Abstract
Free full text

Genomic differentiation within East Asian Helicobacter pylori
Abstract
The East Asian region, including China, Japan and Korea, accounts for half of gastric cancer deaths. However, different areas have contrasting gastric cancer incidences and the population structure of
Helicobacter pylori
in this ethnically diverse region is yet unknown. We aimed to investigate genomic differences in
H. pylori
between these areas to identify sequence polymorphisms associated with increased cancer risk. We analysed 381 H
.
pylori
genomes collected from different areas of the three countries using phylogenetic and population genetic tools to characterize population differentiation. The functional consequences of SNPs with a highest fixation index (Fst) between subpopulations were examined by mapping amino acid changes on 3D protein structure, solved or modelled. Overall, 329/381 genomes belonged to the previously identified hspEAsia population indicating that import of bacteria from other regions of the world has been uncommon. Seven subregional clusters were found within hspEAsia, related to subpopulations with various ethnicities, geographies and gastric cancer risks. Subpopulation-specific amino acid changes were found in multidrug exporters (hefC), transporters (frpB-4), outer membrane proteins (hopI) and several genes involved in host interaction, such as a catalase site, involved in H2O2 entrance, and a flagellin site mimicking host glycosylation. Several of the top hits, including frpB-4, hefC, alpB/hopB and hofC, have been found to be differentiated within the Americas in previous studies, indicating that a handful of genes may be key to local geographic adaptation.
H. pylori
within East Asia are not homogeneous but have become differentiated geographically at multiple loci that might have facilitated adaptation to local conditions and hosts. This has important implications for further evaluation of these changes in relation to the varying gastric cancer incidence between geographical areas in this region.
H
.
pylori
genomes collected from different areas of the three countries using phylogenetic and population genetic tools to characterize population differentiation. The functional consequences of SNPs with a highest fixation index (Fst) between subpopulations were examined by mapping amino acid changes on 3D protein structure, solved or modelled. Overall, 329/381 genomes belonged to the previously identified hspEAsia population indicating that import of bacteria from other regions of the world has been uncommon. Seven subregional clusters were found within hspEAsia, related to subpopulations with various ethnicities, geographies and gastric cancer risks. Subpopulation-specific amino acid changes were found in multidrug exporters (hefC), transporters (frpB-4), outer membrane proteins (hopI) and several genes involved in host interaction, such as a catalase site, involved in H2O2 entrance, and a flagellin site mimicking host glycosylation. Several of the top hits, including frpB-4, hefC, alpB/hopB and hofC, have been found to be differentiated within the Americas in previous studies, indicating that a handful of genes may be key to local geographic adaptation.
H. pylori
within East Asia are not homogeneous but have become differentiated geographically at multiple loci that might have facilitated adaptation to local conditions and hosts. This has important implications for further evaluation of these changes in relation to the varying gastric cancer incidence between geographical areas in this region.
Data Summary
All supporting data have been provided within the article or through supplementary data files. Public genome data were retrieved from the National Center for Biotechnology Information GenBank (Tables S2 and S3, available in the online version of this article). Newly sequenced genomes were deposited into GenBank with BioProject number PRJNA482300.upplementary material can
Introduction
Helicobacter pylori
has co-evolved with human beings for at least 100 000 years and has strong association with the occurrence of gastric (stomach) cancer [1].
H. pylori
are transmitted most effectively within households and have therefore experienced low migration rates compared to many other members of the human microbial flora. As a result, we can expect the pattern of differentiation to reflect historical human-migration patterns. The population structure of
H. pylori
worldwide has been classified into seven major groups that indeed correlate with ancient human migrations [2], of which the hpEastAsia includes at least three subgroups: hspEAsia, hspIndigenousAmerica [3] and hspMaori. The hspEAsia subgroup is thought to be ubiquitous within East Asian countries with high gastric cancer incidence, including China, Japan and Korea. These countries together make up 1/5 of the world population but account for half the global mortality from the disease [4, 5]. The hspEAsia strains have documented higher virulence than other subpopulations and have diverged from the Western strains in several proteins including virulence factors [6, 7]. Many genes have also diverged within this region [6].
years and has strong association with the occurrence of gastric (stomach) cancer [1].
H. pylori
are transmitted most effectively within households and have therefore experienced low migration rates compared to many other members of the human microbial flora. As a result, we can expect the pattern of differentiation to reflect historical human-migration patterns. The population structure of
H. pylori
worldwide has been classified into seven major groups that indeed correlate with ancient human migrations [2], of which the hpEastAsia includes at least three subgroups: hspEAsia, hspIndigenousAmerica [3] and hspMaori. The hspEAsia subgroup is thought to be ubiquitous within East Asian countries with high gastric cancer incidence, including China, Japan and Korea. These countries together make up 1/5 of the world population but account for half the global mortality from the disease [4, 5]. The hspEAsia strains have documented higher virulence than other subpopulations and have diverged from the Western strains in several proteins including virulence factors [6, 7]. Many genes have also diverged within this region [6].
The prevalence of gastric cancer shows geographic and ethnic variations also within East Asia. Some north and southeast areas of China such as Fujian have higher incidence [8], whereas some west regions, such as Yunnan, have low incidence (Fig. 1) [9, 10]. China also shows diverse distribution of population ethnicities; in southwest Yunnan province, more than 20 ethnicities exist. South Korea has been reported with the highest incidence in the world [11, 12] and similar is true for the Japanese main islands (including Hokkaido) [13], while in Okinawa, where the ethnic composition is different, the incidence is low [13, 14]. Previous studies [8, 15] on the high-risk regions have suggested that diet, lifestyle and H. pylori properties may contribute to the high risk. However, despite the complex human migrations and evolutionary history in these areas, variation of H. pylori within the hspEAsia subpopulation has been poorly explored.
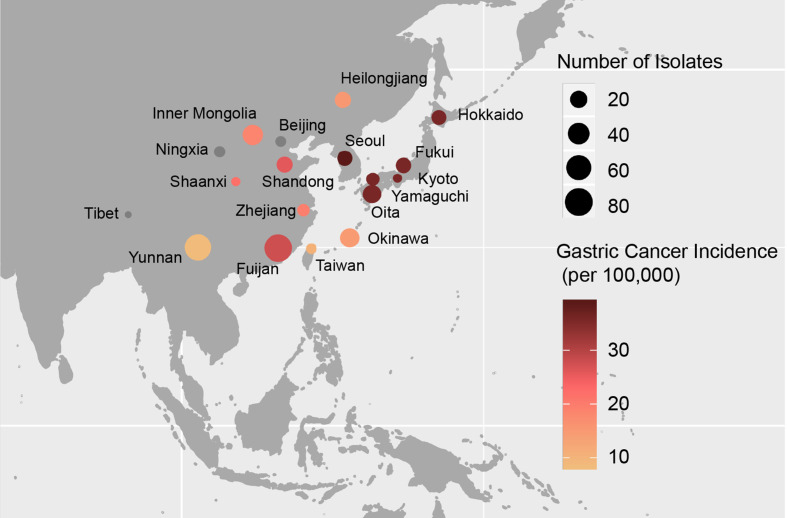
Geographic distribution of the East Asian H. pylori strain dataset of this study. Shade of the circle fill indicates gastric cancer incidence. Grey fill indicates missing incidence data. All are age-standardized gastric cancer rates from 2014 by world population except that for Okinawa, only the data from 2016 is available.
