Abstract
Lay summary
Two novel fungal species were cultured and characterized from four cases of suspected ringworm in cats at an animal shelter in CA, US. These species were genetically distinct from other dermatophytes and are herein described as Arthroderma lilyanum sp. nov. and Arthroderma mcgillisianum sp. nov.Free full text

Two novel species of Arthroderma isolated from domestic cats with dermatophytosis in the United States
Associated Data
Abstract
Dermatophytosis is a superficial fungal infection of keratinized tissues that can occur in humans and other animals. In domestic cats, the majority of cases are caused by Microsporum canis and can spread to other animals and humans via arthrospores. Between 2019 and 2021, 164 cases of suspected dermatophytosis were recorded in animals from a high-volume shelter in California. Samples (hair, nail, and skin scraping) were collected for routine screening from these individuals. One hundred and twenty-six of these were diagnosed as M. canis by culture and internal transcribed spacer (ITS) sequence. In four suspected dermatophytosis cases occurring in kittens in 2019, cultures grown at 20°C yielded fungi with colony morphology more similar to Arthroderma species than Microsporum. Morphologic and microscopic examinations were conducted, and gene segments for the ITS, β-tubulin, and translation elongation factor 1-alpha (TEF1) regions were sequenced from DNA extracted from these cultures. Sequences were aligned to other dermatophytes using maximum likelihood and neighbor-joining trees and were compared to previously described fungal species to assess nucleotide homology. We identified two previously undescribed fungal species, herein proposed as Arthroderma lilyanum sp. nov. and Arthroderma mcgillisianum sp. nov. M. canis co-cultured in two of the four cases. Other physiologic tests supported this diagnosis. These species have significance as potential pathogens and should be considered as rule-outs for dermatophytosis in cats. The potential for infection of other species, including humans, should be considered.
Lay summary
Two novel fungal species were cultured and characterized from four cases of suspected ringworm in cats at an animal shelter in CA, US. These species were genetically distinct from other dermatophytes and are herein described as Arthroderma lilyanum sp. nov. and Arthroderma mcgillisianum sp. nov.
Introduction
Dermatophytes are filamentous fungi that can cause superficial fungal infections in humans and other animals. These fungi are classified as geophilic, zoophilic, and anthropophilic.1,2 While humans can be infected by numerous dermatophyte species, dermatophytosis in cats and dogs is generally caused by a few specific fungal species.3 Over 90% of infections in cats are caused by Microsporum canis, a zoonotic fungus.4Trichophyton spp., Arthroderma insingulare (formerly Trichophyton terrestre), Arthroderma uncinatum (formerly Trichophyton ajelloi), A. vanbreuseghemii (formerly Trichophyton mentagrophytes), Nannizzia gypsea (formerly Microsporum gypseum), Nannizzia persicolor (formerly Microsporum persicolor), and Nannizzia nana (formerly Microsporum nanum) have been reported as rare causes of feline dermatophytosis.3–5 These infections frequently occur in high density populations such as animal shelters, and are often transmitted among animals in outbreak situations and may cause zoonotic infection in animal care providers.6 Diagnosis is often symptomatic and presumptive, and thus there is poor understanding of regional differences and variations in etiology and potential causative agents for this common disease.
During routine dermatophytosis screening at an animal shelter in California in 2019, three of 164 cats presenting with typical clinical signs of dermatophytosis, and a fourth asymptomatic cat with a positive Wood's lamp screening test, were found to be infected with two dermatophytes belonging to the genera Arthroderma. Two of the three animals with clinical signs were also M. canis positive. We describe genetic features of these two novel species and assess potential source of infection.
Methods
Animal care
Animals involved in this study were a part of a larger investigation involving dermatophytosis cases in domestic cats across the United States. All protocols were approved by Colorado State University clinical review board (VCS #2019-223) and biosafety committee (#145399) and were granted an Institutional Animal Care and Use Committee (IACUC) waiver prior to initiation of this study.
Case presentations
Case 1: A stray 6-week-old intact female domestic shorthair kitten living outside was presented to San Jose Animal Center on May 25, 2019. Upon physical examination, one lesion (between 1.3 and 2.5 cm) with scales, crusts and alopecia was identified on the abdomen. A modified UV lamp test (Wood's lamp) was performed to detect fluorescence often associated with M. canis, and was found to be positive (Table 1). This patient had no prior medical history, and anti-fungal treatment was not performed before hair sample collection. The patient was adopted by a local rescue organization and was lost to medical follow-up.
Table 1.
Demographic information for four cases presenting with novel fungal species. All four cases occurred in stray (i.e. unowned) cats that were younger than 6 months. Lesion presentation was similar between Case 1 and 2. Case 4 presented asymptomatically but was from a litter found to be positive during Wood's lamp screening. DSH = domestic shorthair. N/A = not applicable.
| Clinical Parameters | Case 1 | Case 2 | Case 3 | Case 4 |
|---|---|---|---|---|
| Age | 6 weeks | 6 weeks | 5 months | 2 months |
| Sex/neuter status | Female intact | Male intact | Male intact | Male intact |
| Breed | DSH | DSH | DSH | Tabby |
| Date of presentation | 05/25/2019 | 07/10/2019 | 07/18/2019 | 09/24/2019 |
| Number of lesions | 1 | 1 | 5 | 0 |
| Lesion types | Scales, crusts, alopecia | Scales, crusts, alopecia | Scales, crusts, alopecia | N/A |
| Lesion location | Abdomen | Head | Head, neck, abdomen, back, legs | N/A |
| Lesion size | 1.3–2.5 cm | 1.3–2.5cm | 1.3–2.5 cm | N/A |
| Wood's Lamp results | Positive | Positive | Positive | Positive |
| Presumptive diagnosis | Arthroderma lilyanum | Arthroderma lilyanum and Microsporum canis | Arthroderma mcgillisianum and Microsporum canis | Arthroderma mcgillisianum |
Case 2: A stray 6-week-old intact domestic shorthair male kitten living outside was presented to San Jose Animal Center on July 10, 2019. Upon physical examination, one lesion described as scales, crusts, and alopecia (between 1.3 and 2.5 cm) was identified on the head (Table 1). The kitten was screened using a Wood's lamp and had a positive fluorescence. This patient had no prior medical history, and anti-fungal treatment was not performed before hair sample collection. The patient was adopted by a local rescue organization and was lost to medical follow-up.
