establishment and validation of CF patient-derived nasal epithelial stem cell cultures under CRC conditions
Nasal epithelial cells were obtained from affected CF patients with variable CFTR gene defects (two mutated CFTR genes) and from carrier individuals (heterozygous for CFTR gene pathogenic variants) through cytology brushing. Nasal brushing cells were cultured under CRC conditions as described in the supplementary material and based on our previously established protocols [13]. Although the number of freshly collected cells varied in different samples (generally ~5×104, but ranging from 1×103 to 2.75×105 cells), all samples gave rise to cell clones in culture, leading to the generation of monolayers of adherent cells with epithelial morphology (100% efficiency of culture establishment) (supplementary figure S1a). They proliferated with a high to moderate growth rate for a prolonged time, guaranteeing a long duration of cultures and generating very large amounts of cells (supplementary figure S1b). The calculated mean yield of total cells expanded in a time frame of 2 months from the brushing collection of culture has been estimated as >2.5×108; moreover, this number was certainly increased when more abundant starting material was dedicated to culture (not shown), in line with results from other groups [19, 20].
CRC cells displayed an epithelial cell phenotype, high viability (supplementary figure S2a) and high clonogenic potential, with 96±3% clone-forming efficiency under CRC conditions (supplementary material). They expressed antigens of airway (basal) stem cells (nerve growth factor receptor (NGFR): mean 82.5±5%; integrin α-6 (ITGA6): mean 96.4±2%; tumour-associated calcium signal transducer 2 (TROP2): mean 91.8±5% positive cells), in agreement with our and others’ previous results on healthy subject- or CF patient-derived cells, expanded from bronchoalveolar or nasal epithelia (figure 1a and supplementary figure S2b) [13, 15, 16]. Stem cell antigen expression was heavily reduced when cells were grown under culture conditions used for primary airway cells (bronchial epithelial growth medium (BEGM)) (supplementary figure S2c). We optimised two alternative CRC-differentiation protocols: cultivation of CRC in ALI culture (a condition mimicking the respiratory tissue microenvironment with epithelial cells lining the surface of tissue in contact with air on top of specific transwell membranes) and a simplified differentiation protocol with cells cultured in ALI medium, in standard plates, as monolayers of submerged cultures. Both growth conditions determined decreased expression of the basal stem cell-related antigens (NGFR, ITGA6, cytokeratin 14 (CK14)) and upregulation of mature respiratory cell markers, confirming the possibility of obtaining full in vitro differentiation of the CRC cells (figure 1b). Specifically, the ALI-culture condition generated respiratory tissue (visible at low magnification in the central image of figure 1c) containing mature cellular elements such as acetylated α-tubulin-positive and FoxJ-positive ciliated cells as well as Muc5B-positive mucus-producing secretory cells (figure 1c left and right). Full functional differentiation and polarisation obtained by culturing the basal cells under ALI-culture conditions is additionally proved by the movement of cilia at the air-exposed side of the tissue (video 1) and cilia and mucin-granules distribution at the apical side visible in figure 1c right (Z) and video 2.
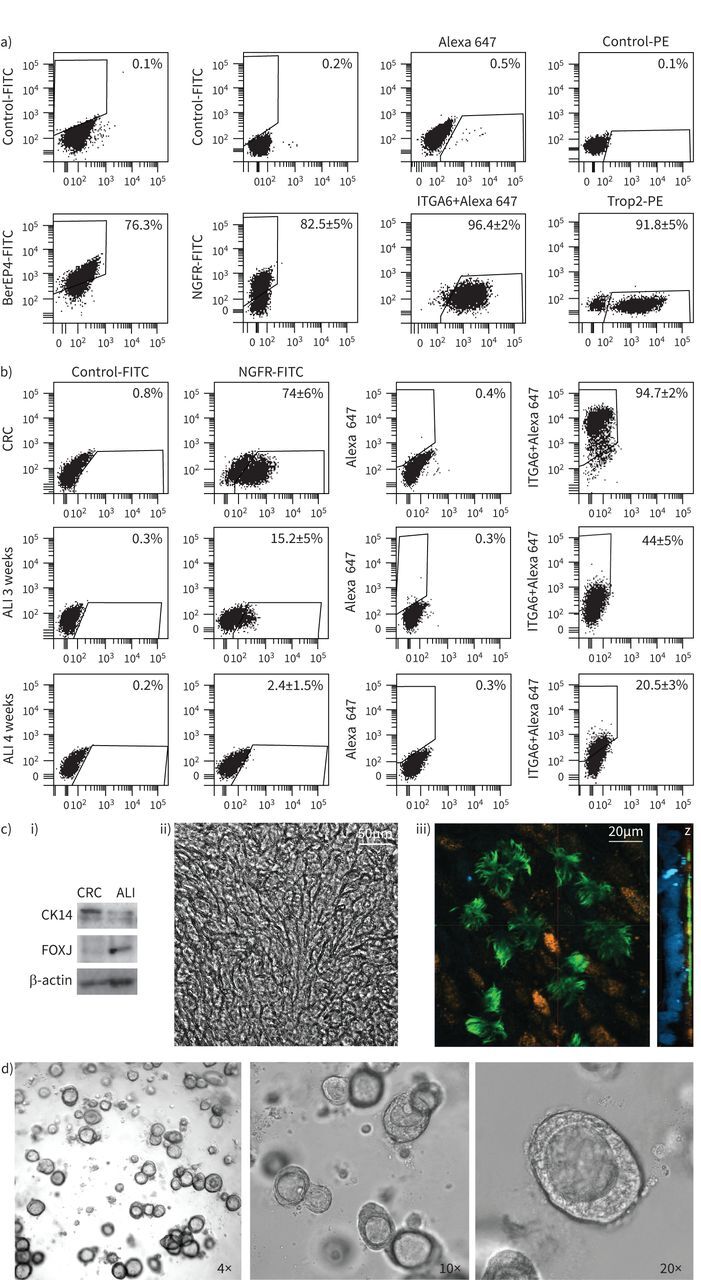
