Abstract
Premise
Many cultivated coffee varieties descend from Coffea canephora, commonly known as Robusta coffee. The Congo Basin has a century-long history of Robusta coffee cultivation and breeding, and is hypothesized to be the region of origin of many of the cultivated Robusta varieties. Since little is known about the genetic composition of C. canephora in this region, we assessed the genetic diversity of wild and cultivated C. canephora shrubs in the Democratic Republic of the Congo.Methods
Using 18 microsatellite markers, we studied the genetic composition of wild and backyard-grown C. canephora shrubs in the Tshopo and Ituri provinces and multiple accessions from the INERA Yangambi Coffee Collection. We assessed genetic clustering patterns, genetic diversity, and genetic differentiation between populations.Results
Genetic differentiation was relatively strong between wild and cultivated C. canephora shrubs, and both gene pools harbored multiple unique alleles. Strong genetic differentiation was also observed between wild populations. The level of genetic diversity in wild populations was similar to that of the INERA Yangambi Coffee Collection, but local wild genotypes were mostly missing from that collection. Shrubs grown in the backyards were genetically similar to the breeding material from INERA Yangambi.Conclusions
Most C. canephora that is grown in local backyards originated from INERA breeding programs, while a few shrubs were obtained directly from surrounding forests. The INERA Yangambi Coffee Collection could benefit from an enrichment with local wild genotypes to increase the genetic resources available for breeding purposes and to support ex situ conservation.Free full text
Genetic diversity of wild and cultivated Coffea canephora in northeastern DR Congo and the implications for conservation
Abstract
Premise
Many cultivated coffee varieties descend from Coffea canephora, commonly known as Robusta coffee. The Congo Basin has a century‐long history of Robusta coffee cultivation and breeding, and is hypothesized to be the region of origin of many of the cultivated Robusta varieties. Since little is known about the genetic composition of C. canephora in this region, we assessed the genetic diversity of wild and cultivated C. canephora shrubs in the Democratic Republic of the Congo.
Methods
Using 18 microsatellite markers, we studied the genetic composition of wild and backyard‐grown C. canephora shrubs in the Tshopo and Ituri provinces and multiple accessions from the INERA Yangambi Coffee Collection. We assessed genetic clustering patterns, genetic diversity, and genetic differentiation between populations.
Results
Genetic differentiation was relatively strong between wild and cultivated C. canephora shrubs, and both gene pools harbored multiple unique alleles. Strong genetic differentiation was also observed between wild populations. The level of genetic diversity in wild populations was similar to that of the INERA Yangambi Coffee Collection, but local wild genotypes were mostly missing from that collection. Shrubs grown in the backyards were genetically similar to the breeding material from INERA Yangambi.
Conclusions
Most C. canephora that is grown in local backyards originated from INERA breeding programs, while a few shrubs were obtained directly from surrounding forests. The INERA Yangambi Coffee Collection could benefit from an enrichment with local wild genotypes to increase the genetic resources available for breeding purposes and to support ex situ conservation.
Coffee is one of the most valuable crops in the world and is the second‐most exported product of developing countries (Pendergast, 2009). Most cultivated coffee varieties descend from two wild Coffea species (Rubiaceae): Coffea arabica L. (Arabica coffee) and C. canephora Pierre ex A.Froehner (Robusta coffee), of which the latter represents close to 44% of the global coffee production (data provided by ICO, statistical service). Whereas Arabica coffee was introduced by Arabian merchants in Yemen for cultivation ca. 1000 years ago (Smith, 1985), the commercial cultivation of Robusta coffee is less than 150 years old. In the late 19th century, local cultivation of Coffea canephora was reported from Gabon, Angola, and Uganda (P. Stoffelen, personal observations of herbarium labels; Chevalier, 1929). However, widespread colonial cultivation of Robusta coffee started only in the early 20th century. The introduction and promotion of “Coffea robusta” as a robust coffee species by the Belgian horticulturist Linden in 1900 is probably key for the success of Robusta coffee, as the commercial name suggests. Linden's introduction was done using seeds of wild plants from Sankuru Province in the Democratic Republic of the Congo (DR Congo). This material was sent to Java, where it was crossed with other Robusta lineages from Lower Congo and Uganda and elsewhere. After the arrival of Robusta coffee in Java in the early 20th century, Java developed into an important breeding and distribution center of Robusta coffee (Ferrão et al., 2019). Before the European colonization of Africa, Coffea canephora was only grown locally (Jaroget and Descroix, 2002), mainly in the northeastern and southwestern part of its natural distribution area.
In the early 1900s, the first Robusta coffee research and breeding stations were also installed in Central Africa (e.g., the Botanical Garden in Eala). In DR Congo, the INEAC (Institut National pour l'Etude Agronomique du Congo Belge) was created in 1933 to develop a program for scientific research focused on agriculture and forestry, with a network of research stations throughout the country (Jaroget and Descroix, 2002). Yangambi (Tshopo Province, northeastern DR Congo) became the principal research station of the INEAC, in general and for Robusta coffee (Leplae, 1936). In the years following the second World War, DR Congo and Uganda took over Java's role as principal research and breeding centers for C. canephora (Jaroget and Descroix, 2002). From there, plants and seeds were distributed to other regions. In this context, INEAC Yangambi also assembled a large Robusta coffee gene bank. In 1962, 2 years after the Independence of Congo, INEAC changed to become INERA (Institut National des Etudes et Recherches Agronomiques). After this change, the research and breeding activities at INERA Yangambi were gradually reduced and during the last decades, many accessions of the gene bank were lost. In 2016, the Robusta Coffee Collection of the INERA Yangambi held 94 different genetic lines, of which seven were elite breeding lines (6 Lula lines and 1 Java line; F. Vandelook and P. Stoffelen, personal observations). Currently, important Robusta research centers are situated in Brazil, Vietnam, Uganda, and India. Valuable Robusta genetic resources, including material originating from the INEAC station in Yangambi, are held in collections in Cameroon, Ivory Coast, India, and Madagascar (Cubry et al., 2013; Bramel et al., 2017).
In contrast to C. arabica (Aerts et al., 2013), vast amounts of untouched wild genetic diversity of C. canephora are expected to have remained across its wide distribution range. Wild C. canephora in the rainforests of West and Central Africa, from Guinea to Uganda occupies the largest distribution area among all Coffea species (Noirot et al., 2016). A recent molecular study demonstrated the presence of eight clearly delineated genetic clusters in wild Coffea canephora populations (Merot‐L'anthoene et al., 2019). One of these genetic clusters roughly encompasses the northeastern part of the Congo Basin, including the Yangambi area (Tshopo Province). The cluster covers a large area northeast of the Congo River and the city of Kisangani. Vegetation in this area is characterized by both old‐growth and disturbed forests (Gilson, 1956) in which wild C. canephora populations are present as understory shrubs, often sympatric with Coffea liberica and Coffea dactylifera (F. Vandelook, personal observations). Coffea canephora shrubs typically grow at low density in small, disconnected populations (Musoli et al., 2009). Information on population genetic diversity and structure of wild C. canephora is scarce and, as far as we know, not available for the DR Congo.