Case 3: A stray 5-month-old intact male domestic shorthair kitten living outside was presented to San Jose Animal Center on July 18, 2019. More than five lesions with alopecia and scales/crusts ranging between 1.3 and 2.5 cm were distributed on the head, neck, abdomen, back, and legs (Table 1). This patient also had a concurrent upper respiratory infection during examination. Wood's lamp testing was performed, and the patient had a positive fluorescence. The patient was adopted by a local rescue organization and was lost to medical follow-up.
Case 4: A stray 2-month-old intact tabby male kitten living outside was presented to San Jose Animal Center on September 24, 2019, with five littermates. Case 4 and its littermates were screened for dermatophytosis upon intake via Wood's lamp and were positive. No lesions were present for Case 4 upon physical examination (Table 1). This patient has no prior medical history, and anti-fungal treatment was not performed before hair sample collection. The patient was adopted by a local rescue organization and was lost to medical follow-up.
Sample collection
Following clinical examination, hair samples were collected via MacKenzie toothbrush method.7 Patients were not routinely screened for other infections via skin scraping as ectoparasites were an uncommon diagnosis and lesions were characteristic of dermatophytosis. All samples were sent to Colorado State University. Upon receipt, hair samples were stored at 4°C, and nail and skin samples were stored at -20°C until utilized for down streaming processing.
Characterization of fungal isolates
Initial colony isolation: Hair samples collected on the toothbrushes were inoculated onto a biplate of Sabouraud dextrose agar (SDA) with chloramphenicol and gentamicin, and Dermatophyte Test Medium (Hardy Diagnostics, Santa Maria, CA, USA) and incubated in the dark at room temperature (range, 20–25°C) for 4 weeks.8 Plates were monitored for growth every 24–48 h. Color (code and terminology) of fungal colonies was recorded using a color identification textbook.9 To increase sporulation, isolates of the novel taxa were grown on Potato Dextrose Agar (PDA) (Hardy Diagnostics, Santa Maria, CA, USA) at room temperature until colonies reached a sufficient colony size to be utilized for microscope slides.
Microscopic evaluation: 2–3 week old cultures were touched lightly with the sticky side of acetate tape, mounted on a microscope slide, and stained with Lactophenol Cotton Blue Stain (Hardy Diagnostics, Santa Maria, CA, USA).10 A minimum of 30 conidia was measured on PDA-grown colonies, and the length and width were recorded.
Temperature range testing: Each isolate was tested for ability to grow at 30, 33, and 37°C on SDA. Two plates were inoculated using one-point inoculation of each of the two isolates that were not co-cultured with M. canis and grown at each of the three temperatures for 10–24 days or until colonies were formed. Colony diameter was measured at day 7.
Urease production: Production of urease was tested by growing the isolates that were not co-cultured with M. canis on urea agar slants (Becton, Dickinson, and Company BBL, Franklin Lakes, NJ, USA) at room temperature. Swabs of the isolates were streaked across the urea agar slants and monitored for color changed every 24 h.
Hair perforation test: 5 ml of UltraPure DNase/RNase free distilled water (Invitrogen, Carlsbad, CA, USA) was inoculated with autoclaved specific pathogen free cat hair. Fungal isolates that were not co-cultured with M. canis were added to the cat hair water via sterile cotton swab of the SDA culture plate. Inoculates were grown at room temperature in the dark for up to four weeks with hair removed approximately twice weekly for microscopic evaluation.10 Hair was examined on a microscope slide with Lactophenol Cotton Blue Stain (Hardy Diagnostics, Santa Maria, CA, USA) and cover slide at 1000x magnification.10
Mating type gene characterization: We tested strains using methods reported for identifying Microsporum and Trichophyton mating type genes.11 This methodology has been used as an alternative to traditional mating type cross experiments.11 DNA was extracted from each unique isolate as described below and subjected to PCR using primers that target the two mating type gene loci: alpha-box and high motility-group (HMG).11 Prototypical T. mentagrophytes (American Type Culture Collection 44687), and clinical isolates from a domestic cat of T. mentagrophytes and M. canis were used as positive controls. Amplification was assessed by gel electrophoresis followed by DNA fragment visualization using SYBR Safe DNA gel stain (ThermoFisher Scientific, Waltham, MA, USA).
DNA extraction and conventional PCR
DNA was extracted from SDA plate using a previously published method12 modified for culture samples. Briefly, a sterile cotton swab collected fungal cells from the plate and added to a 1.5 ml tube with 100 μl of DNA extraction buffer (60 mM sodium bicarbonate, 250 mM potassium chloride, and 50 mM Tris, balanced to pH 9.5). The swab was removed, and the tube was incubated at 95°C for 10 min. 100 μl of 2% bovine serum albumin was then added to the tube. The DNA was stored at −80°C until utilized for down streaming processing.