In the present work, we analysed a collection of genomes of H. pylori strains from various places in these three countries. Our phylogenetic and population genetic analysis revealed presence of pronounced regional and ethnical population structure within hspEAsia, and specific sequence differences differentiating these subpopulations. Most of these highly region-specific variants were found on proteins involved in host interaction. Furthermore, placement of the variants on protein structure provided insight into molecular mechanisms underlying regional adaptation.
Methods
Strain collection across East Asia
We collected 357 H
.
pylori
isolates from China, Japan and South Korea, isolated between 1999 and 2018, including 11 provinces of China and six regions of Japan (Tables S1,S2, Fig. 1). We used data from 2014 to show regional risk levels in China and compared it with data from Japan and Korea [8, 9, 11–14]. Our sample of 77 Yunnan isolates includes strains from four ethnic minorities and the Han majority individuals.
H
.
pylori
isolates from China, Japan and South Korea, isolated between 1999 and 2018, including 11 provinces of China and six regions of Japan (Tables S1,S2, Fig. 1). We used data from 2014 to show regional risk levels in China and compared it with data from Japan and Korea [8, 9, 11–14]. Our sample of 77 Yunnan isolates includes strains from four ethnic minorities and the Han majority individuals.
Genome sequencing
Genomic DNA was extracted using the Qiagen DNeasy Mini Kit and genomes were sequenced using Illumina or PacBio sequencers. For Illumina sequencing, paired-end libraries were created and sequenced using the Illumina Hiseq and Miseq platform. For PacBio sequencing, quality assessment of genomic DNA, SMRTbell library preparation and data evaluation were performed, and sequenced using the PacBio RS II platform. Sequences were deposited into GenBank with BioProject number PRJNA482300. Combining publicly available genomes (available in January 2018 when the study started) with these newly sequenced genomes, we collected a dataset consisting of 381 H
.
pylori
genomes for further analysis (Table S2).
H
.
pylori
genomes for further analysis (Table S2).
Genomic comparison
We used snippy [16] to perform a whole-genome alignment with XZ274 as the reference genome and extracted 225 942 variable core-genome sites. For the phylogenetic analysis, the dataset of 381 strains were combined with 1–3 reference sequences for each of the major H. pylori populations (Table S3).
Population structure and phylogenetic analysis
The concatenated whole-genome SNPs and coordinates were used to prepare the haplotype file for ChromoPainter and fineSTRUCTURE analyses following the instructions from website (http://www.paintmychromosomes.com). In total, 13 highly clonal sequences were removed prior to the analysis, resulting in a comparison of 370 genomes. Using each genome as both donor and recipient haplotypes, we used ChromoPainter to calculate the number of genetic chunks exported from a donor to a recipient and generated a co-ancestry matrix. Then the co-ancestry matrix file was imported into fineSTRUCTURE by setting the burn-in and Markov chain Monte Carlo (MCMC) chain of 100 000 iterations to generate clusters for all the individual strains [17, 18]. We also generated a phylogenetic tree using FastTree (http://www.microbesonline.org/fasttree) [19], which was labelled using iTol (https://itol.embl.de) [20].
000 iterations to generate clusters for all the individual strains [17, 18]. We also generated a phylogenetic tree using FastTree (http://www.microbesonline.org/fasttree) [19], which was labelled using iTol (https://itol.embl.de) [20].
Definition of subgroups and calculation of Fst between subgroups
Subgroups were defined according to the population structure analysed by fineSTRUCTURE (the left-side tree). We named the subgroups assigned by fineSTRUCTURE as ‘Sg’ and also according to the geographic origin of the majority of a clade (Table S2). This criterion for subgroup definition is relatively a rough classification. To identify the SNPs attributed to the divergence of subgroups more accurately, we used a more stringent criterion for a further definition of the subgroups. Only those isolates assigned into a singular cluster in the fineSTRUCTURE tree were defined to form a subset of a subgroup and used for Fst calculation. For example, for the large sampling areas such as China southwest isolates, only those from Yunnan Mosuo and Pumi ethnicities, which clustered into a singular clade with low levels of admixture/recombination, were defined as a subgroup, ‘YunnanMP’. For China southeast, only those from Fujian Changle that clustered into a singular clade were defined as a subgroup, ‘Fuijan’. Subgroups with mixed ethnicities or high levels of admixture were not used in the calculations.
For each SNP site, we calculated a fixation index (Fst) using PopGenome R package [21] for each subgroup such that Fst (sg1)=pairwise calculation of Fst of sg1 versus all other subgroups. We also compared isolates from China with those from Japan and South Korea and the two lower-incidence regions, Yunnan and Okinawa, versus high incidence regions (the remainder of hspEAsia strains).
Mapping high-Fst SNPs on protein structure
We located each of the SNPs with the highest Fst values on the reference genome of XZ274, a Tibetan strain, to identify its gene and effect on amino acid sequence. If the gene was missing or atypical in this strain, we used strain F57, a Japanese strain, or 26 695, a European strain, instead as shown in Table S4. We mapped the amino acids on solved
H. pylori
protein structure or on protein structure homology-modelled by SwissModel and its repository for 26695 (https://swissmodel.expasy.org/repository). We analysed and presented them by PyMOL [22].
695, a European strain, instead as shown in Table S4. We mapped the amino acids on solved
H. pylori
protein structure or on protein structure homology-modelled by SwissModel and its repository for 26695 (https://swissmodel.expasy.org/repository). We analysed and presented them by PyMOL [22].
Results
East Asian H. pylori population structure is associated with geography and host ethnicity
To analyse the genetic structure of H. pylori in China, Japan and South Korea, we constructed a phylogenetic tree (Fig. 2a) and clustered the strains using fineSTRUCTURE [17, 18] (Fig. 2b). The two methods gave broadly concordant clustering and indicated differentiation at multiple scales. By including reference genomes from the other main H. pylori populations, we found that the 52 most differentiated strains do not belong to hspEAsia and have had distinct evolutionary histories. One main cluster of these comprised isolates from individuals of Mongolian ethnicity (Sg4, Fig. 2b), which in the tree grouped between the references of hspIndigenousAmerica (previously called hspAmerind) and hpAsia2 populations. We also identified a cluster of Okinawan strains diverging after hpEurope, and before hpAsia2 and hpEastAsia (Fig. 2a), also shown in the lowest line of the co-ancestry matrix (Fig. 2b). This likely corresponds to group C in MLST and STRUCTURE analysis of Okinawan strains [23]. Because these populations are best analysed in the context of broader regional variation, we focused our remaining analysis on variation within the 329 hspEAsia isolates in our sample.