The Congo Basin is hypothesized to be the region of origin of many cultivated Robusta coffee genotypes (Dulloo et al., 1998; Cubry et al., 2013). Consequently, populations of C. canephora native to this region contain a valuable part of the wild gene pool, but the extent of this genetic reservoir remains unknown. In Uganda, comparison of cultivated Robusta accessions with wild C. canephora populations indicated a significantly higher genetic diversity among cultivated accessions than in wild populations (Musoli et al., 2009). A recent study has shown that cultivated accessions in Uganda are genetically very similar to wild populations from southwestern Uganda (Kiwuka et al., 2021), suggesting a common genetic origin, a recent introduction to cultivation, and limited breeding. Coffea canephora is mainly cultivated in plantations in Uganda, but not in the Congolese Tshopo and Ituri provinces. Although Coffea canephora plantations were found throughout the Tshopo and Ituri provinces in the 20th century, these plantations have disappeared over the last decades. Currently, C. canephora shrubs are mostly grown in small‐scale backyard garden systems consisting of only a few shrubs for domestic use.
Although the DR Congo potentially harbors an enormous reservoir of genetic diversity of C. canephora and has a century long history of breeding and cultivation, virtually nothing is known about the genetic composition of either the wild or the cultivated C. canephora in this region. Therefore, we present for the first time a study focused on the genetic diversity of C. canephora in the DR Congo, in which we apply population genetics methods on wild and backyard‐grown shrubs in the Tshopo and Ituri provinces and on multiple accessions in the INERA Yangambi Coffee Collection. The following questions were addressed: (1) Are backyard‐grown coffee shrubs genetically different from nearby wild shrubs? (2) How does the genetic diversity compare between cultivated and wild shrubs? (3) How much (local) genetic diversity is preserved in the Coffee Collection of the INERA Yangambi? (4) What is the level of genetic differentiation among wild Coffea canephora populations? (5) What are the implications for the conservation of C. canephora genetic resources in the DR Congo?
MATERIALS AND METHODS
Taxon sampling and DNA extraction
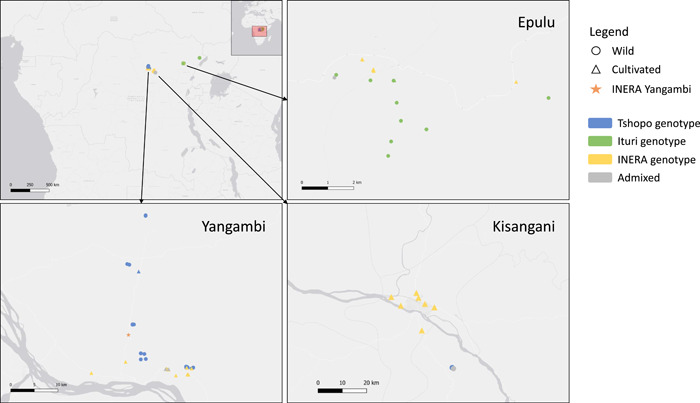
Leaf samples of wild and cultivated Coffea canephora shrubs were collected at multiple localities in the DR Congo (Figure 1). Natural populations were sampled in the Yangambi and Yoko reserves (both in Tshopo Province) and in Epulu and Djugu (both in Ituri Province). Cultivated specimens were collected from backyards in Yangambi and Kisangani (both in Tshopo Province) and Epulu, very often fairly close to the wild shrubs. The INERA Yangambi Coffee Collection was sampled exhaustively (45 samples). In total, 195 leaf samples were collected (Appendix S1) and dried with silica gel for molecular analyses. Genomic DNA was isolated using a cetyltrimethylammonium bromide (CTAB) protocol (Doyle and Doyle, 1990) with an additional sorbitol washing step (Janssens et al., 2006).
Microsatellite primer selection and genotyping
Microsatellite loci were amplified using 18 primer pairs previously used on wild and cultivated C. canephora samples from Uganda (Kiwuka et al., 2021). To reduce the cost of primers, multiplex PCRs were done using an M13‐like labelling protocol as described by Schuelke (2000). Therefore, a unique Q‐tail sequence (i.e., Q1 after Schuelke [2000] and Q2, Q3, or Q4 after Culley et al. [2008]) was added to the 5′ end of the original reverse primers (Appendix S2). The PCR mix (final volume of 15.75 µL) consisted of 7.5
µL) consisted of 7.5 µL Type‐it Multiplex PCR Master Mix (QIAGEN, Hilden, Germany), 3
µL Type‐it Multiplex PCR Master Mix (QIAGEN, Hilden, Germany), 3 µL Q solution (5×), 0.3
µL Q solution (5×), 0.3 µL unlabeled forward primer (10
µL unlabeled forward primer (10 µM), 0.1
µM), 0.1 µL Q‐tailed reverse primer (10
µL Q‐tailed reverse primer (10 µM), 0.3
µM), 0.3 µL of a primer (10
µL of a primer (10 µM) composed of the same universal Q1–Q4 sequence with 6‐FAM, NED, VIC, and PET fluorescent dye, respectively, attached to the 5′ end, 1
µM) composed of the same universal Q1–Q4 sequence with 6‐FAM, NED, VIC, and PET fluorescent dye, respectively, attached to the 5′ end, 1 µL DNA extract, and H2O. Multiplex PCR conditions were as follows: initial denaturation at 95°C (3
µL DNA extract, and H2O. Multiplex PCR conditions were as follows: initial denaturation at 95°C (3 min); 25 cycles of denaturation at 95°C (30
min); 25 cycles of denaturation at 95°C (30 s), annealing at 57°C (45
s), annealing at 57°C (45 s), elongation at 72°C (1
s), elongation at 72°C (1 min); 10 cycles of denaturation at 95°C (30
min); 10 cycles of denaturation at 95°C (30 s), annealing at 53°C (45
s), annealing at 53°C (45 s), elongation at 72°C (60
s), elongation at 72°C (60 s); and a final extension step at 72°C (10
s); and a final extension step at 72°C (10 min).
min).
Genotyping was done on an ABI 3730 DNA Analyzer (Thermo Fisher Scientific; Applied Biosystems, Waltham, MA, USA) with 1.5 µL PCR product, 12
µL PCR product, 12 µL Hi‐Di Formamide (Thermo Fisher Scientific; Applied Biosystems) and 0.3
µL Hi‐Di Formamide (Thermo Fisher Scientific; Applied Biosystems) and 0.3 µL MapMarker 500 labelled with DY‐632 (Eurogentec, Seraing, Belgium). Allele calling and locus bin setting was done using the Microsatellite Plugin 1.4.6 in Geneious 9.1.6 (Kearse et al., 2012).
µL MapMarker 500 labelled with DY‐632 (Eurogentec, Seraing, Belgium). Allele calling and locus bin setting was done using the Microsatellite Plugin 1.4.6 in Geneious 9.1.6 (Kearse et al., 2012).