This unpurified DNA was used in conventional PCR for the internal transcribed spacer (ITS), β-tubulin, and translation elongation factor 1-alpha (TEF1) regions. The ITS, β-tubulin, and TEF1 regions were amplified using previously published primers (Table 2).13–15 The following protocol was utilized for ITS PCR: 10 μl of 2× KAPA Taq ReadyMix with dye (Roche, Indianapolis, IN, USA), 6 μl UltraPure DNase/RNase free distilled water (Invitrogen, Carlsbad, CA, USA), 1 μl forward primer, 1 μl reverse primer and 2 μl of DNA template solution. The reaction was run in a C1000 Touch thermal cycler (Bio-Rad, Hercules, CA, USA) for one cycle at 95°C for 3 min, 34 cycles at 95°C for 30 s, 60°C for 30 s, 72°C for 1min, and one cycle at 72°C for 5 min. The protocol was modified for amplifying the β-tubulin and TEF1 regions by adjusting the annealing temperatures to 55 and 56°C, respectively. PCR products were run on a 1.5% agarose gel, and a band at 600, 500, and 1000 bp was visualized for ITS, β-tubulin, and TEF1 PCRs, respectively. PCR products were purified using ExoSAP-IT kit (Applied Biosystems, Carlsbad, CA, USA) and Sanger sequenced at a commercial laboratory (Psomagen, Rockville, MD, USA). Each region was sequenced a minimum of two times.
Table 2.
Primers used for targeting internal transcribed spacer (ITS), β-tubulin, and translation elongation factor 1-alpha (TEF1) regions.
| Target region | Primer | Sequence | Reference |
|---|---|---|---|
| ITS | ITS5F | 5′- GGAAGTAAAAGTCGTAACAAGG | [8] |
| ITS4R | 5′- TCCTCCGCTTATTGATATGC | ||
| β-tubulin | Bt2a | 5′- GGTAACCAAATCGGTGCTGCTTTC | [9] |
| Bt2b | 5′- ACCCTCAGTGTAGTGACCCTTGGC | ||
| TEF1 | 983F | 5′-GCYCCYGGHCAYCGT-GAYTTYAT | [10] |
| 2218R | 5′-ATGACACCRA-CRGCRACRGTYTG |
Phylogenetic analysis
The ITS and β-tubulin sequences from all four isolates were aligned with 51 isolates of other dermatophytes (Supplementary Table 1) using MUSCLE Alignment (Geneious v10.0.9) with a maximum of 8 iterations. Available sequences were downloaded from NCBI's GenBank database (https://www.ncbi.nlm.nih.gov/). These alignments were utilized for constructing a maximum likelihood tree (Geneious v10.0.9, RAxML plugin v8.2.11)16 with 1000 bootstrap replicates. Additionally, a neighbor-joining tree was constructed (Geneious v10.0.9) with 1000 bootstrap replicates and CBS269.89 Guarromyces ceretanicus was classified as the outgroup. Bootstrap values greater than 50% were displayed in phylogenetic trees.
The sequences for ITS, β-tubulin, and TEF1 for the four cases were separately aligned with available sequences for A. redellii and A. quadrifidum (Supplementary Tables 1 and 2) of the same regions using MUSCLE Alignment (Geneious v10.0.9) with a maximum of 8 iterations. Only one sequence was available on NCBI's database for β-tubulin and TEF1 from A. redellii and one sequence for the full length of TEF1 from A. quadrifidum (Supplementary Tables 1 and 2). These genes were utilized for analysis based on a previous report for distinguishing species within the Arthroderma genus.17
Results
Initial fungal isolation
Culture of the hair sample from Case 1 exclusively grew a single fungus on the biplate. Colonies first appeared after 3 days of incubation at room temperature without bacterial contamination. Biplate cultures from hair samples from Cases 2-4 grew multiple saprophytes and were re-cultured from the hair samples a second time. During regrowth a single fungus was identified from Case 4 starting 3 days post-incubation at room temperature. For Cases 2 and 3, colonies of M. canis first appeared 3-4 days after incubation at room temperature, followed by growth of a second white, fluffy fungus (Figure 1K) that outgrew M. canis. The singular fungi from Cases 1 and 4, and overgrowth fungi from Cases 2 and 3 had colony morphology similar to Trichophyton spp. and Arthroderma spp. (Figure 1, Table 3).18–20 Additional morphologic and physiologic characterization are described below.