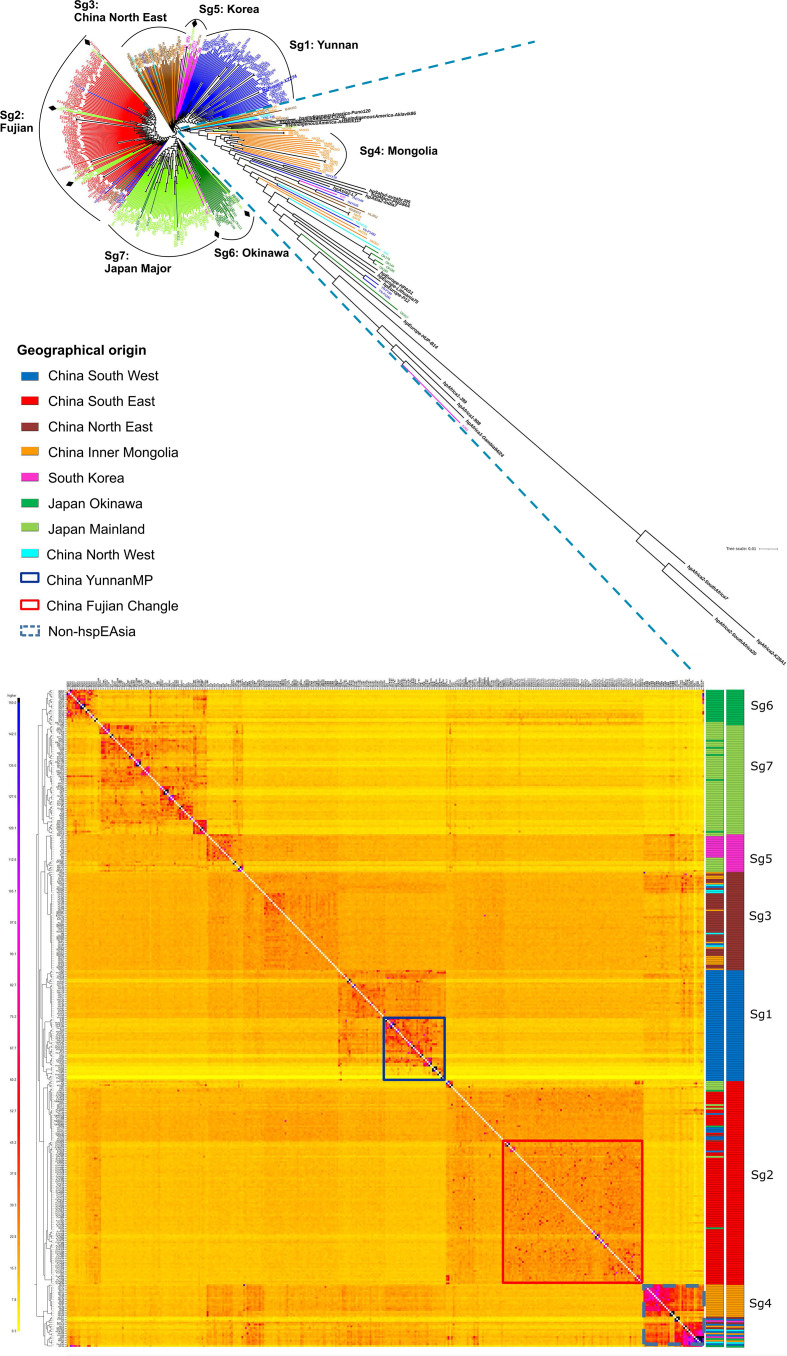
Phylogenetic and population genetic analysis of East Asia H. pylori strains. (a) Phylogenetic tree of East Asia genomes combined with reference sequences from global H. pylori populations. Diamonds indicate Japanese and Korean isolates that are dispersed in other subgroups. (b) Co-ancestry matrix of East Asia isolates calculated using fineSTRUCTURE after removing isolates with highly clonal relatedness. The tree inferred on the left of the co-ancestry matrix indicates the relationships between groups with distinct ancestry profile as inferred by fineSTRUCTURE. On the right side, the first column shows the geographic origin of each isolate. The second column indicates each subgroup with a distinct colour.
HspEAsia isolates are relatively homogeneous but show fine-scale differentiation that is strongly correlated with geography. The majority of Japanese isolates clustered together (Sg7), with Okinawan hspEAsia strains forming a distinct subpopulation (Sg6). The latter likely corresponds to group A in MLST [23]. Two strains from Hokkaido cluster with the Sg6 Okinawan strains. This is likely related to the evolutionary structure of Japanese people and of the Okinawa people [24]. The native ethnicity remains in Hokkaido (as Ainu ethnicity) and in Okinawa.
South Korean isolates also form a distinct subpopulation, which cluster together with isolates from the Northeast of China, concordant with its geographic location. A few Japanese strains are in the Korean cluster while a few Korean strains are in the main Japanese cluster and branching around the same time as the Okinawa cluster (Fig. 2a,b). This may reflect immigration from Korea to Northern Kyushu and Okinawa. Within China, there is clear differentiation between the Northeast, Southeast and Southwest areas. Within the Southwest, a subpopulation containing strains from Yunnan Mosuo and Pumi ethnicities could also be observed. A Tibetan isolate clustered with the Pumi population, consistent with their migration and mixture history. Within the Southeast, there was also a subpopulation (Sg2), specific to Fujian Changle, a region of high gastric cancer incidence (Fig. 2, table S1). The Southeast cluster also includes multiple clusters of Japanese/Okinawa strains (Fig. 2a,b) likely reflecting later immigration to Japan/Okinawa.
Many genetic variants show strong differentiation by subpopulations
In order to explore the genetic basis of local differentiation, we calculated fixation index, Fst, between the hspEAsia subpopulations identified by fineSTRUCTURE. To obtain more reliable SNPs associated with subgroup separation, we used the YunnanMP subcluster of Sg1 and Fujian subcluster of Sg2, along with the remaining five subgroups defined by the fineSTRUCTURE analysis. For most of the subpopulations, more than 99 % of SNPs were weakly differentiated, with Fst less than 0.3 (Fig. S1). The Korean subpopulation had a smaller sample size and showed the weakest Fst values. We defined clustered nucleotide polymorphisms (CNPs) as occurring when two or more SNPs with high Fst were found in the same gene. CNPs likely result from the co-inheritance of different SNPs on the same gene fragment.
% of SNPs were weakly differentiated, with Fst less than 0.3 (Fig. S1). The Korean subpopulation had a smaller sample size and showed the weakest Fst values. We defined clustered nucleotide polymorphisms (CNPs) as occurring when two or more SNPs with high Fst were found in the same gene. CNPs likely result from the co-inheritance of different SNPs on the same gene fragment.