Genetic population structure and admixture
We used the Bayesian clustering algorithm implemented in structure software v. 2.3.4 (Pritchard et al., 2000) to infer population structure and to assess levels of admixture among cultivated and wild C. canephora plants collected in northeastern DR Congo. The following parameters were used: burn‐in period and number of MCMC replicates after burn‐in both set at 100,000, admixture model, independent allele frequency model, maximum number of clusters set between K =
= 1 and K
1 and K =
= 10, and 10 iterations for each K. As recommended by Wang (2017), an alternative ancestry prior α was used, which improves individual assignments and inference of the number of clusters K, even if sampling is highly unbalanced (Wang, 2017). This alternative was especially useful in our case, since few samples from Djugu and Yoko were included, and the INERA Yangambi Coffee Collection contains rare specimens from underrepresented localities (Appendix S1). Therefore, the ancestry prior α for each cluster was assumed to be distinct and α was set to an initial value of 0.25 (equals 1/K, with K
10, and 10 iterations for each K. As recommended by Wang (2017), an alternative ancestry prior α was used, which improves individual assignments and inference of the number of clusters K, even if sampling is highly unbalanced (Wang, 2017). This alternative was especially useful in our case, since few samples from Djugu and Yoko were included, and the INERA Yangambi Coffee Collection contains rare specimens from underrepresented localities (Appendix S1). Therefore, the ancestry prior α for each cluster was assumed to be distinct and α was set to an initial value of 0.25 (equals 1/K, with K =
= 4 based on preliminary clustering runs). By declaring recessive null alleles for all loci in structure, null allele frequencies were estimated and accounted for. The most optimal number of genetic clusters was determined by plotting the log‐likelihood of the data Ln P(D) against the number of clusters K (Pritchard et al., 2000) using STRUCTURE HARVESTER (Earl and vonHoldt, 2012), and by assessing the stability of replicate runs for each K (10 iterations per K).
4 based on preliminary clustering runs). By declaring recessive null alleles for all loci in structure, null allele frequencies were estimated and accounted for. The most optimal number of genetic clusters was determined by plotting the log‐likelihood of the data Ln P(D) against the number of clusters K (Pritchard et al., 2000) using STRUCTURE HARVESTER (Earl and vonHoldt, 2012), and by assessing the stability of replicate runs for each K (10 iterations per K).
The genetic diversity among the wild and cultivated specimens was summarized using a principal component analysis (PCA) and visualized as a scatterplot with the R (R Core Team, 2011) packages adegenet (Jombart, 2008) and ade4 (Chessel et al., 2007).
Genetic diversity and differentiation
To compare genetic diversity among wild and cultivated C. canephora shrubs and among the different sampling locations, the following genetic diversity indices were calculated: number of alleles (NA), number of effective alleles (N e), rarefied allelic richness (AR), expected heterozygosity (H e), observed heterozygosity (H o), and inbreeding coefficient (FIS). Pairwise genetic differentiation was assessed by calculating F ST between wild and cultivated C. canephora shrubs and between wild populations from different geographic areas. Furthermore, allele frequencies for the 18 microsatellite loci were calculated and compared among the wild and cultivated specimens to assess the genetic similarity of cultivated and wild C. canephora shrubs. All calculations were done using SPAGeDi 1.5d (Hardy and Vekemans, 2002).
RESULTS
Genetic population structure and admixture
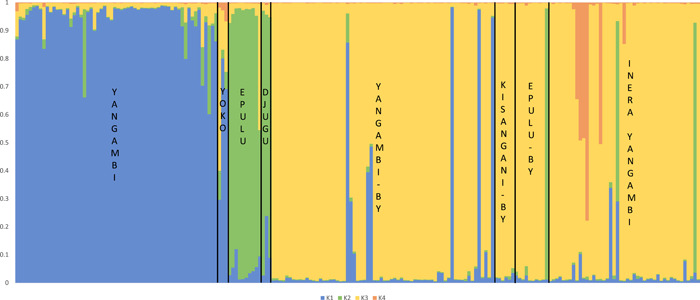
Four genetic clusters were inferred for our C. canephora data set (Figure 2) using the Bayesian clustering algorithm implemented in structure. The plotted log‐likelihood of the data Ln P(D) against the number of clusters K showed that the increase in average Ln P(D) was highest between K =
= 1 and K
1 and K =
= 2, while the highest average Ln P(D) was observed at K
2, while the highest average Ln P(D) was observed at K =
= 4 (Appendix S3). Variability in Ln P(D) among the 10 iterations was relatively low across all K‐values. In this clustering analysis, wild populations were first separated from the cultivated plants sampled in backyards and from the INERA Yangambi collection (at K
4 (Appendix S3). Variability in Ln P(D) among the 10 iterations was relatively low across all K‐values. In this clustering analysis, wild populations were first separated from the cultivated plants sampled in backyards and from the INERA Yangambi collection (at K =
= 2) (Appendix S4). Subsequently, wild populations were subdivided based on geographic origin, separating the specimens collected in Epulu and Djugu (Ituri Province) from specimens collected in Yangambi and Yoko (Tshopo province) (at K
2) (Appendix S4). Subsequently, wild populations were subdivided based on geographic origin, separating the specimens collected in Epulu and Djugu (Ituri Province) from specimens collected in Yangambi and Yoko (Tshopo province) (at K =
= 3). Lastly, six accessions from the INERA Yangambi collection were separated (at K
3). Lastly, six accessions from the INERA Yangambi collection were separated (at K =
= 4). Of these six accessions, four were identified as ‘Petit Kwilu’, a variety originating from the Mayombe (western DR Congo and Congo‐Brazzaville, Cabinda and coastal Gabon). These six distinct individuals showed increasing levels of admixture at K
4). Of these six accessions, four were identified as ‘Petit Kwilu’, a variety originating from the Mayombe (western DR Congo and Congo‐Brazzaville, Cabinda and coastal Gabon). These six distinct individuals showed increasing levels of admixture at K =
= 5 to K
5 to K =
= 10 (Appendix S4).
10 (Appendix S4).
Some of the specimens collected from backyards in Yangambi and Epulu had genotypes that matched the wild genotypes found in the local forest populations (four and one specimen, respectively) (Figures 1 and 2). By contrast, no (introduced) wild genotypes were found in the backyards in Kisangani. Overall, six specimens had a “hybrid” wild–cultivated genotype: one in Yoko, one in Epulu, three in Yangambi backyards, and one in the INERA Yangambi collection (SY66). One accession (L51Y65) from the INERA Yangambi collection had a genotype that matched the wild genotypes from Ituri Province, and one accession (NA; no accession number available) showed a mix of the wild genotypes from Ituri and Tshopo. The structure assignment probabilities also indicated low levels of admixture between wild populations from the Tshopo and the Ituri Province (Figure 2).
The PCA (Appendix S5) showed similar clustering as obtained with structure, with the first axis (PC1) mostly separating wild from cultivated specimens. Along the second axis (PC2), the wild populations were separated depending on their geographic origin (Ituri vs. Tshopo Province), and the six distinct accessions collected from INERA Yangambi (which were separated at K =
= 4 in the structure cluster analysis) were separated from the rest of the cultivated specimens. The third axis (PC3) again separated the wild populations based on their geographic origin, highlighting the relatively high genetic diversity found in the wild populations. The first three principal components explained 6.09%, 3.38%, and 2.86% of the variance.
4 in the structure cluster analysis) were separated from the rest of the cultivated specimens. The third axis (PC3) again separated the wild populations based on their geographic origin, highlighting the relatively high genetic diversity found in the wild populations. The first three principal components explained 6.09%, 3.38%, and 2.86% of the variance.