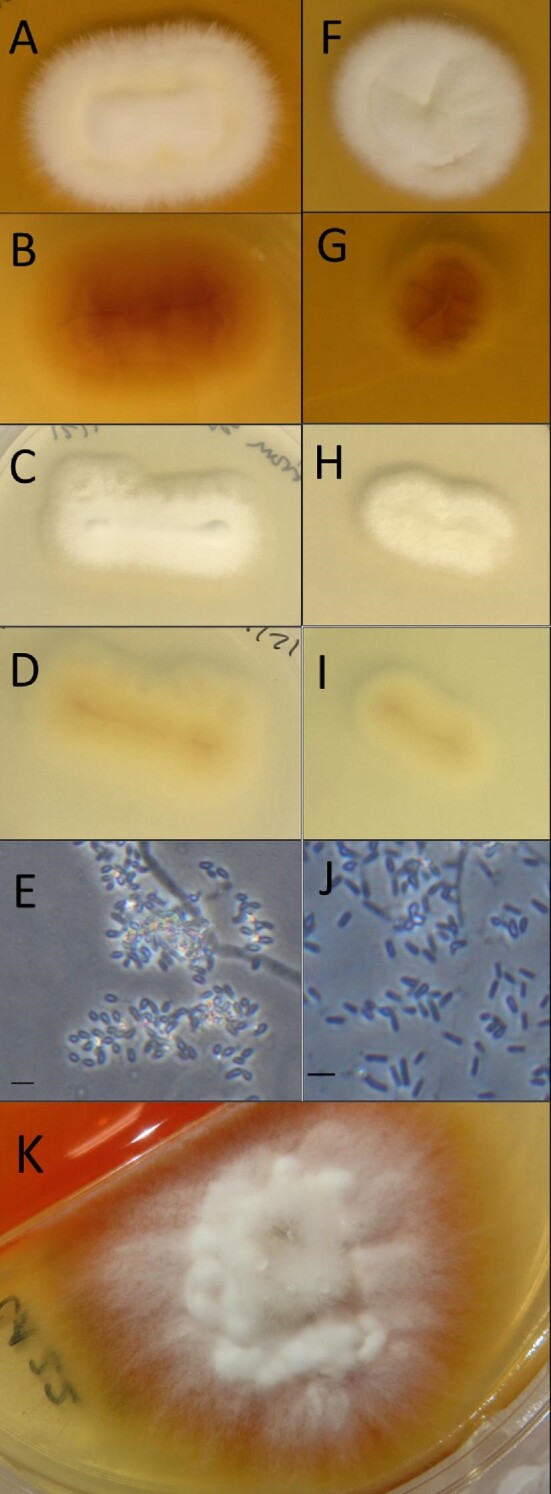
Arthroderma lilyanum (ex-type living culture) – (A) colony surface grown on SDA, (B) colony reverse grown on SDA, (C) colony surface grown on PDA, (D) colony reverse grown on PDA, (E) conidia and hyphae under dark field at 400X magnification. Bar = 10 μm. Arthroderma mcgillisianum (ex-type living culture) – (F) colony surface grown on SDA, (G) colony reverse grown on SDA, (H) colony surface grown on PDA, (I) colony reverse grown on PDA, (J) conidia observed under dark field at 400X magnification. Bar = 10 μm. Co-culture of A. mcgillisianum and M. canis – (K) colony surface grown on SDA. SDA plates were incubated for 10 days for single isolates and 9 days for co-culture colonies, and PDA plates for 7 days, at room temperature in the dark.
Table 3.
Description of Arthroderma lilyanum and Arthroderma mcgillisianum, and similar genetic or clinical presentation fungi. CA = California. TN = Tennessee. TX = Texas. WI = Wisconsin. * = culture characteristics of fungi grown on Sabouraud dextrose agar (SDA). ** = major reservoir species listed, there are reports of other animals harboring this fungus.
| Fungal species | Culture characteristics* | Micromorphology | Habitat | Distribution |
|---|---|---|---|---|
| Arthroderma lilyanum | • Growth at 20–25°C, 30°C• No growth at 33°C, 37°C• Surface color: lily white• Reverse color: oxide yellow• Texture: cottony | • Hyaline, septate hyphae• Spiral hyphae rare• Ellipsoidal microconidia• Hair perforation test: negative• Urease positive | Domestic cats | USA: CA |
| Arthroderma mcgillisianum | • Growth at 20–25°C, 30°C• No growth at 33°C, 37°C• Surface color: satin white• Reverse color: apricot to butter yellow• Texture: cottony | • Hyaline, septate hyphae• Ellipsoidal microconidia• Hair perforation test: negative• Urease positive | Domestic cats | USA: CA |
| Microsporum canis 18 | • Growth at 20–25°C• Surface color: greyish- to tannish-white• Reverse color: deep ochraceous-yellow• Texture: woolly | • Tear-shaped microconidia• Spindle-shaped macroconidia• Hair perforation test: positive• Urease positive | Domestic cats, dogs, rabbits** | Worldwide |
| Arthroderma quadrifidum 19 | • Growth at 25°C• Surface color: white to pale yellowish• Reverse color: yellowish brown• Texture: fluffy to powdery | • Cylindrical macroconidia• Elongate pyriform microconidia• Pale yellow, septate hyphae | Soil, feathers, hair | Worldwide |
| Arthroderma redellii 26 | • No growth at 25°C• Surface color: white• Reverse color: yellow• Texture: velutinous | • Hyaline, septate hyphae• Thallic conidia | Bats (Myotis lucifugus and Myotis velifer) | USA: TN, TX, WI |
Phylogenetic analysis
Two previously uncharacterized Arthroderma spp. were isolated from four cats
ITS and β-tubulin maximum likelihood and neighbor-joining trees for all four isolates placed them within the genus Arthroderma (Figures 2 and 3). Dermatophytes speciated from Cases 1 and 2 are most closely related to Arthroderma redellii as analyzed by both ITS maximum-likelihood (Figure 2) and the neighbor-joining tree (Figure 3A). While the isolate associated with Case 1 has high nucleotide alignment with A. redellii ITS and TEF1 (both over 99%), it had a lower nucleotide alignment for β-tubulin (less than 98.5%) and had different growth characteristics than reported for A. redellii (Tables 3 & 4). Case 2 non-M. canis dermatophyte isolate had 98.5% or greater alignment for all three A. redellii genes (Table 4). Case 1 and 2 ITS and β-tubulin amplicons were > 99% identical, suggesting the same or very similar species (Table 5).
