Abstract
Free full text
Assessing the risk of human‐to‐wildlife pathogen transmission for conservation and public health
Abstract
The SARS‐CoV‐2 pandemic has led to increased concern over transmission of pathogens from humans to animals, and its potential to threaten conservation and public health. To assess this threat, we reviewed published evidence of human‐to‐wildlife transmission events, with a focus on how such events could threaten animal and human health. We identified 97 verified examples, involving a wide range of pathogens; however, reported hosts were mostly non‐human primates or large, long‐lived captive animals. Relatively few documented examples resulted in morbidity and mortality, and very few led to maintenance of a human pathogen in a new reservoir or subsequent “secondary spillover” back into humans. We discuss limitations in the literature surrounding these phenomena, including strong evidence of sampling bias towards non‐human primates and human‐proximate mammals and the possibility of systematic bias against reporting human parasites in wildlife, both of which limit our ability to assess the risk of human‐to‐wildlife pathogen transmission. We outline how researchers can collect experimental and observational evidence that will expand our capacity for risk assessment for human‐to‐wildlife pathogen transmission.
Abstract
Human‐to‐wildlife pathogen transmission has played an important role in the epidemiology of a number of pathogens in the past, and is likely continuing to play an important role in the Anthropocene. Despite much concern surrounding the transmission of human pathogens into animal populations, particularly during the SARS‐CoV‐2 pandemic, there has been little critical assessment of the evidence base and the threat of these pathogens for conservation and public health. Here, we compile an evidence base of 97 published human‐to‐wildlife pathogen transmission events, and we discuss the sampling biases that have likely played an important role in producing the observed patterns, discussing how to collect further data to aid in risk assessment in the near future.
INTRODUCTION
Multi‐host pathogens are becoming a dominant feature of the Anthropocene. Driven by deforestation, land use conversion, and climate change, pathogens are increasingly being transmitted from animals into human populations, presenting a significant threat to public health (Jones et al., 2013; Plowright et al., 2015; Woolhouse & Brierley, 2018). Recently, concerns have been raised about transmission of pathogens from humans into wild animals (Edwards & Santini, 2020; Oreshkova et al., 2020; Prince et al., 2021; Shi et al., 2020; Wang et al., 2020). For a variety of reasons this process, known as “(zoo)anthroponosis,” “reverse zoonosis,” or more colloquially as “spillback,” could pose a difficult problem for wildlife conservation and public health efforts in the near future.
Concerns about human‐to‐wildlife pathogen transmission have grown throughout the ongoing COVID‐19 pandemic (Briggs & Helen, 2020; Edwards & Santini, 2020; Gorman, 2020; Olival et al., 2020; Prince et al., 2021; Santini & Edwards, 2020). SARS‐CoV‐2 has been transmitted from humans to a variety of animals including household cats and dogs, big cats, gorillas, and—perhaps most notably—has established epizootic transmission in mink farms on multiple continents (Garigliany et al., 2020; Gibbons & Ann, 2021; Molenaar et al., 2020; Munnink et al., 2020; OIE‐World Organisation for Animal Health, 2020; Oreshkova et al., 2020; Patterson et al., 2020; Sailleau et al., 2020; Segalés et al., 2020; Sit et al., 2020). Only recently, SARS‐CoV‐2 was transmitted back into wild mink populations in Spain and white‐tailed deer in North America (Aguiló‐Gisbert et al., 2021; Hale et al., 2021; Kuchipudi et al., 2021). Further, because SARS‐CoV‐2 is most closely related to sarbecoviruses infecting Rhinolophus bats, some fear that the virus might become established in bat populations outside of Asia and form a novel reservoir, complicating efforts to prevent future resurgence in humans (Banerjee et al., 2021; Hedman et al., 2021; Olival et al., 2020; Zhou et al., 2020). Additionally, if it causes clinical disease, introduction of SARS‐CoV‐2 might threaten the conservation of vulnerable bat species (Cook et al., 2021; Olival et al., 2020). So far, these potential risks have had far‐reaching consequences including widespread moratoria on bat research (Aizenman, 2020; Donahue, 2020) and extensive disease‐related cullings of mink (Dobson et al., 2020), while provoking broader discussions about prohibiting wildlife farming. Nevertheless, little is understood about the overall frequency of broader spillback events and their underlying drivers.
Human‐to‐wildlife pathogen transmission has doubtless played an important role in our epidemiological history. Studies have documented human‐sourced influenza strains in free‐ranging seals and skunks, zoonotic enteropathogens and respiratory viruses in human‐habituated gorillas and chimpanzees, and Zika virus in a selection of sylvatic wildlife (Britton et al., 2019; Favoretto et al., 2019; Goldberg et al., 2007; Kaur et al., 2008; Nizeyi et al., 2001; Obanda et al., 2013; Osterhaus et al., 2000; Rwego et al., 2008; Terzian et al., 2018). Additionally, evidence indicates that humans introduced certain pathogens to wildlife populations in which they are now enzootic, including yaws, yellow fever virus, and plague (Bryant et al., 2007; Dean et al., 2018; Mubemba et al., 2020). Nevertheless, there has been little critical analysis concerning the magnitude of the threat, and the evidence base has yet to be assessed and put on the same footing as its inverse (i.e. the more frequently characterised animal‐to‐human transmission events, or anthropozoonosis, colloquially referred to as ‘spillover’). This fact ultimately limits our understanding of the realised and potential impacts of spillback on conservation and public health (Ryan & Walsh, 2011).