Genetic diversity and differentiation
Both the number of alleles (NA) and the effective number of alleles (N
e) were highest for the wild populations (NA =
= 7.94, N
e
7.94, N
e =
= 3.94), followed by the INERA Yangambi Coffee Collection (NA
3.94), followed by the INERA Yangambi Coffee Collection (NA =
= 7.39, N
e
7.39, N
e =
= 3.37) and the backyard samples (NA
3.37) and the backyard samples (NA =
= 6.89, N
e
6.89, N
e =
= 3.09) (Table 1). The allelic richness (AR, among 12 gene copies k), expected heterozygosity (H
e) and observed heterozygosity (H
o) were highest in the INERA Yangambi collection (AR
3.09) (Table 1). The allelic richness (AR, among 12 gene copies k), expected heterozygosity (H
e) and observed heterozygosity (H
o) were highest in the INERA Yangambi collection (AR =
= 4.27, H
e
4.27, H
e =
= 0.64, H
o
0.64, H
o =
= 0.53), followed by the wild populations (AR
0.53), followed by the wild populations (AR =
= 4.25, H
e
4.25, H
e =
= 0.63, H
o
0.63, H
o =
= 0.47) and the backyard samples (AR
0.47) and the backyard samples (AR =
= 3.83, H
e
3.83, H
e =
= 0.59, H
o
0.59, H
o =
= 0.50). Since a relatively large part of the genetic diversity in the INERA Yangambi collection might originate from the six distinct accessions (LAF159, S23, S19, L6, L251Y128, and NA) that were separated in the PCA and the cluster analysis, genetic diversity indices were also calculated without the respective six accessions. As a result, the estimates of genetic diversity were lower for the INERA Yangambi collection (NA
0.50). Since a relatively large part of the genetic diversity in the INERA Yangambi collection might originate from the six distinct accessions (LAF159, S23, S19, L6, L251Y128, and NA) that were separated in the PCA and the cluster analysis, genetic diversity indices were also calculated without the respective six accessions. As a result, the estimates of genetic diversity were lower for the INERA Yangambi collection (NA =
= 6.61, N
e
6.61, N
e =
= 3.18, AR
3.18, AR =
= 4.01, H
e
4.01, H
e =
= 0.62, and H
o
0.62, and H
o =
= 0.50). These estimates were all slightly lower than the genetic diversity measures estimated in the wild populations (except for H
o). Among the wild populations, genetic diversity was higher in Tshopo Province (NA
0.50). These estimates were all slightly lower than the genetic diversity measures estimated in the wild populations (except for H
o). Among the wild populations, genetic diversity was higher in Tshopo Province (NA =
= 6.83, N
e
6.83, N
e =
= 3.57, AR
3.57, AR =
= 4.04, H
e
4.04, H
e =
= 0.61, and H
o
0.61, and H
o =
= 0.48) than in Ituri Province (NA
0.48) than in Ituri Province (NA =
= 4.78, N
e
4.78, N
e =
= 3.45, AR
3.45, AR =
= 3.84, H
e
3.84, H
e =
= 0.59, and H
o
0.59, and H
o =
= 0.46). The number of alleles (NA and N
e) might be higher in Tshopo Province because of the larger sample size, while AR and H
e were not affected by sample size. Individual inbreeding coefficients (F
IS) were significant for all groups and ranged between 0.16 (backyard specimens) and 0.26 (wild populations) (Table 1).
0.46). The number of alleles (NA and N
e) might be higher in Tshopo Province because of the larger sample size, while AR and H
e were not affected by sample size. Individual inbreeding coefficients (F
IS) were significant for all groups and ranged between 0.16 (backyard specimens) and 0.26 (wild populations) (Table 1).
Table 1
Genetic diversity variables for wild and cultivated Coffea canephora populations
| Population | n | NA | N e | AR (k = = 12) 12) | H e | H o | F IS |
|---|---|---|---|---|---|---|---|
| Wild | 69 | 7.94 | 3.94 | 4.25 | 0.64 | 0.47 | 0.26 |
| Tshopo Province | 56 | 6.83 | 3.57 | 4.04 | 0.61 | 0.48 | 0.22 |
| Ituri Province | 13 | 4.78 | 3.45 | 3.84 | 0.59 | 0.46 | 0.24 |
| Backyards | 81 | 6.89 | 3.09 | 3.83 | 0.59 | 0.50 | 0.16 |
| Backyards – 5ind | 76 | 6.33 | 2.98 | 3.68 | 0.58 | 0.50 | 0.13 |
| INERA Yangambi Collection | 45 | 7.39 | 3.37 | 4.27 | 0.64 | 0.53 | 0.18 |
| Collection – 6ind | 39 | 6.61 | 3.18 | 4.01 | 0.62 | 0.50 | 0.20 |
| All | 195 | 10.67 | 4.03 | 4.57 | 0.67 | 0.50 | 0.26 |
Notes: n, number of individuals analyzed; NA, number of alleles; N
e, effective number of alleles (Nielsen et al., 2003); AR (k =
= x), allelic richness or number of alleles among x gene copies; H
e, expected heterozygosity corrected for sample size; H
o, observed heterozygosity; F
IS, individual inbreeding coefficient. F
IS was significantly larger than 0 for all populations. Backyards – 5ind, all the cultivated backyard individuals without the five individuals with a wild genotype; Collection – 6ind, all the cultivated individuals from the INERA Yangambi Coffee Collection without the six genetically distinct individuals (mostly ‘Petit Kwilu’ accessions).
x), allelic richness or number of alleles among x gene copies; H
e, expected heterozygosity corrected for sample size; H
o, observed heterozygosity; F
IS, individual inbreeding coefficient. F
IS was significantly larger than 0 for all populations. Backyards – 5ind, all the cultivated backyard individuals without the five individuals with a wild genotype; Collection – 6ind, all the cultivated individuals from the INERA Yangambi Coffee Collection without the six genetically distinct individuals (mostly ‘Petit Kwilu’ accessions).
The allele frequencies calculated for the 18 microsatellite loci in the wild and cultivated specimens showed that all loci harbored alleles that were unique to at least one of both categories (Table 2): three loci harbored alleles that were unique to cultivated specimens (R325, SSR209, R342), while the other 15 loci harbored unique alleles for both cultivated and wild specimens. For one locus (SSR196, 14 alleles), only unique alleles were observed, either to wild or to cultivated specimens. Among all loci, we found 52 alleles that were unique to wild specimens and 48 alleles unique to cultivated specimens. Nine of those 48 alleles were only present in the six cultivated specimens with a distinct genotype, collected from INERA Yangambi collection. Furthermore, we found five alleles among all loci that were only present in wild specimens and in those six distinct cultivated specimens, but not in any of the other cultivated specimens.