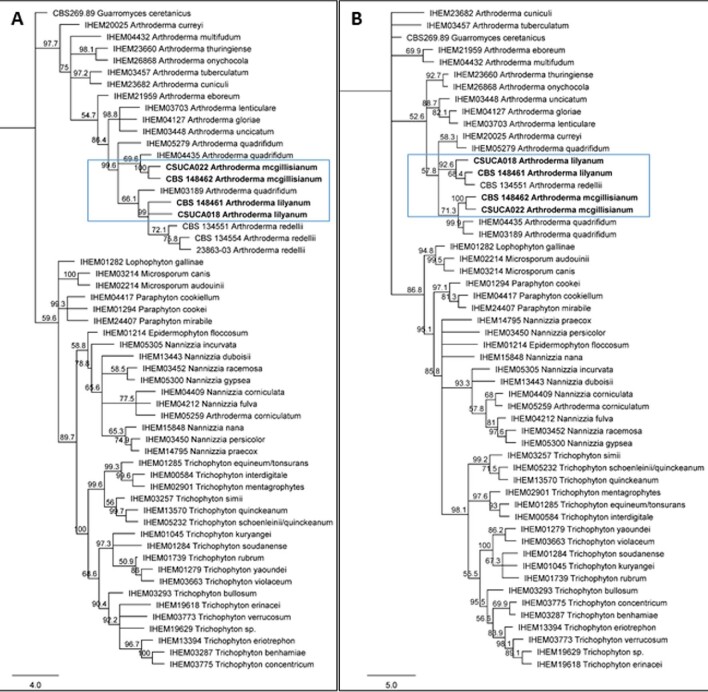
Consensus trees for maximum likelihood analysis of novel dermatophytes. A) internal transcribed spacer (ITS) analysis. B) β-tubulin. All four dermatophyte isolates are placed with the genus Arthroderma (blue box indicates location of new isolates). Using 1000 iterations for RAxML analysis, bootstrap support values ≥50% are placed above the branches. Westerdijk Fungal Biodiversity Centre (CBS), Belgian Co-ordinated Collections of Micro organisms/Institute of Hygiene and Epidemiology Mycology (BCCM/IHEM), Center for Forest Mycology Research, or Colorado State University strain number is placed before isolate name. Scale bar represents mean number of nucleotide substitutions per site.
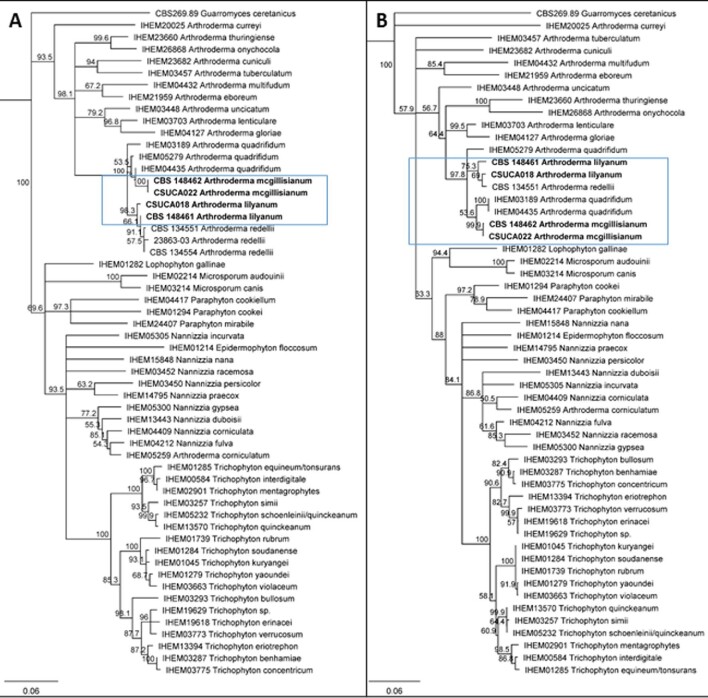
Neighbor-joining analysis of novel dermatophytes. (A) internal transcribed spacer (ITS) region. B) β-tubulin. All novel dermatophytes were placed with the genus Arthroderma (blue box indicates new isolates). Using 1000 iterations, bootstrap support values ≥50% are placed above the branches. Westerdijk Fungal Biodiversity Centre (CBS), Belgian Co-ordinated Collections of Micro-organisms/Institute of Hygiene and Epidemiology Mycology (BCCM/IHEM), Center for Forest Mycology Research, or Colorado State University strain number is placed before isolate name. Scale bar represents substitutions per nucleotide site.
Table 4.
Nucleotide homology illustrates two novel dermatophyte strains. Case 1 and 2 have high homology with A. redellii (over 98% for internal transcribed spacer (ITS), partial β-tubulin, and translation elongation factor 1-alpha (TEF1) regions). They are distinguished from A. redellii phenotypically since they grew readily at room temperature. Cases 3 and 4 are less than 98.5% homologous to A. quadrifidum and A. redellii across ITS, partial β-tubulin, and TEF1 regions. N/A = sequence data not available.
| Arthroderma quadrifidum IHEM05279 | Arthroderma quadrifidum IHEM03189 | Arthroderma quadrifidum IHEM04435 | Arthroderma quadrifidum UAMH2941 | Arthroderma redellii CBS 134551 | Arthroderma redellii CBS 134554 | Arthroderma redellii 23863-03 | ||
|---|---|---|---|---|---|---|---|---|
| Case 1 | ITS | 96.3% | 96% | 96% | N/A | 99.1% | 99.5% | 99.4% |
| β-tubulin | 93.4% | 93.1% | 93.1% | N/A | 98.1% | N/A | N/A | |
| TEF1 | N/A | N/A | N/A | 97.8% | 99.7% | N/A | N/A | |
| Case 2 | ITS | 96.1% | 95.8% | 95.8% | N/A | 98.9% | 99.4% | 99.2% |
| β-tubulin | 94.1% | 93.8% | 93.8% | N/A | 98.5% | N/A | N/A | |
| TEF1 | N/A | N/A | N/A | 98.2% | 99.3% | N/A | N/A | |
| Case 3 | ITS | 96.1% | 95.1% | 96.8% | N/A | 96.9% | 97.2% | 97.1% |
| β-tubulin | 96.3% | 96.1% | 96.1% | N/A | 94.6% | N/A | N/A | |
| TEF1 | N/A | N/A | N/A | 98.4% | 98.4% | N/A | N/A | |
| Case 4 | ITS | 96.2% | 95.1% | 96.8% | N/A | 96.9% | 97.2% | 97.1% |
| β-tubulin | 96.3% | 96% | 96% | N/A | 94.6% | N/A | N/A | |
| TEF1 | N/A | N/A | N/A | 98.5% | 98.5% | N/A | N/A | |
Table 5.