Cystic fibrosis (CF)-“culture reprogramming condition” (CRC) characterisation and generation of differentiated respiratory cell models. Results from one representative preparation (CF2 according to table 1) are reported. a) Flow cytometric analysis of CF-CRC for epithelial/basal-like antigens: epithelial cell adhesion molecule (EpCAM)/BerEP4, integrin α-6 (ITGA6), tumour-associated calcium signal transducer 2 (TROP2), nerve growth factor receptor (NGFR). Passage 2 cells have been used for these experiments. b) Flow cytometry analysis of the basal cell markers NGFR and ITGA6 in CF-CRC (passage 3) and 3–4 weeks differentiated cells in air–liquid interface (ALI) culture condition. c) i) Immunoblot showing the acquisition of the differentiation marker of mature ciliated cells FOXJ and the loss of basal stem cell marker cytokeratin 14 (CK14), after ALI differentiation; β-actin is shown for equal loading. ii) Picture of ALI-cultured cells showing the tissue appearance at 10× magnification. iii) Immunofluorescence and confocal analysis for differentiation markers acetylated α-tubulin (green) and mucin5B (orange) after ALI-culture differentiation. Magnification is 60× and orthogonal projection (Z) shows tissue thickness with nuclei (stained in blue with 4′,6-diamidino-2-phenylindole) at the bottom and differentiation antigens at the apical side. d) Representative images of CRC-derived organoids showing their density, distribution in the well and morphology (4× magnification) and the three-dimensional structure with the organoid wall and the presence of the internal lumen visible at higher magnification (10× and 20×). All experiments described have been performed in triplicate.
Through inclusion in matrigel and after 3 weeks’ culture in ALI medium, CRC cells generated organoids, functionally differentiated respiratory tissue, resembling mini-airways with three-dimensional structure and morphology. They gradually exhibited the formation of an internal lumen and contained specialised functionally mature cellular elements (i.e. the ciliated cells endowed with actively moving cilia oriented towards the lumen of organoid are visible by optical microscopy examination) (figure 1d and video 3).
CRC cultures maintained the stem cell phenotype after repeated passages in vitro, even after cryopreservation, guaranteeing the possibility to largely expand and store these cells for subsequent studies. Basal stem marker expression, although decreasing gradually after prolonged cell passages (suggesting the progressive propensity of cells to partially differentiate after prolonged time in culture), remained moderate and much higher than that observed in cells differentiated in ALI culture (supplementary figure S2). These results demonstrated a partial but low extent of cell differentiation under prolonged CRC conditions in line with the reduction of proliferation rate described for late-passage cells (supplementary figure S1b). However, cells maintained stem-cell properties after several passages, such as the ability to generate differentiated organoids (functional organoids with lumen and beating cilia obtained from CRC cells at passage 6 are visible in video 4).
Validation of CFTR gene/transcript diagnostic alterations in CRC
CFTR genetic defects, previously identified during diagnostic analysis of patient blood cells and patient brushing genetic material, were assessed in DNA and RNA extracted from CRC cultures, based on previously used protocols [21–23]. CFTR DNA and RNA sequence analysis allowed to confirm the genetic alteration previously detected in affected patients, as well as, in some cases, to refine genetic diagnosis through identification of previously undetected mutations due to scarcity of biological material (table 1). Subsequent DNA or RNA analysis for the newly identified alterations confirmed their presence in the carrier parent's genotype, delineating their relevance for the specific CF phenotype and proving the mechanism of disease inheritance. Genetic defects were maintained during repeated cell passages of CF-CRC cells (analyses of CFTR sequence at second or sixth passage of cultured cells provided identical results) and no other genetic alterations arose, demonstrating that cultured cells maintained genetic integrity during prolonged culture, and at the same time guaranteeing the reliability of diagnostic material obtained after cell expansion. Thus, the CRC approach represents a precious tool to contribute to improve diagnostic procedures by providing large amounts of patient-derived material.