Table 2
The number (No.) of alleles per locus that were unique to wild or cultivated specimens. The number between brackets () indicates the number of alleles that were unique to the six genetically distinct individuals from the INERA Yangambi Coffee Collection (mostly ‘Petit Kwilu’ accessions)
| Locus | No. of alleles | No. of alleles unique to wild specimens | No. of alleles unique to cultivated specimens |
|---|---|---|---|
| R338 | 13 | 2 | 4 (1) |
| SSR146 | 9 | 2 | 3 (1) |
| R278 | 11 | 5 | 1 |
| R339 | 17 | 6 | 1 |
| R336 | 14 | 2 | 4 |
| R325 | 5 | 0 | 3 |
| SSR196 | 14 | 8 | 6 (1) |
| R268 | 7 | 3 | 2 (1) |
| R168 | 11 | 3 | 1 |
| R189 | 9 | 1 | 2 |
| SSR497 | 19 | 7 | 1 |
| R175 | 7 | 2 | 1 |
| SSR495 | 8 | 2 | 3 (1) |
| R250 | 15 | 2 | 6 |
| R148 | 9 | 4 | 1 |
| SSR209 | 8 | 0 | 4 (3) |
| R342 | 4 | 0 | 1 |
| SSR533 | 11 | 3 | 4 (1) |
| Total | 191 | 52 | 48 |
Pairwise genetic differentiation (F
ST) (Table 3) was highest between the wild populations and the specimens collected from backyards (F
ST =
= 0.144) or from the INERA Yangambi collection (F
ST
0.144) or from the INERA Yangambi collection (F
ST =
= 0.135). Genetic differentiation was low between the specimens collected from backyards and from the INERA Yangambi collection (F
ST
0.135). Genetic differentiation was low between the specimens collected from backyards and from the INERA Yangambi collection (F
ST =
= 0.004). Pairwise genetic differentiation between the wild populations from the Tshopo and the Ituri Province was slightly lower than the differentiation between all wild populations and cultivated accessions in backyards and the INERA Yangambi Coffee collection (F
ST
0.004). Pairwise genetic differentiation between the wild populations from the Tshopo and the Ituri Province was slightly lower than the differentiation between all wild populations and cultivated accessions in backyards and the INERA Yangambi Coffee collection (F
ST =
= 0.119).
0.119).
Table 3
Genetic differentiation (F ST) between wild and cultivated specimens of Coffea canephora, and among wild populations from Tshopo and Ituri provinces in northeastern DR Congo
| Wild | Backyards | |
|---|---|---|
| Backyards | 0.144 | |
| INERA Collection | 0.135 | 0.004 |
| Tshopo | ||
| Ituri | 0.119 | |
DISCUSSION
The distribution of the genetic diversity of wild and cultivated C. canephora in Tshopo and Ituri provinces revealed some unexpected patterns. Firstly, the agreement in the genetic constitution of the INERA Yangambi accessions and the vast majority of the shrubs in the backyard gardens, indicates that most people growing C. canephora locally, received their material directly or indirectly from INERA breeding programs. Secondly, the cultivated shrubs, both from backyards and the INERA collection, are genetically clearly distinct from the local wild gene pool, showing relatively large genetic differentiation. Levels of genetic diversity are similar for the INERA and the wild populations, but both wild and cultivated specimens harbor numerous unique alleles. Finally, the wild gene pool from Tshopo and Ituri provinces is not represented in the INERA Yangambi collection. These observations have important conservation implications as will be discussed below.
Genetic diversity, structure, and origin of wild Coffea canephora
Genetic diversity of C. canephora was slightly higher in Tshopo Province as compared to the shrubs sampled in Ituri, although expected and observed heterozygosity were fairly similar. This result can be explained by the broader geographic sampling in Tshopo Province, which included wild populations from both the Yangambi and the Kisangani region (including Yoko). The expected heterozygosity in Tshopo and Ituri provinces (H e ~0.60) matched that of the most diverse populations in Uganda (Kiwuka et al., 2021) and that of some of the most‐diverse undisturbed C. arabica stands (Aerts et al., 2013). Both studies used SSR markers with 19 and 24 microsatellite loci, respectively, as compared to 18 in our study. The values of expected heterozygosity are at the upper end of those estimated for diversity of an outcrossing perennial plant using microsatellite markers (0.47–0.68; Nybom, 2004). The pronounced self‐incompatibility system in C. canephora (Lashermes et al., 1996) seems to ensure the maintenance of high levels of heterozygosity within populations.
Relatively strong genetic differentiation (F
ST =
= 0.119) was observed between wild populations from Tshopo province and Epulu, which are separated about 400
0.119) was observed between wild populations from Tshopo province and Epulu, which are separated about 400 km from each other. A more pronounced genetic structure is found amongst wild C. canephora populations in Uganda, which can be explained by the more pronounced (historic and/or present‐day) population fragmentation in the area (Kiwuka et al., 2021). The tropical rainforest in the Congo Basin is still unfragmented, and we can expect that C. canephora shrubs are distributed somewhat continuously throughout this rainforest. However, past glaciations (e.g., during the Pleistocene) drastically reduced the rainforest cover (Maley, 1996; Anhuf, 2000; Gomez et al., 2009; Hardy et al., 2013), which caused genetic differentiation between populations in isolated forest refugia due to genetic drift, bottlenecks and inbreeding. Such past barriers to gene flow could (partly) explain the observed differentiation between the populations in Tshopo and Ituri, possibly in combination with present‐day dispersal barriers. While little is known about pollinator specificity in wild C. canephora shrubs and the distances the pollinators can cover, it has been suggested that long‐distance seed dispersal of the red Coffea berries by birds and perhaps mammals could potentially reach 100 km, thus contributing significantly to gene flow across large distances (Charrier, 1971; Berthaud, 1986). However, this claim of long‐distance dispersal has been disputed due to the fact that birds living in the rainforest understory commonly have a sedentary habit and a rapid gut passage (Theim et al., 2014; Grant et al., 2019).
km from each other. A more pronounced genetic structure is found amongst wild C. canephora populations in Uganda, which can be explained by the more pronounced (historic and/or present‐day) population fragmentation in the area (Kiwuka et al., 2021). The tropical rainforest in the Congo Basin is still unfragmented, and we can expect that C. canephora shrubs are distributed somewhat continuously throughout this rainforest. However, past glaciations (e.g., during the Pleistocene) drastically reduced the rainforest cover (Maley, 1996; Anhuf, 2000; Gomez et al., 2009; Hardy et al., 2013), which caused genetic differentiation between populations in isolated forest refugia due to genetic drift, bottlenecks and inbreeding. Such past barriers to gene flow could (partly) explain the observed differentiation between the populations in Tshopo and Ituri, possibly in combination with present‐day dispersal barriers. While little is known about pollinator specificity in wild C. canephora shrubs and the distances the pollinators can cover, it has been suggested that long‐distance seed dispersal of the red Coffea berries by birds and perhaps mammals could potentially reach 100 km, thus contributing significantly to gene flow across large distances (Charrier, 1971; Berthaud, 1986). However, this claim of long‐distance dispersal has been disputed due to the fact that birds living in the rainforest understory commonly have a sedentary habit and a rapid gut passage (Theim et al., 2014; Grant et al., 2019).