To functionally interpret differentiation between the populations, we focused on the SNPs with the top 20 Fst values in each subgroup and then removed SNPs with Fst <0.5. This resulted in a list of 56 genes, several of which occurred more than once (Table S4, bold, summarized in Table 1). In the main text, we focus on the genes with the strongest evidence for differentiation. Eight of these had one SNP with Fst >0.6 and appeared in at least two top 20 lists in different pairwise comparisions. Another four had at least two SNPs with Fst >0.6. Of these, HofC has multiple non-synonymous SNPs with Fst up to 0.86, marking it out as also being a particularly strong candidate for being differentiated by natural selection (Table 2).
and appeared in at least two top 20 lists in different pairwise comparisions. Another four had at least two SNPs with Fst >0.6. Of these, HofC has multiple non-synonymous SNPs with Fst up to 0.86, marking it out as also being a particularly strong candidate for being differentiated by natural selection (Table 2).
Table 1.
High Fst SNPs distributed in each hspEAsia subgroup and involved genes
|
Subgroup |
Gene |
Annotation |
Fst |
SNP coordinate |
|---|---|---|---|---|
|
Sg1 (YunnanMP) |
hefC |
Inner pump of a multidrug efflux system |
0.891 |
827 |
|
porB |
Pyruvate-ferredoxin oxidoreductase subunit beta |
0.800 |
1 | |
|
hefC |
Inner pump of a multidrug efflux system |
0.757 |
827 | |
|
flaA |
Flagellin A |
0.748 |
833 | |
|
gltA |
Citrate synthase |
0.731 |
1 | |
|
tlpD |
Chemotaxis sensor |
0.716 |
836 | |
|
katA |
Catalase |
0.714 |
507 | |
|
gltA |
Citrate synthase |
0.706 |
1 | |
|
gltA |
Citrate synthase |
0.706 |
1 | |
|
flaA |
Flagellin A |
0.705 |
833 | |
|
frpB-4 |
TonB-dependent outer membrane Ni importer |
0.694 |
1 | |
|
oppD |
Oligopeptide permease ATPase protein |
0.669 |
283 | |
|
oppD |
Oligopeptide permease ATPase protein |
0.663 |
283 | |
|
porB |
Pyruvate-ferredoxin oxidoreductase subunit beta |
0.649 |
1 | |
|
hopI |
Outer membrane protein HopI |
0.638 |
1 | |
|
gltA |
Citrate synthase |
0.626 |
1 | |
|
Sg2 (Fujian) |
hofC |
Outer membrane protein involved in adhesion and diffusion of cations including antibiotics |
0.855 |
496 |
|
hofC |
Outer membrane protein involved in adhesion and diffusion of cations including antibiotics |
0.850 |
496 | |
|
hofC |
Outer membrane protein involved in adhesion and diffusion of cations including antibiotics |
0.800 |
496 | |
|
katA |
Catalase |
0.756 |
507 | |
|
hopB/alpB |
Outer membrane protein, Omp21 |
0.704 |
983 | |
|
flaA |
Flagellin A |
0.651 |
833 | |
|
Sg3 (China North East) |
frpB-4 |
TonB-dependent outer membrane Ni importer |
0.752 |
1 |
|
frpB-4 |
TonB-dependent outer membrane Ni importer |
0.752 |
1 | |
|
frpB-4 |
TonB-dependent outer membrane Ni importer |
0.738 |
1 | |
|
frpB-4 |
TonB-dependent outer membrane Ni importer |
0.738 |
1 | |
|
frpB-4 |
TonB-dependent outer membrane Ni importer |
0.713 |
1 | |
|
frpB-4 |
TonB-dependent outer membrane Ni importer |
0.706 |
1 | |
|
frpB-4 |
TonB-dependent outer membrane Ni importer |
0.701 |
1 | |
|
frpB-4 |
TonB-dependent outer membrane Ni importer |
0.701 |
1 | |
|
frpB-4 |
TonB-dependent outer membrane Ni importer |
0.637 |
1 | |
|
Sg5 (Korea) |
hypC |
hydrogenase expression/formation protein |
0.652 |
959 |
|
Sg6 (Okinawa) |
rhoD |
Rhodanese, a cyanide-detoxifying enzyme |
0.646 |
1 |
|
Sg7 (Japan_Major) |
frpB-4 |
TonB-dependent outer membrane Ni importer |
0.658 |
1 |
 Criteria for showing high Fst of each subpopulation in this table is based on the following:
Criteria for showing high Fst of each subpopulation in this table is based on the following:
Fst > 0.6.
Sg4 (Mongolia) is exclued from the table as it does not belong to the hspEAsia subgroup.
SNPs of high/low and China_All are not listed because they were specific comparisons, not representing subpopulations.
Table 2.
Genes with a SNP with Fst >0.5 in the top 20 lists
|
Criterion |
Gene |
Locus tag |
Figure |
Category |
Annotation |
Residue function |
|---|---|---|---|---|---|---|
|
Fst >0.6+ |
hefC |
HP0607 |
3(b) |
Efflux pump |
Inner membrane component of the multidrug HefABC efflux pump |
Channel entrance |
|
frpB-4 |
HP1512 |
4(a) |
TonB-dependent importer |
Outer membrane nickel importer |
Ligand binding | |
|
flaA |
HP0601 |
6(b) |
Motility |
Flagellin A of flagella, involved in immune system evasion |
Glycosylation for host mimicry and more | |
|
katA |
HP0875 |
6(a) |
Sensor |
Catalase sensing and destroying H2O2 |
Channel entrance; Dimer-dimer interface | |
|
hopI |
HP1156 |
Outer membrane protein |
Outer membrane protein HopI |
| ||
|
hopB/alpB |
HP0913 |
Outer membrane protein |
Hop family adhesin HopB/AlpB/Omp21. |
| ||
|
tlpD |
HP0599 |
6(c) |
Sensor |
Chemotaxis sensor sensing and destroying HOCl |
Active site | |
|
exbB-2 |
HP1339 |
S3C |
TonB-dependent importer |
Energizer/motor in the inner membrane driven by proton |
Channel entrance | |
|
Fst >0.6 two or more SNPs |
hofC |
HP0486 |
Outer membrane protein |
Hof family protein implicated in adhesion and antibiotics diffusion |
| |
|
porB |
HP1111 |
Energy metabolism |
Subunit of pyruvate:ferredoxin oxidoreductase, part of the microaerophilic metabolic pathway leading to acetyl~CoA |
Subunit interaction | ||
|
gltA |
HP0026 |
5(b) |
Energy metabolism |
Citrate synthase, the first enzyme in TCA cycle incorporating acetyl~CoA |
Subunit interaction | |
|
oppD |
HP0250 |
5(a) |
Importer |
Oligopeptide ABC transporter, ATPase subunit |