TABLE 1
Genotypes of patients included in the study
| Legacy name (old nomenclature) | HGVS name | ||
| DNA level | Protein level | ||
| CRC-CF#1 | [F508del;I1027T]/F508del | c.[1521_1523delCTT;3080T>C];[1521_1523delCTT] | p.[(Phe508del;Ile1027Thr)];[(Phe508del)] |
| CRC-CF#2 | F508del/F508del | c.[1521_1523delCTT];[1521_1523delCTT] | p.[(Phe508del)];[(Phe508del)] |
| CRC-CF#3 | F508del/L558S | c.[1521_1523delCTT];[1673T>C] | p.[(Phe508del)];[(Leu558Ser)] |
| CRC-CF#4 | [G576A;R668C]/wt | c.[1727G>C;2002C>T];[=] | p.[(Gly576Ala;Arg668Cys)];[(=)] |
| CRC-CF#5 | c.[1521_1523delCTT];[1210-34TG[10]_1210-34TG[4]ins317] | ||
| CRC-CF#6 | G85E/wt | c.[254G>A];[=] | p.[(Gly85Glu)];[(=)] |
| CRC-CF#7 | c.[1521_1523delCTT];[1210-34TG[10]_1210-34TG[4]ins358] | ||
| CRC-CF#8 | (TG)11T5/L1065P | c.[1210-34TG[11];1210-12T[5]];[3194T>C] | |
| CRC-CF#9 | (TG)12T5/wt | c.[1210-34TG[12];1210-12T[5]];[=] | |
| CRC-CF#10 | [F508del;I1027T]/F508del | c.[1521_1523delCTT;3080T>C];[1521_1523delCTT] | p.[(Phe508del;Ile1027Thr)];[(Phe508del)] |
| CRC-CF#11 | c.[1210-34TG[10]_1210-34TG[4]ins354];[=] | ||
| CRC-CF#12 | c.[1521_1523delCTT];[1210-34TG[10]_1210-34TG[4]ins354] | ||
| CRC-CF#13 | F508del/F508del | c.[1521_1523delCTT];[1521_1523delCTT] | p.[(Phe508del)];[(Phe508del)] |
| CRC-CF#14 | [621+3A>G;F1052V]/wt | c.[489+3A>G;3154T>G];[=] | |
The identification of each preparation of “culture reprogramming condition” (CRC) cells is indicated, as used throughout the text. Genotypes are indicated in old (legacy) and new (Human Genome Variation Society (HGVS)) nomenclature at both DNA and, whenever possible, protein level. For recently discovered disease-causing variants, only the HGVS name is used. wt: wild-type allele.
The analysed genotypes included four F508del homozygotes (table 1; CRC-CF#1, 2, 10 and 13, two of them with another CFTR variation in cis with one of the F508del), four compound heterozygotes for the F508del and another CFTR pathogenic variant (table 1; CRC-CF#3, 5, 7 and 12), one compound heterozygote for non-F508del pathogenic variants (table 1; CRC-CF#8) and five carriers (table 1; CRC-CF#4, 6, 9, 11 and 14). Two out of the three large nucleotide insertions (table 1; CRC-CF#5, 7) are known to produce anomalous splicing of CFTR mRNA with the complete skipping of exon 10 (legacy name exon 9) according to our previous results [24]. The third large nucleotide insertion (table 1; CRC-CF#12) is a novel CFTR pathogenic variant, similar to those already described, but with a different number of inserted nucleotides. It produces the same effect of a complete exon 10 skipping (our unpublished results). The skipping of exon 10 of CFTR is known to produce a nonfunctional protein for the failure of full glycosylation, misfolding and clearance in the endoplasmic reticulum, with a pattern resembling that of F508del [25, 26]. Consequently, these three large insertions are CFTR pathogenic variants, probably of class II.
CFTR expression in fresh brushings, CRC cells, standard culture cells, and ALI-differentiated cells
We next evaluated CFTR expression in CF-CRC and CF-CRC-derived differentiated cells with the aim to determine which culture condition could provide high CFTR-expressing cells for functional studies of its basal/residual or pharmacologically restored activity. Real-time PCR analysis showed that CFTR was highly transcribed in all brushing samples, its expression was limited in CRC-cultured cells, as expected for nonspecialised stem cells and slightly increased under standard BEGM culture. Finally, CFTR was strongly expressed in CRC-derived ALI-culture differentiated cells (figure 2a and andb).b). In line with transcript expression, immunoblot analysis revealed very low amounts of CFTR protein in CRC or standard-culture cells (BEGM) and prominent amounts in ALI-medium differentiated cells (figure 2c and andd).d). Therefore, ALI culture seems the more suitable approach to model CF in vitro for CFTR studies. Parallel evaluation of differentiated ciliated cell marker (FOXJ) (figure 2d) in the same samples proved the occurrence of cell differentiation.