Pairwise homology across three dermatophytes identified in this study. Case 2 and 3 have 100% nucleotide agreement for the internal transcribed spacer (ITS), partial β-tubulin, and translation elongation factor 1-alpha (TEF1) regions. Case 1 had less than 98% identity within ITS, β-tubulin, and TEF1 to Cases 3 and 4, while having over 99% nucleotide agreement with Case 2 for ITS and TEF1 regions.
| Case 1 | Case 2 | Case 3 | Case 4 | ||
|---|---|---|---|---|---|
| Case 1 | ITS | - | 99.2% | 97.1% | 97.1% |
| β-tubulin | - | 97.6% | 94.3% | 94.2% | |
| TEF1 | - | 99% | 97.8% | 97.8% | |
| Case 2 | ITS | 99.2% | - | 96.9% | 96.9% |
| β-tubulin | 97.6% | - | 94.8% | 94.8% | |
| TEF1 | 99% | - | 98.1% | 98.6% | |
| Case 3 | ITS | 97.1% | 96.9% | - | 100% |
| β-tubulin | 94.3% | 94.8% | - | 100% | |
| TEF1 | 97.8% | 98.1% | - | 100% | |
| Case 4 | ITS | 97.1% | 96.9% | 100% | - |
| β-tubulin | 94.2% | 94.8% | 100% | - | |
| TEF1 | 97.8% | 98.6% | 100% | - | |
Genetic analysis of Case 3 and 4 indicated 100% nucleotide alignment for ITS, β-tubulin, and TEF1 (Table 5) and were closely related by phylogenetic analysis (Figures 2 and 3). These cases had less than 98% homology; in particular, β-tubulin homology distinguished Cases 3 and 4 from Cases 1 and 2 (less than 95% identity, Table 5). When compared to A. redellii and A. quadrifidum, both Case 3’s and 4’s dermatophytes have low nucleotide alignment with the TEF1 region; the highest agreement is less than 98.5% (Table 4).
M. canis and T. mentagrophytes mating type gene strains were amplified by PCR specific for mating type genes for these species. No amplification products were detected for any of the novel Arthroderma spp. using the Trichophyton specific primers. Two of the novel species that grew as pure isolates (Cases 1 and 4) were negative for bands using Microsporum specific primers. A single product of approximately 420 base-pairs was amplified from the two cases (2 and 3) that has been originally co-cultured with M. canis (Supplementary Figure 1). We conducted Sanger sequencing on these products (Psomagen, Rockville, MD, USA) and via comparison with the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/), determined that these bands were 100% identical to M. canis alpha-box (data not shown). We therefore conclude these products reflected DNA from M. canis DNA present in co-cultured materials and were not primary Arthroderma spp. fragments.
Species description
Arthroderma lilyanum: Moskaluk, sp. nov. Figure 1. MycoBank: MB841894
Holotype: US, California, San Jose Animal Care Center, hair of a domestic cat, 25 May 2019, collected by Dr. Katherine Tyson, isolated by A. Moskaluk, dried culture on SDA (holotype, Westerdijk Fungal Biodiversity Centre CBS 148461); ex-living culture CSU CA002.1, GenBank ITS: OK623475, β-tubulin: OL342744, TEF1: OL342746.
Etymology: The species name honors Lily Moskaluk for her invaluable support during this study.
Diagnosis: This species can infect domestic cats, causing clinical signs similar to other dermatophytes that infect cats, including producing a positive fluorescence on Wood's lamp testing. This species is morphologically similar to Arthroderma redellii, but can grow well at room temperature.
Holotype culture characteristics: Colonies exhibited growth on SDA after 3 days at room temperature. Colony diameter was 17.5 mm after 7 days and 49 mm after 14 days grown on SDA at room temperature. Colonies had flat topography, cottony texture with surface color being lily white (1A1) and reverse color being oxide yellow (5C7) in the center with the color progressing to amber yellow (4B6) at the periphery. Colonies exhibited a similar growth rate on PDA at room temperature with colonies first visible after 2 days. Colony diameter was 22 mm after 7 days on PDA at room temperature. Colonies had flat topography with lacy margins, granular texture with surface color being milk white (1A2) and reverse color being light orange (5A4) at the center progressing to orange white (5A2) at the margins. Cultures grew on SDA at 30°C and were 11 mm at 7 days, which was smaller than that observed at room temperature (17.5 mm, as noted above). No growth was observed at 33°C for 10 days and at 37°C for 24 days. Cultures were urease positive after 2 days of growth on urea agar slants at room temperature.