Cofactor binding | |
|
Fst >0.5 |
ompA-18 |
HP1125 |
Outer membrane protein |
OmpA family peptidoglycan-associated lipoprotein |
Ligand binding | |
|
fixP |
HP0147 |
S4C |
Energy metabolism |
Subunit of cytochrome c oxidase in aerobic respiration |
Channel entrance | |
|
hypC |
HP0899 |
Hydrogenase expression/formation protein |
(Synonymous change) | |||
|
hefD |
HP0971 |
3(c) |
Efflux pump |
Outer membrane component of the HefABC multidrug efflux pump |
Protein binding | |
|
copA |
HP1503 |
S2E |
Exporter |
Copper(I) exporter |
Protein stability | |
|
mscS-1 |
HP0284 |
Sensor |
Mechano-sensor sensing tension in the membrane |
Scaffolding in the periplasm | ||
|
metQ |
HP1564 |
S2D |
Importer |
d-methionine ABC transporter, methionine-binding subunit |
Ligand binding | |
|
frpB-1 |
HP0876 |
S3A |
TonB-dependent importer |
Outer membrane haem importer |
Ligand binding | |
|
fecA-1 |
HP0686 |
S3B |
TonB-dependent importer |
Outer membrane iron(III) dicitrate importer |
Ligand binding; Channel (plug) | |
|
exbD-2 |
HP1340 |
S3C |
TonB-dependent importer |
Inner membrane energizer/motor driven by proton |
Proton channel entrance | |
|
hpaA |
HP0797 |
S3F |
Outer membrane protein |
Neuraminyllactose-binding hemagglutinin |
Subunit interaction | |
|
omp |
HP0358 |
Outer membrane protein |
Putative outermembrane protein |
| ||
|
panD |
HP0034 |
S4A (i) |
Micronutrient synthesis |
Vitamin B5 synthesis |
Interaction between cleaved peptides | |
|
bioD |
HP0029 |
Micronutrient synthesis |
Vitamin B7 synthesis |
| ||
|
tgt |
HP0281 |
S4A (iii) |
Micronutrient synthesis |
Q-base synthesis; Q-base on tRNA affects translation accuracy |
Active site | |
|
rhoD |
HP1223 |
S4B |
Detox |
Rhodanese detoxifying cyanide generated in microbiome |
| |
|
rkiP |
HP0218 |
S2B |
Oncoprotein |
Mimic of human RKIP tumour suppressor |
Interaction with signal peptide | |
|
hcpX |
– |
S2A |
Effector |
SLR family with repeated alpha-helix pairs |
Human protein binding | |
|
dsbI |
HP0595 |
S3D |
Secretion |
S-S formation |
Active site | |
|
jag |
HP1451 |
Secretion |
Regulator of VirB11/Cag-alpha gate of Cag secretion system |
Cofactor binding | ||
|
lpxE |
HP0021 |
S2C |
Membrane lipid modifier |
Lipid A 1-phosphatase to hide it from the innate immune response |
Substrate binding; active site | |
|
cfaS |
HP0416 |
S3E |
Membrane lipid modifier |
Cyclopropane-fatty-acyl-phospholipid synthase for acid protection |
Substrate binding | |
|
fur |
HP1027 |
S4D |
Transcription factor |
Regulator of Fe/Ni import, redox balance and acid response |
Cofactor binding |
To predict the effect of differentiated variants on H. pylori biology and pathogenesis, we functionally annotated the genes and interpreted the impact of differentiated amino acids on the protein structures, solved or based on homology modelling. The majority of genes containing the most differentiated SNPs could be grouped into four major categories; (i) transporters, (ii) outer membrane proteins, (iii) metabolism and (iv) host interaction.
Transporters
The Hef multidrug efflux pump
H. pylori carries four gene clusters that each encode a set of RND superfamily of multidrug efflux pump corresponding to TolC-AcrA-AcrB of E. coli (Fig. 3). They pump out endogenous bile salts and ceragenins as well as various antibiotics [25–27]. The single most differentiated SNP in our analysis is in hefC (HP0607) in the YunnanMP subpopulation, with an Fst=0.89. This N86S is also the most differentiated between the lower cancer incidence regions (Yunnan and Okinawa) and the remainder. This residue corresponds to the gate to channel III for planar aromatic cations in the E. coli homologue [28] and the regional adaptation may therefore remodel this gate to export chemicals of this type at different ratios. Also, the outer component of the efflux pump, HefD, showed YunnanMP specific residues in the equatorial domain that may be involved in the interaction with the inner component to open the aperture.
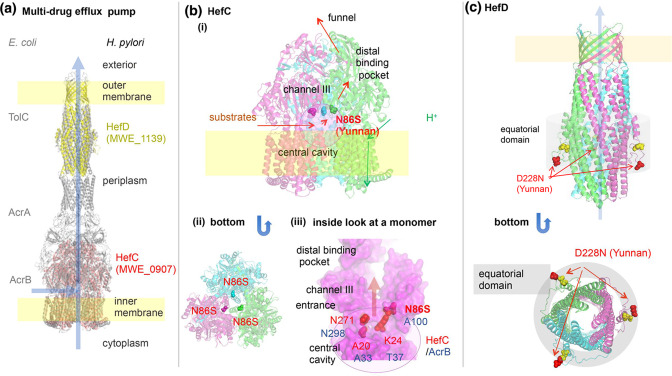
Inferred structure and differentiated amino acid sites of multidrug efflux pump related proteins. (a) TolC-AcrA-AcrB of E. coli and its H. pylori homologues. (b) HefC (MWE_0907), modelled on PDB 3W9I (MexB of Pseudomonas aeruginosa ). (iii) HefC modelled on PDB 3AOD (AcrB of E. coli). Yunnan-specific N86S is at the entrance of channel III. (c) HefD (MWE_1139), modelled on PDB 5BUN (ST50 from Salmonella enterica subsp. enterica serovar Typhi). Yellow spheres indicate Mongol-differentiated E219D.
The TonB-dependent nickel importer FrpB-4
A gene containing multiple highly differentiated SNPs, especially in North Eastern China, is frpB-4 (HP1512), encoding an outer membrane transporter of nickel of some form [29]. It is a member of TonB-dependent transporter family, which forms a trimer of 22-stranded beta barrels each filled with a ‘plug’ (Fig. 4a,b). H. pylori reference strain 26695 carries four frpB homologues: frpB-1 (HP0876), frpB-2/3 (HP0916/5) and frpB-4 (HP1512).
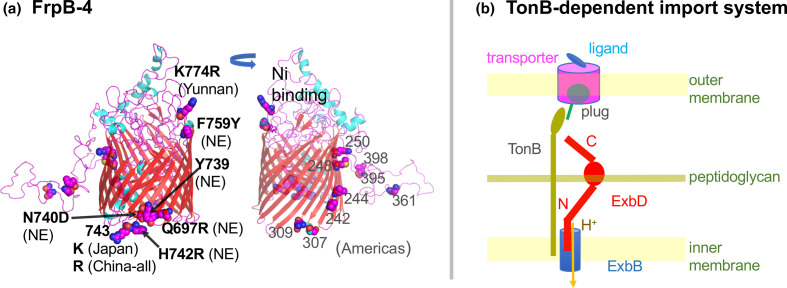
(a) FrpB-4 (MWE_1700) modelled on PDB 4AIQ (FrpB of N. meningitidis). The left figure shows East Asia-specific sites. NE means north east China. The right one shows Americas-specific sites. (b) TonB-dependent transport system. When the transporter in the outer membrane catches a ligand, it pushes out its plug for TonB to pull and open the channel. This movement of TonB is energized by ExbBD motor in the inner membrane driven by H+ flow.