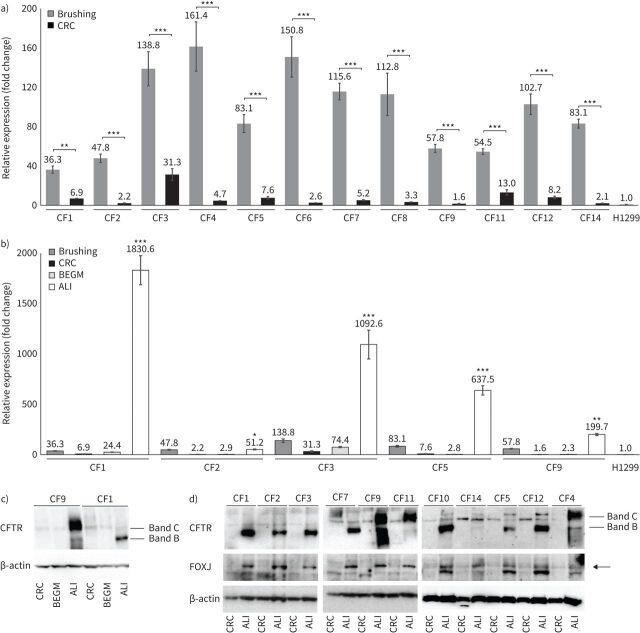
Cystic fibrosis transmembrane conductance regulator (CFTR) expression analysis in cystic fibrosis (CF)-“culture reprogramming condition” (CRC). Preparations from CF1 to CF14 are according to table 1. a) CFTR mRNA expression levels (mean±sd) measured by real-time PCR comparing fresh brushing samples with CF-CRC cultured in CRC conditions (CRC); ANOVA p<0.0001, Bonferroni's post-test, ***: p<0.0001 for each brushing versus CRC, with the exception of CF1, for which **: p<0.01. b) CFTR mRNA expression levels (mean±sd) measured by real-time PCR comparing CF-CRC undifferentiated (CRC) with differentiated in bronchial epithelial growth medium (BEGM) and air–liquid interface (ALI) conditions; ANOVA p<0.0001, Bonferroni's post-test, ***: p<0.0001 for each ALI versus all the other conditions of the same preparation, with the exception of CF2 for which ALI versus brushing is nonsignificant and for the other differences *: p<0.05, and CF9 for which ALI versus brushing **: p<0.01. In both a) and b), the relative expression (fold change) (calculated as described in the supplementary material, CFTR expression analysis) is referred to the expression in the H1299 cells. c) Immunoblot showing CFTR protein levels in CF-CRC cultured in CRC conditions (CRC), standard primary culture conditions (BEGM) and ALI conditions (according to table 1: CF9 is a carrier, CF1 is a homozygote F508del); β-actin is shown for equal loading. d) Immunoblot showing CFTR protein levels in CF-CRC cultured in CRC (CRC) or ALI conditions (according to table 1: CF1, CF2, CF3, CF5, CF7, CF10 and CF12 are homozygotes or compound heterozygotes, whereas CF4, CF9, CF11 and CF14 are carriers); FOXJ immunoblot is shown for validation of proper in vitro differentiation. For both c) and d), the mature (band C) and immature (band B) CFTR protein are shown; β-actin is shown for equal loading. All experiments have been performed in triplicate.
Wild-type and F508del-mutated CFTR proteins could be distinguished in immunoblot, based on their different molecular weights (high molecular weight band C of wild-type functional CFTR and low molecular weight band B of defective misfolded protein) (figure 2c and anddd).
Pharmacological rescue of CFTR conformation: biochemical assay (immunoblot)
Next, we evaluated the possibility to use CRC-derived CF models for assessment of CFTR basal/residual function as well as for determination of the ability of pharmacological agents to increase/rescue CFTR activity. At the biochemical level this was accomplished through detection of the basal or drug-enhanced relative amount of band C, associated with correct conformation of CFTR.
We compared the efficacy of the clinically approved CFTR correctors lumacaftor, tezacaftor and elexacaftor, each indicated for a specific patient subgroup. Correctors were used as single agents or in combination, including the Trikafta corrector combination elexacaftor/tezacaftor. Initially, we tested a homozygous F508del patient, in order to verify the increment of CFTR maturation expected from Trikafta in comparison with previously approved drugs as proved in clinical trials and to validate the ALI-culture model approach (figure 3a). Next, we used this biochemical assay to determine the efficacy of drugs in the three unexplored rare compound heterozygous complex genotypes bearing the F508del and the three large nucleotidic insertions (F508del/ins), probably belonging to mutational class II (figure 3b–d).
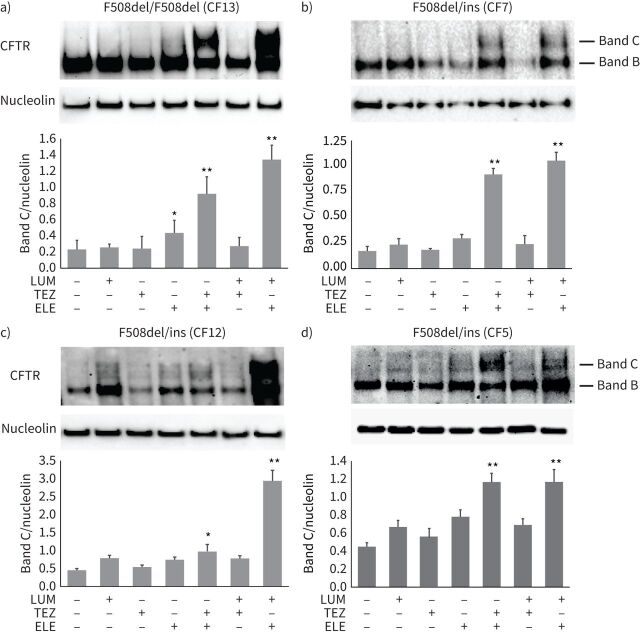
Evaluation of cystic fibrosis transmembrane conductance regulator (CFTR) protein maturation rescue induced by modulators. Immunoblot analysis of expression levels and molecular mass of CFTR protein, in the different control or drug-treated cystic fibrosis (CF)-“culture reprogramming condition” (CRC)-derived air–liquid interface (ALI)-differentiated samples, are shown. Nucleolin is shown for equal loading. a) F508del/F508del homozygous genotype; b), c) and d) F508del/ins (ins: one of the rare alleles with the insertion of variable length) compound heterozygotes, with genotypes according to table 1. For each panel, the densitometric quantification (mean±sd) is shown below the immunoblot. The treatments by the drugs are indicated at the bottom. LUM: lumacaftor; TEZ: tezacaftor; ELE: elexacaftor; band C: mature CFTR; band B: immature CFTR. Mean±sd of three independent experiments is shown. *: p<0.05, **: p<0.01.