Diversity and origin of the INERA Yangambi collection
The majority of the accessions in the INERA Yangambi collection are referred to as Lula varieties (Appendix S1) and presumably originate from the Lula Research Station near Kisangani. The wild origin of the Lula variety remains unclear, but given the high level of distinct alleles in the wild and cultivated shrubs, it seems unlikely that the origin is to be found in our sampling region, i.e., Tshopo and Ituri. Nonetheless, the Lula varieties are assumed to have originated from the Congo Basin and probably root back to the early introduction of “Coffea robusta” by Linden from Sankuru, but additional sampling and research are needed to trace the region of origin. Cultivated material from the former INEAC Yangambi collection is still present in the CNRA collection in the Ivory Coast (Cubry et al., 2013), but this cultivated material dates back to 1935 (Bodard, 1965). The CNRA C. canephora material originating from the former INEAC was shown to be closely related to C. canephora growing in Uganda (Cubry et al., 2013; Leroy et al., 2014). However, further studies are required to assess the relationships between the Lula variety currently present in the INERA Yangambi Coffee Collection and the material distributed to CNRA during the INEAC period and the Ugandan gene pool. The relatively high genetic variation in the INERA Yangambi Coffee Collection, as expressed by the high levels of heterozygosity and allelic richness, can be explained by the diverse origin of several rare genetic lines in the collection. Four Petit Kwilu accessions, originating from the Mayombe Region (western DR Congo, Congo and Gabon), in the collection were genetically clearly distinct from the Lula varieties, as was confirmed by previous studies (e.g., Leroy et al., 2014). In addition, accessions originating from the North Kivu, Haute Zaire, and Equateur provinces, with one representative each, were also present in the collection (Appendix S1), but note that the provenances in the collection are not well documented. Very little (local) wild genetic diversity seems to be preserved in the INERA Yangambi Coffee Collection. The introduction of the local wild genetic diversity into the INERA field collection is a relatively easy, but very important, way to enrich Robusta coffee genetic resources in the collection. The availability of local genetic resources in the collection could potentially be a useful source for breeding of C. canephora shrubs that are adapted to local soil and climatic conditions. In addition, ex situ conservation of local genetic resources, which are threatened by deforestation, change in forest structure, and the disappearance of seed dispersers (Sellan et al., 2017; van Vliet et al., 2018; Kyale Koy et al., 2019) can complement in situ conservation efforts.
Backyard garden cultivation
Genetic differentiation between C. canephora shrubs in the INERA Yangambi Coffee Collection and the shrubs in backyards was very low. The vast majority of the coffee plants in backyards are most likely Lula varieties originating from the INERA breeding program, that have been distributed to local villagers. Even the cultivated plants in the Ituri province were closely related to the Lula variety. Since coffee grown in backyards in the study region is used mainly for home consumption and for leaf decoctions (Campa et al., 2012), the dominance of non‐local Lula varieties was somewhat surprising. The lower genetic diversity of backyard shrubs can be explained by the fact that seven Lula elite lines, originating from the INERA Yangambi collection, are mainly used for germplasm production and distribution (T. Ebele, personal observations). Since breeding activities at the INERA Yangambi have been reduced drastically over the last decade, we can expect that the genetic resources that have been distributed have remained relatively uniform. The large gene pool of wild C. canephora shrubs in the Tshopo and Ituri provinces was very poorly represented in the backyard cultivation systems of local villagers, despite the fact that they are sometimes separated by a kilometer or less. Moreover, genetic differentiation was highest between the wild and the backyard gene pool, and only 5 of 81 samples (6.2%) collected in backyards could be assigned a local wild origin. This observation contrasts somewhat with the situation for C. canephora in Uganda, where gene pools of cultivated and wild plants are much more mixed (Musoli et al., 2009; Kiwuka et al., 2021). This lower genetic differentiation between cultivated and wild gene pools in Uganda is likely due to more extensive use of local wild genetic resources during Robusta cultivation and to a more recent and less extensive selection process.
Due to the close vicinity of cultivated C. canephora shrubs (sometimes less than 1 km) and the limited domestication (wild shrubs are morphologically quite similar to cultivated material), cultivated shrubs would be expected to considerably impact the integrity of the wild gene pool. Indications of crop–wild introgression in coffee were found in a study of C. arabica populations in Ethiopia (Aerts et al., 2013) and Coffea canephora in Uganda (Kiwuka et al., 2021), yet we found little evidence for gene flow from the cultivated gene pool into the wild gene pool in the NE Congo Basin. Any attempt to explain this contrasting observation would be speculative at this point, and a broader sampling is needed to confirm this observation. Five putative crosses between wild local shrubs and cultivated C. canephora shrubs were, however, observed in backyards of Yangambi, Yoko, and Epulu. The origin of such “hybrids” appears to be unclear, yet the most reasonable explanation would be a crossing event from wild and cultivated C. canephora growing together in backyards.
CONCLUSIONS
The present findings show that the cultivated Coffea canephora accessions from INERA Yangambi and the vast majority of the shrubs in the backyard gardens in northeastern DR Congo are genetically very similar. These results indicate that most people growing C. canephora locally received their material directly or indirectly from INERA breeding programs, while a few shrubs were obtained directly through collections from surrounding forests. Furthermore, the cultivated shrubs from the backyards and the INERA collection are genetically distinct from the local wild gene pool, showing relatively large genetic differentiation. and both gene pools harbor multiple unique alleles. The introduction of the local wild genetic diversity into the INERA field collection is a great way to increase the genetic resources available for breeding purposes and to support the ex situ conservation of C. canephora. High‐throughput sequencing in future studies would be beneficial to characterize putative introgression and gene flow between cultivated shrubs and local wild populations, often growing in close proximity. Such genetic studies can be complemented with studies focused on pollen and seed dispersers, since little is known about gene‐dispersal mechanisms in C. canephora and in tropical understory shrubs in general.
AUTHOR CONTRIBUTIONS
F.V., S.V.A., P.S., and S.B.J. conceived and designed the study. S.V.A., F.V., P.S., and S.B.J. wrote the manuscript. Y.B. did the molecular work. S.V.A. analyzed the data. F.V., S.N., J.A.A., B.K., I.M.M., S.V.A., and P.S. collected data and samples. All authors read and approved the final version of the manuscript.
Supporting information
Appendix S1. List of wild and cultivated Coffea canephora accessions from northeastern Democratic Republic of the Congo included in the study.
Appendix S2. Sequences for the microsatellite primers used in the present study, including fluorescent Q‐tail and multiplex information.
Appendix S3. Likelihood of the microsatellite data set as a function of the assumed number of genetic clusters (K) according to the Bayesian clustering algorithm implemented in structure.
Appendix S4. Bar plots for K =
= 2 to K
2 to K =
= 4, and K
4, and K =
= 10 representing the assignment probabilities (y‐axis) inferred using structure, for the complete Coffea canephora microsatellite dataset.
10 representing the assignment probabilities (y‐axis) inferred using structure, for the complete Coffea canephora microsatellite dataset.
Appendix S5. Principal component analysis of the genetic diversity in the Coffea canephora microsatellite data set.
ACKNOWLEDGMENTS
We thank Wim Baert (Meise Botanic Garden) and Pieter Asselman (UGent) for support with molecular work. We are grateful to the Ministère de L'Environnement et Développement Durable and Institut National pour l'Étude et la Recherche Agronomiques (INERA), Democratic Republic of Congo for their help with obtaining collecting permits. We thank Associate Editor Daniel Potter and an anonymous reviewer for their helpful comments. This study was financially supported by EU XIth Development Fund (FORETS project), the Foundation for the promotion of biodiversity research in Africa (SBBOA, www.sbboa.be), Research Foundation—Flanders (Research project G090719N), the Belgian Science Policy (BELSPO; grant B2/191/P1/COFFEEBRIDGE). Fieldwork was financially supported by Institut de Recherche pour le Développement (IRD). S.V.A. received funding from the Belgian American Educational Foundation (www.baef.be); J.D. received funding from the Research Foundation—Flanders (FWO; 1125221N).