Description of micromorphology: Vegetative mycelium consisted of hyaline, septate hyphae. Spiral hyphae were rarely observed. No racquet hyphae were observed. No ascomata were observed. Few conidia were observed from cultures grown on SDA. Numerous conidia were present from cultures grown on PDA. Conidia ranged from 2.3 to 4.9 μm length (mean ± standard deviation: 3.3 ± 0.7 μm) and 1.4-2.1 μm width (mean ± standard deviation: 1.7 ± 0.2 μm) with a length to width ratio ranging from 1.2 to 3.2 (mean ± standard deviation: 2 ± 0.5). Arthrospores were occasionally present. Hair perforation test was negative at 24 days.
Habitat and distribution: Found on domestic cats in California, US.
Additional cultures examined: US, California, San Jose Animal Care Center, hair of a domestic cat, 10 July 2019, collected by Dr. Katherine Tyson, isolated by A. Moskaluk, living culture CSU CA018, GenBank ITS: OK597194, β-tubulin: OL342745, TEF1: OL342747.
Arthroderma mcgillisianum: Moskaluk, sp. nov. Figure 1. MycoBank: MB841895
Holotype: US, California, San Jose Animal Care Center, hair of a domestic cat, 24 September 2019, collected by Dr. Sharon Ostermann, isolated by A. Moskaluk, dried culture on SDA (holotype, CBS 148462); ex-living culture CSU CA033.1, GenBank ITS: OK631727, β-tubulin: OL342748, TEF1: OL342750.
Etymology: The species name honors the late Gladys “Beth” McGillis, honorably discharged Leading Aircraftwoman of the Royal Canadian Air Force for her inspiration of women in STEM and dedication to feline medicine.
Diagnosis: This species can infect domestic cats, causing clinical signs similar to other dermatophytes that infect cats, including producing a positive fluorescence on Wood's lamp testing. This species is morphologically similar to Arthroderma quadrifidum.
Holotype culture characteristics: Colonies exhibited growth on SDA after 3 days at room temperature. Colony diameter was 11 mm after 7 days and 33 mm after 14 days grown on SDA at room temperature. Colonies had flat topography, cottony texture with surface color being satin white (2A1) and reverse color being apricot yellow (5B6) in the center progressing to butter yellow (4A5) at the margins. Colonies exhibited a slightly faster growth rate on PDA at room temperature with colonies first visible after 2 days. Colony diameter was 17.5 mm after 7 days on PDA at room temperature. Colonies had flat topography with lacy margins, granular texture with surface color being snow white (1A1) and reverse color being pale orange (5A3) in the center progressing to orange white (5A2) at the periphery. Growth was similar growth at 30°C to room temperature (colony diameter 12.7 mm at 7 days). No growth was observed at 33°C for 10 days and for 24 days at 37°C. Species was urease positive after 3 days on urea agar slants at room temperature.
Description of micromorphology: Vegetative mycelium consisted of hyaline, septate hyphae. No spiral or racquet hyphae were observed. No ascomata were observed. Numerous conidia were observed from cultures grown on SDA. Conidia ranged from 3.8 to 9.6 μm length (mean ± standard deviation: 6.7 ± 1.3 μm) and 2.1-3.4 μm width (mean ± standard deviation: 2.7 ± 0.3 μm) with a length to width ratio ranging from 1.3 to 3.5 (mean ± standard deviation: 2.5 ± 0.4). Arthrospores were occasionally present. Hair perforation test was negative at 24 days.
Habitat and distribution: Found on domestic cats in California, US.
Additional cultures examined: US, California, San Jose Animal Care Center, hair of a domestic cat, 18 July 2019, collected by Dr. Sharon Ostermann, isolated by A. Moskaluk, living culture CSU CA022, GenBank ITS: OK631735, β-tubulin: OL342749, TEF1: OL342751.
Discussion
Dermatophytosis is a common skin infection that can be caused by a variety of fungal species. In domestic cats, the main agent is M. canis with reports of other dermatophytes causing dermatophytosis from the genera Arthroderma, Nannizzia, and Trichophyton.4,5 Investigators in Turkey found that while M. canis was responsible for over 60% of dermatophytosis cases in cats in Western Turkey, almost 20% of the cases were caused by N. nana, N. gypsea and A. vanbreuseghemii.21 Cats that spend all or part of their time outdoors may be exposed to a variety of dermatophytes through the soil or contact with other animals.3
In 2019, four cats were presented to an animal shelter in California, US for suspected dermatophytosis. While three of these animals had clinical signs consistent with dermatophytosis at the time of examination, hair samples from all four cases grew fungi with morphology similar to Trichophyton spp. and Arthroderma spp. Microscopic features of these isolates were consistent with dermatophyte species. Through maximum likelihood trees and neighbor-joining trees for ITS and β-tubulin, Cases 1 and 2 were determined to be strains of a novel dermatophyte species described herein as Arthroderma lilyanum sp. nov. Cases 3 and 4 were found to be 100% identical in the markers that were investigated and determined to be a novel dermatophyte species described herein as Arthroderma mcgillisianum sp. nov. Mating type gene analysis using reagents that successfully typed M. canis and T. mentagrophytes were unable to phenotype these novel strains. Strains were found to be urease positive and hair perforation negative, consistent with other species of Arthroderma.22–24 Other Arthroderma species that have been reported to be urease positive include Arthroderma chiloniense24 and Arthroderma vanbreuseghemii,23 while negative hair perforation test species include A. chiloniense24 and Arthroderma benhamiae.22 Investigators have reported that hair from domestic cats may be resistant to perforation due to thicker diameter of the different hair layers and less favorable nutritional composition for fungal growth compared to human hair, thus further studies could be conducted to assess ability of these isolates to attach to and degrade other types of hair samples.25
While the sequences of the ITS, β-tubulin, and TEF1 regions for the two A. lilyanum isolates had high nucleotide agreement with A. redellii, the host and growing conditions are different between the species (Table 3). A. redellii has only been isolated from bats in Indiana, Texas, and Wisconsin, with the distinguishing feature of being unable to grow at ambient room temperature.26 As noted here, the strain isolated here grew within the temperature range of 20–30°C. Given the genetic similarities, it is suggestive of these two species sharing a recent ancestor and possibly diverging to better suit their respective hosts.