The ability of CFTR correctors to restore CFTR maturation was evaluated in ALI medium-differentiated CF-CRC cells exposed to correctors for 48 h, through immunoblot quantification of CFTR band C relative proportion in control or drug-treated cells. The F508del homozygous sample, used as positive control, showed a marked relative increase of band C, following tezacaftor/elexacaftor drug treatment (figure 3a). These findings were in line with the higher Trikafta activity found in vitro and in clinical trials [6]. Single correctors as well as the lumacaftor/tezacaftor combination treatments induced a limited or nonsignificant increase in band C level. In contrast, elexacaftor/lumacaftor treatment resulted in robust correction of the CFTR protein, even superior to the tezacaftor/elexacaftor (figure 3).
In F508del/ins samples (CF#5, 7 and 12 in table 1) CFTR protein drug correction occurred at similar extent compared to homozygous F508del/F508del genotypes. In addition, in F508del/ins genotypes, the efficacy of drug combinations was superior than that observed by single correctors, and corrector combinations appeared particularly effective when elexacaftor was used, suggesting higher synergy of this corrector. These results imply a promising therapeutic efficacy of Trikafta for the tested F508del/ins genotypes. In effect, the efficacy of Trikafta in a phase 3 study in patients with a single F508del allele has been demonstrated [5]. However, it is likely that in F508del/ins genotypes here tested, there is also an additional effect of correctors on the allele with the large insertion (probably of mutational class II). We found a general highly encouraging efficacy of the Trikafta correctors elexacaftor/tezacaftor in all variants analysed. In addition, a particularly high effectiveness was observed in cells treated with elexacaftor/lumacaftor combination, suggesting that further investigation in this direction might be indicated.
Pharmacological rescue of CFTR function: forskolin-induced swelling of organoids and fluid re-absorption assay
Although highly indicative of CFTR protein folding/maturation, biochemical quantification of CFTR band C may not be sufficient to determine whether a specific compound is also able to restore CFTR surface localisation and gating function. Thus, in order to substantiate biochemical results, we evaluated the efficacy of CFTR modulators through the forskolin-induced swelling (FIS) assay of organoids, based on the measurement of CFTR activity, as the direct effect of chloride passage through the channel and fluid movement [27]. Following CFTR modulators exposure, organoids generated from CF-CRC bearing specific genotypes responded to drugs with variable measurable swellings (figure 4), whose extents were consistent with the relative band C increase, observed in immunoblot of the corresponding genotypes (figure 3). The homozygote F508del responded to an approximately similar extent as the genotypes with a single F508del allele associated to the other alleles with large insertions, in line with immunoblot assays. Combined drugs proved higher activity than single drugs with a markedly encouraging efficacy of Trikafta (figure 4a). The triple combination elexacaftor/lumacaftor/ivacaftor induced a robust swelling of organoids, comparable (and in some cases superior) to Trikafta, in line with biochemical analysis (figure 4a).
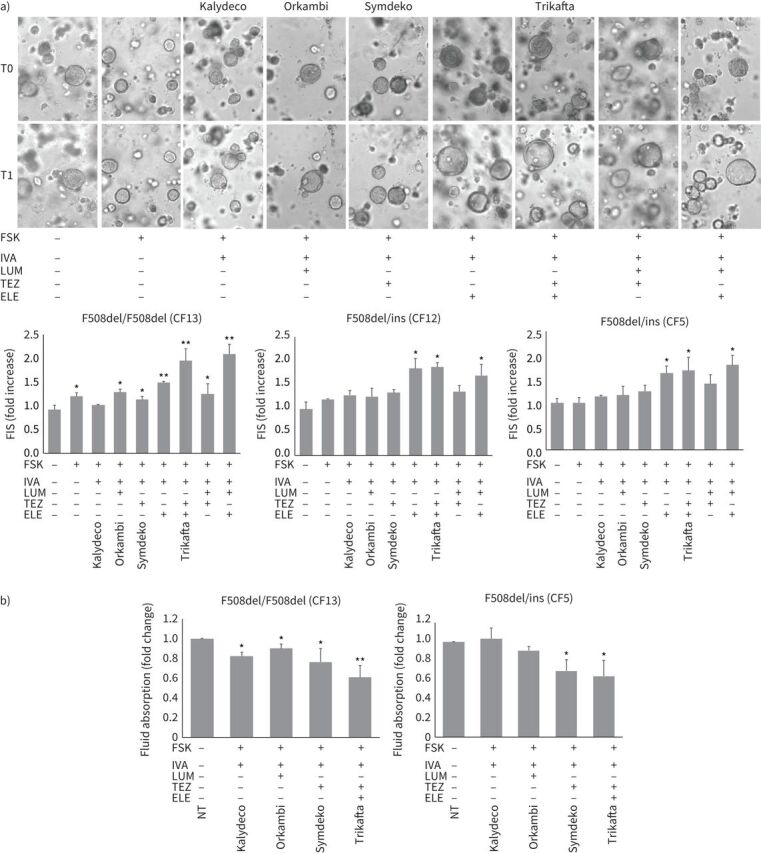
Evaluation of cystic fibrosis transmembrane conductance regulator (CFTR) function recovery induced by modulators. a) Forskolin-induced swelling (FIS) in cystic fibrosis (CF)-“culture reprogramming condition” (CRC)-derived three-dimensional organoids. In the images some representative examples of FIS are reported relative to F508del/F508del homozygous cells (CF13). The same organoids, monitored and measured before (T0) and after (T1) treatments, are shown. In the histograms (lower panels), the response of F508del/F508del homozygous genoptypes (mean of three independent experiments including that corresponding to the images above) and two F508del/ins (ins: one of the rare alleles with the insertion of variable length) compound heterozygous genotypes (according to table 1) is shown. Measures are relative to the increase of organoid area (mean of at least five organoids per sample) after stimulation with the drugs indicated. FSK: forskolin; IVA: ivacaftor; LUM: lumacaftor; TEZ: tezacaftor; ELE: elexacaftor. Commercial names of clinically used drugs are indicated above corresponding images and below histograms. b) Fluid re-absorption assay in CF-CRC-derived air–liquid interface (ALI)-cultures. The results of a F508del/F508del homozygous and a F508del/ins compound heterozygous genotypes (according to table 1) are shown. The absorbed fluid is calculated as the input volume in the apical chamber detracted of the remaining volume/transwell membrane area in cm2 (1.12 cm2)/time in hours (48 h). The ratio corresponding to values of sample treated with the indicated drug versus the value of untreated sample (NT) is reported. Mean±sd of three independent experiments is shown. *: p<0.05, **: p<0.01. Commercial names of clinically used drugs are indicated below the charts.