Ligand binding lets TonB change the conformation of the plug, which opens a channel, a process energized by the ExbBD proton-driven motor [30] (Fig. 4b). Four China North East-specific Fst sites cluster (Fig. 4a), with three sites (739, 740, 742) presumably representing a CNP. The next residue (743) distinguishes between Japan (K) and China (R). Northeast China-differentiated F759Y (F for the remainder and Y for Northeast China) and YunnanMP-differentiated K774R are situated above the barrel in the model and may interact with the ligand. Various regions of the Americas show region-specific amino acid changes in other areas of the protein [31], out of which three are predicted to be in the decoy loop. Taken together, these changes may affect nickel transport and, consequently, urease activity, since the urease enzyme requires nickel for acid acclimation [32]. The changes could be related to regional differences in host nickel metabolism and stomach acidity.
Other transporters
Hof proteins ( Helicobacter -specific outer membrane protein family [33]) are 18-stranded β-barrels homologous to Occ family of Pseudomonas and Campylobacter jejuni MOMP (major outer membrane protein) involved in passive diffusion of cations including antibiotics and in adhesion [34–36]. HofC (HP0486), required for H. pylori colonization in mice [37], contains the most differentiated SNPs in Fujian, a region of high cancer incidence with Fst=0.86. The gene is highly variable in global strains and shows many America-differentiated SNPs and region-differentiated SNPs within the Americas [31], within one narrow region. Fujian-differentiated D186S in HP0486 (166 in MWE_0556) lies at a distance from them. Fujian-differentiated V9A is in its signal peptide.
OppD (HP0250), the cytoplasmic subunit of the oligopeptide ABC transporter, has YunnanMP-differentiated KG306R within the ATP binding Walker motif A (Fig. 5a).
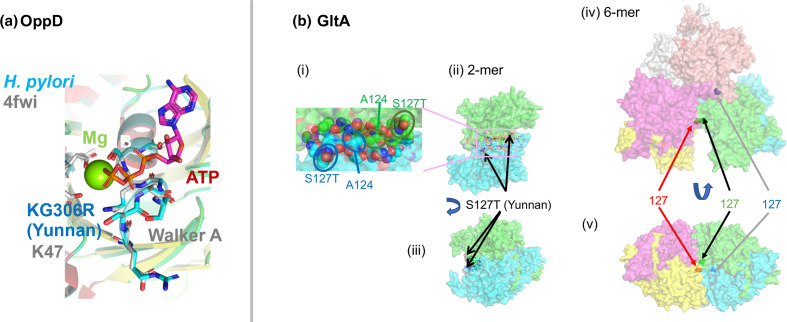
(a) OppD (MWE_0327), ATP-binding subunit of an oligopeptide ABC transporter as modelled on PDB 4FWI ( Thermoanaerobacter tengcongensis ). (b) Citrate synthase, GltA (i) (ii) (iii) MWE_1570 modelled on PDB 3MSU ( Francisella tularensis homolog). (iv) (v) On PDB 2h12 ( Acetobacter aceti ).
Outer-membrane proteins
HopB/ AlpB/ Omp21 (HP0913), an adhesin of the Hop family required for colonization, carries Fujian-differentiated N289D and N286H. According to a previous study, 24 polymorphic sites within 49 bp in AlpB are enriched for Asian ancestry in hspEuropeColombia and 32 polymorphic sites within 65
bp in AlpB are enriched for Asian ancestry in hspEuropeColombia and 32 polymorphic sites within 65 bp were enriched for Asian ancestry in hspAfrica1Nicaragua populations [31].
bp were enriched for Asian ancestry in hspAfrica1Nicaragua populations [31].
Another member of the Hop family of outer-membrane proteins, HopI (HP1156), has a site (467) that distinguishes between Japan (H) and China-all (D) and YunnanMP-differentiated V633L.
Central metabolism
The region-differentiated amino acid changes involve a handful of key metabolic enzymes.
Citrate synthase GltA (HP0026, MWE_1570), is the first enzyme in the TCA cycle catalysing the conversion of acetyl-CoA and oxaloacetate to citrate. Yunnan-differentiated S127T is located between the two identical monomers and is likely involved in their association as well as in dimer–dimer association to form a 6-mer (Fig. 5b). The differentiated SNP might change the quaternary structure. Mutation of A124 in this interface was found in experimental evolution in E. coli [38]. In addition to GltA, two other key metabolic enzymes, PorB (HP1111), a subunit of pyruvate:ferredoxin oxidoreductase, and FixP (HP0147, MWE_0216), a subunit of cytochrome c oxidase, had high Fst values in YunnanMP. PorB, a key enzyme in the microaerophilic metabolism of H. pylori, converts pyruvate to acetyl-CoA, the substrate of GltA, and FixP is a component in aerobic respiration. The Yunnan-differentiated residue is at the proton entrance (Fig. S4C). Together, these changes might affect the metabolic capacity of this regional H. pylori subpopulation.
Host interaction
In addition to the genes listed above, region-specific non-synonymous variants are present in several genes that are annotated as known virulence or host interaction factors.
Catalase KatA (HP0875) (Fig. 6a) detoxifies H2O2 generated by host immune cells. It also binds host vitronectin, thereby protecting against complement-mediated killing [39]. The preferred route for H2O2 is the channel S451-D109-H56-haem (Fig. 6a (ii)) [40]. YunnanMP-differentiated P160H by the entrance S451 drastically changes local conformation and surface electric charge. Fujian-differentiated N248D with −1 change in the electric charge takes place near the dimer–dimer interface (Fig. 6a (i)) likely changing their interaction.