Notes
Vanden Abeele S., Janssens S. B., Asimonyio Anio J., Bawin Y., Depecker J., Kambale B., Mwanga Mwanga I., Ebele T., Ntore S., Stoffelen P., and Vandelook F.. 2021. Genetic diversity of wild and cultivated Coffea canephora in northeastern DR Congo and the implications for conservation. American Journal of Botany 108(12): 2425–2434. 10.1002/ajb2.1769 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
DATA AVAILABILITY STATEMENT
Voucher information and sampling location for the Coffea canephora accessions included in this study are given in Appendix S1 of the Supporting Information. The data set with the corresponding genotypes is available from the Zenodo repository (https://doi.org/10.5281/zenodo.5378527).
REFERENCES
- Aerts R., Berecha G., Gijbels P., Hundera K., Van Glabeke S., Vandepitte K., Muys B., et al. 2013. Genetic variation and risks of introgression in the wild Coffea arabica gene pool in south‐western Ethiopian montane rainforests. Evolutionary Applications 6: 243–252. [Europe PMC free article] [Abstract] [Google Scholar]
- Anhuf D. 2000. Vegetation history and climate changes in africa north and south of the equator (10° S to 10° N) during the last glacial maximum. In P. Smolka , editor; and W. Volkheimer , editor. [eds.], Southern hemisphere paleo‐ and neoclimates: key sites, methods, data and models, 225–248. Springer, Berlin, Germany. [Google Scholar]
- Berthaud J. 1986. Les ressources génétiques pour l'amélioration des caféiers africains diploiedes: évaluation de la richesse génétique des populations sylvestres et de ses mécanismes organisateurs. Conséquences pour l'application. ORSTOM, Paris, France.
- Bodard L. 1965. Historique des caféiers de RCI. [no other information available].
- Bramel P., Krishnan S., Horna D., Lainoff B., and Montagnon C.. 2017. Global conservation strategy for coffee genetic resources. Crop Trust and World Coffee Research, Bonn, Germany.
- Campa C., Mondolot L., Rakotondravao A., Bidel L. P. R., Gargadennec A., Couturon E., La Fisca P., et al. 2012. A survey of mangiferin and hydroxycinnamic acid ester accumulation in coffee (Coffea) leaves: biological implications and uses. Annals of Botany 110: 595–613. [Europe PMC free article] [Abstract] [Google Scholar]
- Charrier A. 1971. Study of pollination in cultivated coffee by labelling pollen with radioactive phosphorus and sulphur. Café Cacao Thé 181–190. [Google Scholar]
- Chessel D., Dufour A., and Thioulouse J.. 2007. The ade4 package ‐ I: One‐table methods. R News 7: 47–52. [Google Scholar]
- Chevalier A. 1929. Les caféiers du globe. Fasc. I. Généralités sur les caféiers. Encyclopédie Biologique 5. P. Lechevalier, Paris, France.
- Cubry P., De Bellis F., Pot D., Musoli P., and Leroy T.. 2013. Global analysis of Coffea canephora Pierre ex Froehner (Rubiaceae) from the Guineo‐Congolese region reveals impacts from climatic refuges and migration effects. Genetic Resources and Crop Evolution 60: 483–501. [Google Scholar]
- Culley T. M., Weller S. G., Sakai A. K., and Putnam K. A.. 2008. Characterization of microsatellite loci in the Hawaiian endemic shrub Schiedea adamantis (Caryophyllaceae) and amplification in related species and genera. Molecular Ecology Resources 8: 1081–1084. [Abstract] [Google Scholar]
- Doyle J. J., and Doyle J. L.. 1990. Isolation of plant DNA from fresh plant tissue. Focus 12: 13–15. [Google Scholar]
- Dulloo M. E., Guarino L., Engelmann F., Maxted N., Newbury J. H., Attere F., and Ford‐Lloyd B. V. 1998. Complementary conservation strategies for the genus Coffea: a case study of Mascarene Coffea species. Genetic Resources and Crop Evolution 45: 565–579. [Google Scholar]
- Earl D. A., and von Holdt B. M.. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4: 359–361. [Google Scholar]
- esri . 2016. ArcGIS‐World light gray base. Esri, Redlands, CA, USA. Website: https://www.arcgis.com/home/item.html?id=ed712cb1db3e4bae9e85329040fb9a49 [accessed 3 March 2021].
- Ferrão L. F. V., Ferrão R. G., Ferrão M. A. G., Fonseca A., Carbonetto P., Stephens M., and Garcia A. A. F.. 2019. Accurate genomic prediction of Coffea canephora in multiple environments using whole‐genome statistical models. Heredity 122: 261–275. [Europe PMC free article] [Abstract] [Google Scholar]
- Gilson P. 1956. Map of soils and vegetation of the Belgian Congo and Ruanda‐Urundi. Explanatory note. 6. Yangambi. Survey 2: Yangambi. Publications de l'Institut National pour l'Etude Agronomique du Congo Belge, 35. Institut National pour l'Etude Agronomique du Congo Belge (INEAC), Yangambi, RD Congo.
- Gomez C., S. Dussert P. Hamon S. Hamon A. De Kochko, and Poncet V.. 2009. Current genetic differentiation of Coffea canephora Pierre ex A. Froehn in the Guineo‐Congolian African zone: cumulative impact of ancient climatic changes and recent human activities. BMC Evolutionary Biology 9: 167. [Europe PMC free article] [Abstract] [Google Scholar]
- Grant E. L., Conroy G. C., Lamont R. W., Reddell P. W., Wallace H. M., and Ogbourne S. M.. 2019. Short distance pollen dispersal and low genetic diversity in a subcanopy tropical rainforest tree, Fontainea picrosperma (Euphorbiaceae). Heredity 123: 503–516. [Europe PMC free article] [Abstract] [Google Scholar]
- Hardy O. J., Born C., Budde K., Daïnou K., Dauby G., Duminil J., Ewédjé E. E. B. K., et al. 2013. Comparative phylogeography of African rain forest trees: a review of genetic signatures of vegetation history in the Guineo‐Congolian region. Comptes Rendus Geoscience 345: 284–296. [Google Scholar]
- Hardy O. J., and Vekemans X.. 2002. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes 2: 618–620. [Google Scholar]
- Janssens S., Geuten K., Yuan Y.‐M., Song Y., Küpfer P., and Smets E.. 2006. Phylogenetics of Impatiens and Hydrocera (Balsaminaceae) using chloroplast atpB‐rbcL spacer sequences. Systematic Botany 31: 171–180. [Google Scholar]
- Jaroget P., and Descroix F.. 2002. Change in Coffea canephora cultivation in Africa, and development issues. Plantations, recherche, développement 54–59. CIRAD, Montepellier, France.