In addition, we used ALI-culture model to set up the fluid-reabsorption assay, to measure basal or drug-potentiated CFTR activity (induction of chloride ions and water efflux in the apical chamber of ALI-culture transwells that counteracts the physiological sodium channel-mediated fluid movement in the opposite direction toward the basal chamber, usually indicated as fluid re-absorption) [28]. The extent of decrease of liquid re-absorption from the apical compartments in specific drug-treated samples compared to control samples indicated that, both in F508del/F508del and F508del/ins genotypes, Trikafta showed high efficacy in restoring CFTR protein function (decreased liquid re-absorption) and its activity was markedly superior to singularly used drugs (figure 4b).
Thus, the different biochemical and functional CFTR assays produced comparable results enforcing reliability of two-dimensional and three-dimensional CF-CRC models and of the different assays, providing confirmatory results of the efficacy of the newly introduced Trikafta triple combination and pushing for further researches to possibly extend its indication to a larger number of CFTR variants.
Discussion
In recent years, a huge research effort in the attempt to increase disease knowledge, identify innovative therapeutic targets and more effective CFTR modulators, together with improvements of symptomatic therapies, have led to markedly increased life expectancy and better quality of life for CF patients. Innovative specific therapies have been developed and approved for the most frequent genetic variants; however, the majority of rare variants of disease remain orphan, and CF remains a progressively devastating disease for most of them [29]. Therapeutic advancements in the direction of personalised treatment have been dramatically hampered by lack of valuable in vitro disease models suitable for experimental therapeutic testing of the huge number of CFTR variants. In fact, rare genetic variants have not been available for research in the pre-clinical experimental setting nor for clinical trials, due to their scarcity and to the inefficiency of in vitro primary culture establishment. Here, we used the CRC approach to generate CF models in vitro with high efficiency. We fully validated and used these models for the evaluation of response to currently used and recently approved drugs in rare disease variants in comparison with the most studied/most frequent F508del homozygous variant.
In agreement with our previous data on bronchial/pulmonary healthy tissue, CRC methodology proved highly efficient in establishment of long-term cultures of AESCs from nasal epithelia of CF patients. CRC cultures were established with 100% efficiency, in spite of the limited amount of nasal brushing samples, and proved able to supply huge amounts of cells [13]. The unprecedented potency of the CRC approach coupled with the fact that nasal epithelium represents an easily accessible source of samples, and that nasal brushing is a relatively low-invasive procedure, might allow pre-clinical experimentation of virtually each CF variant, with strong implications for personalised therapy.
CF-CRC cells proved endowed with airway epithelial stem cell (basal) phenotype and function, being able to extensively proliferate and generate mature respiratory cells of various type when cultured under differentiating conditions. Three different models of differentiation were developed: two-dimensional monolayers of respiratory cells, ALI cultures in transwells generating polarised respiratory tissue, and three-dimensional airway organoids recapitulating the airway tissue architecture. These models were fully validated for differentiation and CFTR expression, before use for experimental assays.
CFTR genetic defects were maintained after cell expansion and patient genotypes did not undergo culture-linked alterations, proving that the CRC approach of cell expansion from CF brushing material may represents a precious tool to contribute improving diagnostic procedures by providing large amounts of patient-derived material.
CFTR was abundantly detected in fresh nasal brushing samples, its expression decreased dramatically in (CRC) culture-expanded cells and reached very high levels in ALI medium-differentiated airway models, proving their suitability for CFTR studies. Biochemical evaluation of CFTR protein folding and maturation was achieved using two-dimensional monolayers through immunoblot, CFTR functional activity was evaluated in three-dimensional organoids (FIS) and in airway tissue generated by standard ALI culture (fluid re-absorption assay).