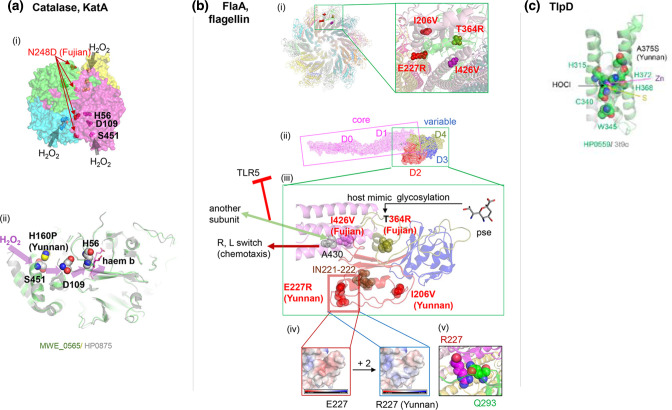
(a) Catalase KatA (HP0875, PDB 2A9E). (i) Dimer of dimer. (ii) Channel for H2O2.Yunnan-specific P160H by its entrance S451 drastically changes local conformation and surface electric charge (mutagenesis in PyMOL). (b) Flagellin, FlaA. (i) A view from the distal end of the 22-mer model of FlaA (MWE_0913) on G508A mutant of
C. jejuni
homologue (PDB 6×80) with four population-specific amino acid changes in two interacting monomers. (ii) Monomer with five domains. (iii) Population-specific amino acid changes. pse: pseudaminic acid. (iv) Surface electric charge change by E227R. (v) R227 interaction with a neighbouring monomer. (c) TlpD, chemotaxis receptor for HOCl. HP0559 modelled on PDB 3T9O, the regulatory CZB domain of DgcZ (E. coli).
acid. (iv) Surface electric charge change by E227R. (v) R227 interaction with a neighbouring monomer. (c) TlpD, chemotaxis receptor for HOCl. HP0559 modelled on PDB 3T9O, the regulatory CZB domain of DgcZ (E. coli).
The flagellar filament made of flagellin FlaA (HP0601) (Fig. 6b), is involved in motility, cell adherence and immune modulation. We have modelled it using the similar C. jejuni homologue [41]. The flagellin has rod-shaped domains forming a hydrophobic core, and the other domains decorating the surface of the filament are hypervariable. The flagellin is glycosylated by pseudaminic acid at several residues to stabilize the flagellum and to mimic host-cell surface, a way for the bacterium to modulate the immune response [42], but Fujian-differentiated T364R eliminates one of these sites (Fig. 6b (iii)). E227R drastically changes the surface electric charge and likely affects its interaction with a neighbouring monomer (Fig. 6b (iv)(v)). Another Fujian-differentiated residue 426 in the conservative core is next to residue 427, which is involved in evasion from TLR5-mediated innate immunity through subunit interaction [41]. Furthermore, residue 430 adjacent to 426 in the 3D structure is somehow involved in switching between R and L conformations for swimming/tumbling in chemotaxis in Campylobacter [43].
TlpD (HP0599, MWE_0916) (Fig. 6c) is a cytosolic chemotaxis sensor required for colonization. TlpD senses HOCl, an antimicrobial produced by neutrophils during inflammation [44]. HOCl oxidizes a conserved cysteine (C340) within a 3His/1Cys Zn-binding motif to inactivate chemo-transduction signalling. YunnanMP-differentiated A375S is right by this motif. Additional proteins in Table 2 are described in the Supplementary Material.
Discussion
Analyses of H. pylori in East Asia have tended to emphasize their homogeneity and uniformly high virulence potential. A single subpopulation of the bacteria, hspEAsia, is prevalent in the region. The hspEAsia strains have been found to be invariably CagPAI positive, with the cagA gene containing the ABD EPIYA motif, which is thought to promote strong binding of the protein to SHP-2 [45]. Our large collection of genomes of H. pylori from multiple regions in China, Japan and Korea confirm these observations. Only a small number of these isolates, of which a majority are from Mongolia and Okinawa, belonged to other H. pylori populations. For the scope of this study, these were excluded from subsequent analyses. All but 13 of the total 381 isolates were cagA positive and 355 have the characteristic ABD EPIYA type, including 316 out of 329 hspEAsia isolates.
Our results add a layer of complexity to the picture of uniformity by demonstrating that there is differentiation of H. pylori strains of the hspEAsia subpopulation between regions in East Asia. Despite a large burden of gastric disease in the region, most H. pylori infections by hspEAsia are asymptomatic, and the gastric cancer incidence varies widely across the region, especially in China. A large part of these differences might be attributable to diet and environment. However, our results imply that there are also bacterial factors differentiating these regions, which may be significant for disease development, especially because the bacteria themselves can adapt to environmental conditions [6].
Geographic differentiation between populations accumulates progressively when migration rates between them are low. Further, adaptation of bacteria to differences in environmental conditions can greatly accelerate the process of differentiation in specific regions of the genome. Our results, in combination with a previous study of genetic variation within the Americas [31] suggest that there are a handful of loci that have undergone rapid differentiation in several regions, and therfore may be considered keys for host adaptation. These include the genes frpB-4, hefC, alpB/hopB and hofC.
Our strategy to identify geographically differentiated SNPs by dividing one population (hspEAsia) into minimal subpopulations, therefore strains with consistent population and strain labels, and comparing fixation index (Fst) site-by-site between these populations reveal numerous loci of differentiation (Table S4). Of these we discuss the 12 with the strongest evidence for being involved in local adaptation in more detail in the main text in this paper. Some of the region-specific SNPs are in genes encoding for proteins that have been implicated in host interaction and virulence in the narrow sense: attack by immune system (catalase, TlpD), host adhesion (HopB/AlpB and several outer membrane proteins), and host surface mimicry (flagellin FlaA). Several of the other genes are transporters that may have implications for antimicrobial resistance, or are involved in nutrient acquisition. These results suggest that various host-adaptive changes in many host-interaction proteins lead to population differentiation. A similar gene set was found when rapid genome changes were investigated in shorter-term, intra-body micro-evolution [46].
Epidemiological and experimental evidence suggests that iron-deficiency increases
H. pylori
virulence and risk of gastric cancer [47]. In our analyses we could see both in iron and nickel metabolism highlighted by regional changes. Apart from the above-mentioned frpB-4, genes encoding for the TonB motor proteins ExbB-2 and ExbD-2, haem transporter FrpB-1 [48] appear in the list of the 34 genes with Fst >0.5 and the central transcription factor ferric uptake regulator, Fur, had regional variants (Supplementary Material). The causal mechanism underlying this association is not clear but it plausibly reflects bacterial response to nutrient limitation. Simply put, it is possible that bacteria adopt more aggressive strategies in interacting with the host and its microbiome when iron and other metals such as nickel, which is necessary for urease function, is in short supply.
and the central transcription factor ferric uptake regulator, Fur, had regional variants (Supplementary Material). The causal mechanism underlying this association is not clear but it plausibly reflects bacterial response to nutrient limitation. Simply put, it is possible that bacteria adopt more aggressive strategies in interacting with the host and its microbiome when iron and other metals such as nickel, which is necessary for urease function, is in short supply.
In other organisms, linkage means that high Fst regions often occur in large blocks, making it difficult to infer which sites are involved in local adaptation. However,
H. pylori
lineages recombine with each other, exchanging substantial fraction of their DNA in individual mixed infections [49]. The size of replacement in one event can be as short as 28 bp [50], with the result that linkage is broken down rapidly. This means that individual nucleotides can rise to high frequencies in specific populations, suggesting that local adaptation can potentially occur on a very exquisite scale.
bp [50], with the result that linkage is broken down rapidly. This means that individual nucleotides can rise to high frequencies in specific populations, suggesting that local adaptation can potentially occur on a very exquisite scale.