- Jombart T. 2008. adegenet: a R package for multivariate analysis of genetic markers. Bioinformatics 24: 1403–1405. [Abstract] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones‐Havas S., Cheung M., Sturrock S., Buxton S., et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [Europe PMC free article] [Abstract] [Google Scholar]
- Kiwuka C., Goudsmit E., Tournebize R., de Aquino S. O., Douma J. C., Bellanger L., Crouzillat D., et al. 2021. Genetic diversity of native and cultivated Ugandan Robusta coffee (Coffea canephora Pierre ex A. Froehner): climate influences, breeding potential and diversity conservation. PLoS One 16: e0245965. [Europe PMC free article] [Abstract] [Google Scholar]
- Kyale Koy J., Wardell D., Mikwa J. F., Kabuanga J. M., Monga Ngonga A. M., Oszwald J., and Doumenge C.. 2019. Dynamique de la déforestation dans la Réserve de biosphère de Yangambi (République démocratique du Congo): variabilité spatiale et temporelle au cours des 30 dernières années. Bois et Forêts des Tropiques 341: 15–28. [Google Scholar]
- Lashermes P., Couturon E., Moreau N., Paillard M., and Louarn J.. 1996. Inheritance and genetic mapping of self‐incompatibility in Coffea canephora Pierre. Theoretical and Applied Genetics 93: 458–462. [Abstract] [Google Scholar]
- Leplae E. 1936. Les plantations de café au Congo Belge. Leur histoire (1881−1935)—leur importance actuelle. Mémoires 8° Tome III fasc. 5 et dernier, Institut Royal Colonial Belge. Brussels, Belgium.
- Leroy T., De Bellis F., Legnate H., Musoli P., A. Kalonji R. G. Loor Solórzano, and Cubry P.. 2014. Developing core collections to optimize the management and the exploitation of diversity of the coffee Coffea canephora . Genetica 142: 185–199. [Abstract] [Google Scholar]
- Maley J. 1996. The African rain forest—main characteristics of changes in vegetation and climate from the Upper Cretaceous to the Quaternary. Proceedings of the Royal Society of Edinburgh, B, Biological Sciences 104: 31–73.
- Merot‐L'anthoene V., Tournebize R., Darracq O., Rattina V., Lepelley M., Bellanger L., Tranchant‐Dubreuil C., et al. 2019. Development and evaluation of a genome‐wide Coffee 8.5K SNP array and its application for high‐density genetic mapping and for investigating the origin of Coffea arabica L. Plant Biotechnology Journal 17: 1418–1430. [Europe PMC free article] [Abstract] [Google Scholar]
- Musoli P., Cubry P., Aluka P., Billot C., Dufour M., De Bellis F., Pot D., et al. 2009. Genetic differentiation of wild and cultivated populations: diversity of Coffea canephora Pierre in Uganda. Genome 52: 634–646. [Abstract] [Google Scholar]
- Nielsen R., Tarpy D. R., and Reeve H. K.. 2003. Estimating effective paternity number in social insects and the effective number of alleles in a population. Molecular Ecology 12: 3157–3164. [Abstract] [Google Scholar]
- Noirot M., Charrier A., Stoffelen P., and Anthony F.. 2016. Reproductive isolation, gene flow and speciation in the former Coffea subgenus: a review. Trees 30: 597–608. [Google Scholar]
- Nybom H. 2004. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molecular Ecology 13: 1143–55. [Abstract] [Google Scholar]
- Pendergast M. 2009. Coffee second only to oil? Is coffee really the second largest commodity? Mark Pendergrast investigates and finds some startling results. Tea & Coffee Trade Journal 4: 38–41. [Google Scholar]
- Pritchard J. K., Stephens M., and Donnelly P.. 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [Europe PMC free article] [Abstract] [Google Scholar]
- QGIS.org. 2021. QGIS geographic information system. QGIS Association. http://www.qgis.org
- R Core Team . 2011. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- Schuelke M. 2000. An economic method for the fluorescent labeling of PCR fragments. Nature Biotechnology 18: 233–234. [Abstract] [Google Scholar]
- Sellan G., Simini F., Maritan A., Banavar J. R., de Haulleville T., Bauters M., Doucet J.‐L., et al. 2017. Testing a general approach to assess the degree of disturbance in tropical forests. Journal of Vegetation Science 28: 659–668. [Google Scholar]
- Smith R. F. 1985. A history of coffee. In M. N. Clifford , editor; and K. C. Willson , editor. [eds.], Coffee, 1–12. Springer, Boston, MA, USA. [Google Scholar]
- Theim T. J., Shirk R. Y., and Givnish T. J.. 2014. Spatial genetic structure in four understory Psychotria species (Rubiaceae) and implications for tropical forest diversity. American Journal of Botany 101: 1189–1199. [Abstract] [Google Scholar]
- van Vliet N., Muhindo J., Kambale Nyumu J., Mushagalusa O., and Nasi R.. 2018. Mammal depletion processes as evidenced from spatially explicit and temporal local ecological knowledge. Tropical Conservation Science 11: 1940082918799494. [Google Scholar]
- Wang J. 2017. The computer program structure for assigning individuals to populations: easy to use but easier to misuse. Molecular Ecology Resources 17: 981–990. [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1002/ajb2.1769
Read article for free, from open access legal sources, via Unpaywall:
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/ajb2.1769
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/119629753
Article citations
Crop-to-wild gene flow in wild coffee species: the case of Coffea canephora in the Democratic Republic of the Congo.
Ann Bot, 133(7):917-930, 01 May 2024
Cited by: 0 articles | PMID: 38441303
Phenotypic Diversity and Genetic Parameters of Coffea canephora Clones.
Plants (Basel), 12(23):4052, 01 Dec 2023
Cited by: 1 article | PMID: 38068686 | PMCID: PMC10707845
Genome-Wide Admixture Mapping Identifies Wild Ancestry-of-Origin Segments in Cultivated Robusta Coffee.
Genome Biol Evol, 15(5):evad065, 01 May 2023
Cited by: 2 articles | PMID: 37079743 | PMCID: PMC10159586
Genetic diversity and structure in wild Robusta coffee (Coffea canephora A. Froehner) populations in Yangambi (DR Congo) and their relation to forest disturbance.
Heredity (Edinb), 130(3):145-153, 03 Jan 2023
Cited by: 1 article | PMID: 36596880 | PMCID: PMC9981769
Phylogeography and conservation gaps of Musa balbisiana Colla genetic diversity revealed by microsatellite markers.
Genet Resour Crop Evol, 69(7):2515-2534, 07 May 2022
Cited by: 2 articles | PMID: 36017134 | PMCID: PMC9393128
Go to all (7) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Crop-to-wild gene flow in wild coffee species: the case of Coffea canephora in the Democratic Republic of the Congo.
Ann Bot, 133(7):917-930, 01 May 2024
Cited by: 0 articles | PMID: 38441303
Genetic diversity and structure in wild Robusta coffee (Coffea canephora A. Froehner) populations in Yangambi (DR Congo) and their relation to forest disturbance.
Heredity (Edinb), 130(3):145-153, 03 Jan 2023
Cited by: 1 article | PMID: 36596880 | PMCID: PMC9981769
Genetic diversity of native and cultivated Ugandan Robusta coffee (Coffea canephora Pierre ex A. Froehner): Climate influences, breeding potential and diversity conservation.
PLoS One, 16(2):e0245965, 08 Feb 2021
Cited by: 9 articles | PMID: 33556074 | PMCID: PMC7870046
An overview on the Brazilian Coffea canephora scenario and the current chemometrics-based spectroscopic research.
Food Res Int, 194:114866, 03 Aug 2024
Cited by: 0 articles | PMID: 39232507
Review

 1
1