Responses to the drugs currently used for specific variants, Orkambi, Symdeko, Kalydeco and Trikafta were assessed in F508del homozygous genotypes as validation of the experimental approach and in heterozygous compound genotypes F508del/ins (see text for explanation) to assess and compare efficacy of different drugs on these variants.
Trikafta showed markedly higher activity compared to other therapies. Moreover, similar efficacy of each treatment was observed in genotypes bearing either two copies (F508del/F508del homozygous) or a single copy (F508del/ins) of F508del. Considering that in F508del/ins genotypes, the ins CFTR allele probably acts as a mutational class II variant, a therapeutic effect in addition on the ins allele is to be considered. These results provide support for the forthcoming treatment of specifically tested genotypes with Trikafta, following the United States template of potential guide of personalised patient treatment based on results derived from “in vitro trials”.
F508del alteration results in lower molecular mass of the immature CFTR protein, detectable in immunoblot, confirming the use of this approach as indicator of nonfunctional CFTR. Based on this assumption, determination of CFTR molecular weight in immunoblot may contribute to the characterisation at protein level of unexplored variants identified through genetic analysis guiding their diagnostic classification, as well as to determine the ability of pharmacological agents to rescue CFTR function (as is the case for the three large insertions analysed here).
The different biochemical and functional CFTR assays used in this study produced comparable results, enforcing reliability of all assays based on AESC-derived models and results.
Of note, the new corrector elexacaftor displayed remarkable efficacy in the enhancement of CFTR activity, compared to lumacaftor or tezacaftor. This might be due to a potential stronger ability to correct CFTR protein, but also to its dual activities, as corrector and potentiator, described recently [30]. Moreover, besides the strong potential of the combination texcaftor/elexacaftor, as expected by these two Trikafta correctors, elexacaftor displayed marked synergy when associated with lumacaftor as well. Thus, the innovative combination of lumacaftor/elexacaftor might warrant further investigation. At the same time the possible efficacy of elexacaftor used as a single agent, or its potential synergy with other compounds should be explored, and might lead to more effective, better tolerated or less expensive treatment options.
Our studies demonstrated that the CRC approach enables massive expansion of AESCs, generating highly suitable CF models for testing CFTR modulators. The extraordinary efficiency of CRC culture establishment and their massive expansion in vitro, coupled with the stability of cultures allow the use of late-passage CRC cells, all aspects missing in other cellular approaches, thus further increasing the cell amount available for experimentation. In fact, CF-CRC cells showed to maintain the differentiation ability and stable antigen expression over several cellular passages (at least up to P6). Conversely, primary airway epithelial cells appeared to be able to maintain the specific airway expression pattern no longer than P3 [31].
The presented cellular approach of CRC-derived ALI cultures is quite novel as applied to CF. However, another similar approach has been described recently for the expansion of human nasal epithelial stem cells, based on dual SMAD inhibition and feeder-free [32]. A comparative study has shown that respiratory cells generated in ALI culture from these SMAD inhibited/feeder-free cells did not differ substantially from ALI cultures generated from standard CRC cells, in terms of structural morphology or baseline global proteomics profile. However, standard CRC-derived ALI cultures displayed increased cilia and CFTR activity, further enhancing the reliability of the standard CRC-approach, used here, for CFTR studies [33].
Finally, given the relative inaccessibility of primary bronchial epithelial cell cultures, our findings support the use of patient-derived nasal epithelial cell cultures for pre-clinical studies of therapeutic interventions. In this context, the approach of theratyping is of enormous translational impact, allowing the identification of specific pathogenetic variants responding to a specific therapy. This methodology has been approved in the United States by the FDA as a unique preliminary step necessary and sufficient for the treatment of patients with the specific responding genotype, by drugs clinically approved for other genotypes [10]. Moreover, the use of personalised organoid technology to guide treatment decisions would be widely accepted by patient community, as reported in a recent Australian report [34]. We are confident that this approach might be assumed as a sort of in vitro personalised clinical trial in the near future, and might contribute towards facilitating the access to more beneficial cure for CF patients, with particular benefit for orphan patients.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary methods and figures ERJ-00908-2021.Supplement
Supplementary video 1 ERJ-00908-2021.Video_1
Supplementary video 2 ERJ-00908-2021.Video_2
Supplementary video 3 ERJ-00908-2021.Video_3
Supplementary video 4 ERJ-00908-2021.Video_4
Shareable PDF
Acknowledgements
We thank Fabio Santavenere (National Center for Innovative Technologies in Public Health, Istituto Superiore di Sanità, Rome, Italy) for support with fibroblast irradiation. This work is dedicated to the memory of Gianni Mastella with gratitude for his outstanding commitment in the cystic fibrosis field.
Footnotes
This article has supplementary material available from erj.ersjournals.com
This article has an editorial commentary: https://doi.org/10.1183/13993003.02735-2021
Author contributions: G. Sette, S. Lo Cicero, A. Eramo and M. Lucarelli designed the experiments; G. Sette, S. Lo Cicero, G. Blaconà, S.M. Bruno, S. Pierandrei and V. Salvati performed the experiments and analysed the data. M. Lucarelli, G. Cimino and B. Fabrizzi selected patients, provided patient brushings and patient clinical diagnosis; G. Sette, S. Lo Cicero, V. Salvati and G. Castelli expanded and cultured patient samples; M. Falchi supported the microscopy analyses; A. Eramo and M. Biffoni supervised cell biobanking. A. Eramo and M. Lucarelli wrote the paper; R. De Maria and M. Biffoni revised the manuscript.