Many of the highly differentiated amino acid changes are close to critical residues of the protein and are plausible candidates to cause important functional changes, based on 3D modelling and previous functional analyses. We have suggested possible functional consequences but validation by targeted experiments and clinical observations is necessary. Although the functional consequences of genomic differentiation of H. pylori within different parts of the world remain to be elucidated, the presence of this differentiation already has potential clinical utility. All else being equal, individuals who are infected by H. pylori that are characteristically found in high gastric cancer incidence regions are likely to be at higher risk than those associated with lower incidence regions, firstly because the bacteria may be more virulent but secondly because infection with the bacteria might also be a marker for exposure to environmental factors that underlie the high disease risk.
Supplementary Data
Funding information
This work was supported by a grant from the State Key Laboratory of Infectious Disease Prevention and Control (SKLID) (2014SKLID102) of the Chinese Centre for Disease Control and Prevention. Supported by National Science and Technology Major Project (2018Z×10712–001) and a joint project 'Isolation and sequence analysis of Helicobacter pylori strains collected from investigations on carriage rate '. K.T. was supported by Swedish Society for Medical research (SSMF). Parts of the bioinformatic analyses were performed on resources provided by Swedish National Infrastructure for Computing (SNIC) through Uppsala Multidisciplinary Centre for Advanced Computational Science (UPPMAX) under projects snic2018-8-24/uppstore2017270, partially funded by the Swedish Research Council through grant agreement no. 2018–05973. This work was also supported in part by MEXT KAKENHI (19K22543, 17H04666, 26113704, 25291080, 221S0002 to I.K., 18K14766 to Y.K.) by NIBB Collaborative Research Program (20-443,21-328) to I.K., by U.S.-Japan Cooperative Medical Sciences Program Collaborative Awards, and by Shanghai Municipal Science and Technology Major Project (No. 2019SHZDZX02 to D.F.).
Acknowledgements
We thank Jonas Korlach and Primo Baybayan for help in SMRT sequencing. We would like to thank the staff of Comparative Genomics Laboratory at National Institute for Genetics, Mishima, Japan for supporting genome sequencing. We thank Dr. Chao Yang, Professor Yujun Cui and Ruifu Yang for the discussions about population genetics of H. pylori, and M. Zwama for discussion on HefC, Mizuki Ohno and Yosuke Kawai for discussion on human genetics.
Author group contributors
TEAMHp comprises Takahiro Bino, Masaki Fukuyo, Rumiko Suzuki, John Harting, Mototsugu Kato, Mutsuko Konno,Yuji Kohara, Christine Lambert, Yohei Minakuchi, Shin Nishiumi, Shuji Shigenobu, Noriko Takahashi, Atsushi Toyoda, Ikuo Uchiyama, Hirokazu Yano and Masaru Yoshida.
Conflicts of interest
Christine Lambert and John Harting are full-time employees at Pacific Biosciences of California, a company developing single-molecule sequencing technologies. The other authors declare that they have no conflict of interest.
Footnotes
Abbreviations: CNP, clustered nucleotide polymorphism; Fst, fixation index; MCMC, Markov Chain Monte Carlo; MOMP, major outer membrane protein; NE, North East; PDB, protein data bank; Sg, subgroup; SNP, single nucleotide polymorphism.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Four supplementary tables, four supplementary figures and one supplementary text.
References
Articles from Microbial Genomics are provided here courtesy of Microbiology Society
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/123456956
Article citations
The Helicobacter pylori Genome Project: insights into H. pylori population structure from analysis of a worldwide collection of complete genomes.
Nat Commun, 14(1):8184, 11 Dec 2023
Cited by: 6 articles | PMID: 38081806 | PMCID: PMC10713588
Study of Helicobacter pylori Isolated from a High-Gastric-Cancer-Risk Population: Unveiling the Comprehensive Analysis of Virulence-Associated Genes including Secretion Systems, and Genome-Wide Association Study.
Cancers (Basel), 15(18):4528, 12 Sep 2023
Cited by: 1 article | PMID: 37760497 | PMCID: PMC10526929
Low Expression of GIGYF1 Inhibits Metastasis, Proliferation, and Promotes Apoptosis and Autophagy of Gastric Cancer Cells.
Int J Med Sci, 20(8):1038-1045, 12 Jun 2023
Cited by: 3 articles | PMID: 37484805 | PMCID: PMC10357435
Repeated out-of-Africa expansions of Helicobacter pylori driven by replacement of deleterious mutations.
Nat Commun, 13(1):6842, 11 Nov 2022
Cited by: 10 articles | PMID: 36369175 | PMCID: PMC9652371
Recombination events drives the emergence of Colombian Helicobacter pylori subpopulations with self-identity ancestry.
Virulence, 13(1):1146-1160, 01 Dec 2022
Cited by: 1 article | PMID: 35838227 | PMCID: PMC9291697
Go to all (6) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioProject
- (2 citations) BioProject - PRJNA482300
Protein structures in PDBe (Showing 7 of 7)
-
(1 citation)
PDBe - 2h12View structure
-
(1 citation)
PDBe - 3AODView structure
-
(1 citation)
PDBe - 3W9IView structure
-
(1 citation)
PDBe - 4FWIView structure
-
(1 citation)
PDBe - 4AIQView structure
-
(1 citation)
PDBe - 5BUNView structure
-
(1 citation)
PDBe - 3T9OView structure
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Evolution in an oncogenic bacterial species with extreme genome plasticity: Helicobacter pylori East Asian genomes.
BMC Microbiol, 11:104, 16 May 2011
Cited by: 94 articles | PMID: 21575176 | PMCID: PMC3120642
Relationship between the diversity of the cagA gene of Helicobacter pylori and gastric cancer in Okinawa, Japan.
J Gastroenterol, 41(7):668-673, 01 Jul 2006
Cited by: 45 articles | PMID: 16933004
Amino acid polymorphisms flanking the EPIYA-A motif of Helicobacter pylori CagA C-terminal region is associated with gastric cancer in east China: experience from a single center.
J Dig Dis, 14(7):358-365, 01 Jul 2013
Cited by: 13 articles | PMID: 23517408
[H. pylori genomics].
Nihon Rinsho, 71(8):1352-1367, 01 Aug 2013
Cited by: 0 articles | PMID: 23967664
Review
Funding
Funders who supported this work.
Chinese Center for Disease Control and Prevention (1)
Grant ID: 2014SKLID102
Ministry of Education, Culture, Sports, Science and Technology (1)
Grant ID: 19K22543
National Major Science and Technology Projects of China (1)
Grant ID: 2018ZX10712-001
Shanghai Municipal Science and Technology Major Project (1)
Grant ID: 2019SHZDZX02
Swedish Research Council (1)
Grant ID: 2018-05973

 10
,
11
,
12
,
13
,
14
,*
10
,
11
,
12
,
13
,
14
,* 