We identified these novel fungal species in California, however, given the low frequency of dermatophyte species identification via culture or genetic sequence analysis in clinical settings,27–29 these species could be more widespread but undiagnosed. Two cases (one isolate for each of the novel species) were co-cultured with M. canis. Given that M. canis colonies grew more rapidly, A. lilyanum and A. mcgillisianum could be missed without fastidious examination of culture plates. While Case 2 (A. lilyanum co-cultured with M. canis) was presented with similar clinical signs as Case 1 (A. lilyanum only), Case 3 (A. mcgillisianum co-cultured with M. canis) had a more severe presentation than Case 4 (A. mcgillisianum only), suggesting that A. mcgillisianum might be less pathogenic as a primary pathogen than when infected in concert with M. canis.
All four of the cases exhibited positive Wood's lamp fluorescence during examination. While M. canis are classically known to exhibit fluoresce under UV light with a blue-green color, other dermatophytes including M. audouinii, M. ferrugineum, M. disortum, N. gypsea, and Trichophyton schoenleinii have also been reported to have positive fluorescence.30 The compound reportedly responsible for M. canis and N. gypsea fluorescence has been reported to be pteridine,31,32 and pteridine and xanthurenic acid derivatives have been reported to contribute to fluorescence of T. schoenleinii.33 It is unclear what benefit or purpose these compounds serve, and what the metabolic pathway for the production of these fluorescent compounds is. It is unknown if A. lilyanum and A. mcgillisianum are geophilic or zoophilic, and whether they are zoonotic or can infect other animals. Determining the natural history, mating scheme, prevalence, fluorescent principle, and geographic distribution of these strains presents interesting questions for future studies.
Supplemental content
Sequence data
Sequence data for ITS, β-tubulin, and TEF1 for Arthroderma lilyanum and Arthroderma mcgillisianum have been deposited into NCBI's GenBank (Supplementary Table 3).
Phylogenetic data
Phylogenetic trees have been deposited into TreeBASE and are freely accessible.
Acknowledgements
We thank Katherine Tyson, Sharon Ostermann, staff, and animals at San Jose Animal Center, CA, US for providing samples and clinical data for each patient; Erick Gagne for assisting with the creation of the phylogenetic trees; Ben Shupe for editing the manuscript; Sally Kuhn and Lauren Darlington for helping with sample preparation; Mary Nehring for laboratory management; Jane Stewart for use of microscopic equipment; Michael Russell for assisting with culture incubation.
Contributor Information
Alex Moskaluk, Microbiology, Immunology, and Pathology, Colorado State University, Fort Collins, CO 80523, USA.
Sue VandeWoude, Microbiology, Immunology, and Pathology, Colorado State University, Fort Collins, CO 80523, USA.
Funding
This work was supported by Morris Animal Foundation [fellowship training D21FE-402]; EveryCat Health Foundation [grant W20-032]; National Institutes of Health [NIH-9-T32 OD0-10437-18 Biomedical Research Training for Veterinarians]; and National Institutes of Health/National Center for Advancing Translational Sciences [NIH/NCATS Colorado CTSA Grant Number TL1 TR002533]. Contents are the authors’ sole responsibility and do not necessarily represent official NIH views.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and the writing of the paper.
References
Articles from Medical Mycology are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/mmy/myac001
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8808258
Citations & impact
Impact metrics
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/123237599
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/mmy/myac001
Article citations
First description of Arthroderma lilyanum in a rabbit with a focal alopecic area of the forelimb.
Med Mycol Case Rep, 40:58-60, 11 May 2023
Cited by: 0 articles | PMID: 37283719 | PMCID: PMC10240506
Name Changes for Fungi of Medical Importance, 2020 to 2021.
J Clin Microbiol, 61(6):e0033022, 28 Mar 2023
Cited by: 2 articles | PMID: 36975779 | PMCID: PMC10281194
Review Free full text in Europe PMC
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Molecular detection and species identification of dermatophytes by SYBR-Green real-time PCR in-house methodology using hair samples obtained from dogs and cats.
Med Mycol, 61(5):myad047, 01 Apr 2023
Cited by: 0 articles | PMID: 37120732
PCR-based methods in detection and identification of dermatophytes in dogs and cats with suspected dermatophytosis in 2021 in Poland.
Pol J Vet Sci, 26(4):629-634, 12 Dec 2023
Cited by: 1 article | PMID: 38088306
Molecular diagnosis of dermatophyte isolates from canine and feline dermatophytosis in Northeast Iran.
Vet Med Sci, 8(2):492-497, 17 Dec 2021
Cited by: 2 articles | PMID: 34919354 | PMCID: PMC8959315
Dermatophytosis in cats: ABCD guidelines on prevention and management.
J Feline Med Surg, 15(7):598-604, 01 Jul 2013
Cited by: 35 articles | PMID: 23813824 | PMCID: PMC11148949
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCATS NIH HHS (2)
Grant ID: TL1 TR002533
Grant ID: TR002533
NIH HHS (2)
Grant ID: NIH-9-T32 OD0-10437-18
Grant ID: T32 OD010437
National Center for Advancing Translational Sciences (1)
Grant ID: TR002533
National Institutes of Health (1)
Grant ID: NIH-9-T32 OD0-10437-18