Conflict of interest: G. Sette has nothing to disclose.
Conflict of interest: S. Lo Cicero has nothing to disclose.
Conflict of interest: G. Blaconà has nothing to disclose.
Conflict of interest: S. Pierandrei has nothing to disclose.
Conflict of interest: S.M. Bruno has nothing to disclose.
Conflict of interest: V. Salvati has nothing to disclose.
Conflict of interest: G. Castelli has nothing to disclose.
Conflict of interest: M. Falchi has nothing to disclose.
Conflict of interest: B. Fabrizzi has nothing to disclose.
Conflict of interest: G. Cimino has nothing to disclose.
Conflict of interest: R. De Maria has nothing to disclose.
Conflict of interest: M. Biffoni has nothing to disclose.
Conflict of interest: A. Eramo has nothing to disclose.
Conflict of interest: M. Lucarelli has nothing to disclose.
Support statement: This work was supported by Italian Cystic Fibrosis Foundation (grant FFC 12/2018 to A. Eramo and M. Lucarelli) and contributions from “Delegazione FFC di Cecina e Rosignano” and “Delegazione FFC di Alberobello”. Support was also from Ricerca Corrente – Ministero della Salute 2019 (FASC. 9ARC to M. Biffoni) and from Sapienza University of Rome (Progetti di Ateneo 2017 and 2018 to M. Lucarelli). The salaries of G. Sette and S.M. Bruno were partially covered by Italian Cystic Fibrosis Foundation grant (FFC 12/2018 to A. Eramo and M. Lucarelli). Funding information for this article has been deposited with the Crossref Funder Registry.
References
Full text links
Read article at publisher's site: https://doi.org/10.1183/13993003.00908-2021
Read article for free, from open access legal sources, via Unpaywall:
https://erj.ersjournals.com/content/erj/58/6/2100908.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/118020909
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1183/13993003.00908-2021
Article citations
Organoids: new frontiers in tumor immune microenvironment research.
Front Immunol, 15:1422031, 29 Jul 2024
Cited by: 0 articles | PMID: 39136020 | PMCID: PMC11317300
Review Free full text in Europe PMC
Alveolar Organoids in Lung Disease Modeling.
Biomolecules, 14(1):115, 16 Jan 2024
Cited by: 1 article | PMID: 38254715 | PMCID: PMC10813493
Review Free full text in Europe PMC
A Fast Scoring of Human Primary Respiratory Epithelia Grown at Air-Liquid Interface (ALI) to Assess Epithelial Morphology in Research and Personalized Medicine Settings.
J Pers Med, 14(1):109, 18 Jan 2024
Cited by: 0 articles | PMID: 38248810 | PMCID: PMC10817428
Laboratory Tools to Predict CFTR Modulator Therapy Effectiveness and to Monitor Disease Severity in Cystic Fibrosis.
J Pers Med, 14(1):93, 13 Jan 2024
Cited by: 0 articles | PMID: 38248793 | PMCID: PMC10820563
Review Free full text in Europe PMC
Emerging In Vitro Models for the Study of Infection and Pathogenesis of Pseudomonas aeruginosa and Testing of Antibacterial Agents.
Methods Mol Biol, 2721:233-239, 01 Jan 2024
Cited by: 0 articles | PMID: 37819526
Go to all (27) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
L1077P CFTR pathogenic variant function rescue by Elexacaftor-Tezacaftor-Ivacaftor in cystic fibrosis patient-derived air-liquid interface (ALI) cultures and organoids: in vitro guided personalized therapy of non-F508del patients.
Respir Res, 24(1):217, 06 Sep 2023
Cited by: 4 articles | PMID: 37674160 | PMCID: PMC10483775
Allosteric folding correction of F508del and rare CFTR mutants by elexacaftor-tezacaftor-ivacaftor (Trikafta) combination.
JCI Insight, 5(18):139983, 17 Sep 2020
Cited by: 116 articles | PMID: 32853178 | PMCID: PMC7526550
ORKAMBI-Mediated Rescue of Mucociliary Clearance in Cystic Fibrosis Primary Respiratory Cultures Is Enhanced by Arginine Uptake, Arginase Inhibition, and Promotion of Nitric Oxide Signaling to the Cystic Fibrosis Transmembrane Conductance Regulator Channel.
Mol Pharmacol, 96(4):515-525, 19 Aug 2019
Cited by: 29 articles | PMID: 31427400
Pharmacological analysis of CFTR variants of cystic fibrosis using stem cell-derived organoids.
Drug Discov Today, 24(11):2126-2138, 04 Jun 2019
Cited by: 11 articles | PMID: 31173911 | PMCID: PMC6856431
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Italian Cystic Fibrosis Foundation (1)
Grant ID: grant FFC 12/2018
Ministero della Salute (1)
Grant ID: Ricerca Corrente. FASC. 9ARC
Sapienza Università di Roma (1)
Grant ID: Progetti di Ateneo 2017, 2018