This research gap leaves three intertwined questions unanswered. First, fundamentally, it is unclear to what degree transmission between humans and animals is a symmetrical process: do the same filters act when a pathogen moves from a human into an animal and vice versa (Plowright et al., 2017), or are some filters direction‐specific? Second, uncertainty exists surrounding which pathogens are most likely to undergo spillback and into which hosts, making it difficult to assess how the rising tide of infectious diseases in humans will affect free‐ranging and captive or habituated wildlife. Finally, it remains to be seen how great a threat human‐to‐wildlife pathogen transmission poses to threatened wildlife populations (Barlow et al., 2016; McKinney et al., 2010). The unknown magnitude of this risk reduces our ability to prioritise efforts to prevent human‐to‐wildlife transmission—particularly where such efforts might directly compete for funding with other health and conservation priorities, or otherwise compromise ongoing animal disease research efforts (e.g. white‐nose surveillance in bat populations).
Here, we critically assess the available evidence for spillback‐related processes and the inferential underpinnings of the relevant studies. First, we propose a conceptual framework for two primary pathways of spillback with meaningful differences in both likelihood and impact (Figure (Figure1).1). We then discuss documented examples of human pathogens transmitted to free‐ranging, captive, or habituated wildlife, and we use these data to highlight common trends in each of the two spillback pathways. We also assess spillback's impact on animal health and the evidence for secondary spillover into humans following establishment in a novel non‐human maintenance reservoir. Finally, we propose an evidential framework by which researchers can assess the risk for human‐to‐wildlife pathogen transmission in the future (Figure (Figure2),2), whether they be emerging (e.g. SARS‐CoV‐2) or well‐established (e.g. influenza).
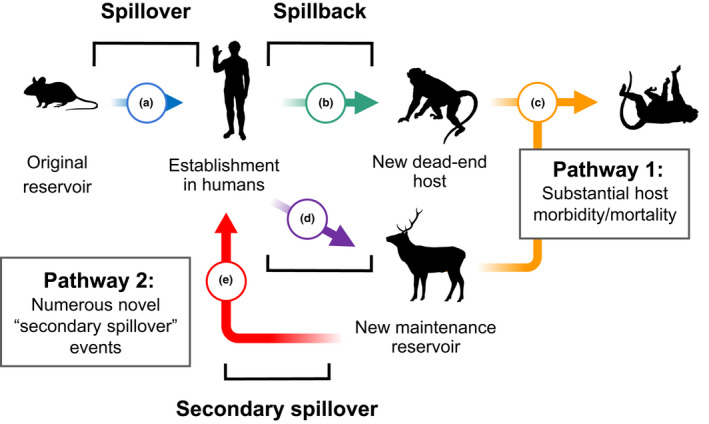
Pathways detailing the two general scenarios in which pathogen transmission from humans to wildlife is problematic. We separate these scenarios into concerns for 1) conservation and 2) public health. In both pathways, spillover into humans (a) results in successful zoonotic establishment; humans, which are competent transmitters of the pathogen, then transmit it onto a new animal population in an instance of “spillback” (b or d). In Pathway 1, the pathogen causes morbidity or mortality, potentially presenting a conservation risk (c). In Pathway 2, the new animal species proves a competent reservoir (d), maintaining the pathogen within the population. This population presents a spillover risk, potentially creating numerous novel infections in humans (e). This maintenance population, or additional wildlife species it shares pathogens with, may also suffer substantial morbidity and mortality, creating concerns for conservation (c). NB the sequence of events represents an idealised and simplified time‐structured scenario, comprising only one of a number of possible transmission paths through a complex metapopulation of hosts; transmission may occur in either direction between the depicted hosts, and the suggested hosts may represent a single species or a community or reservoir complex composed of multiple species. Silhouettes are taken from phylopic.org
THE TWO SPILLBACK PATHWAYS
Narratives surrounding spillback usually focus on one of two negative outcomes (or “Pathways”), which we outline in Figure Figure1.1. In Pathway 1 (Figure 1a–c), spillback events increase the burden of disease in animal populations because some human pathogens can cause morbidity or mortality in animals (Kaur et al., 2008), raising conservation concerns. Because pathogens can exhibit extreme virulence in host species that are distantly related to their original hosts (Farrell & Davies, 2019), if humans expose a vulnerable animal population to their pathogens, the conservation impacts could be severe—similar to threats posed by infectious diseases that spread from domesticated animals to endangered wildlife (Pedersen et al., 2007).
The second major concern is the potential for “secondary spillover” from animals back into human populations, threatening public health. This possibility, which has been frequently discussed in reference to SARS‐CoV‐2, is illustrated in Pathway 2 (Figure 1d–e) (Edwards & Santini, 2020; Gorman, 2020; Santini & Edwards, 2020). Where the pathogen newly infects a competent host species, it may achieve enzootic circulation, eventually being transmitted back into human populations. In this case, the wild host may represent a significant barrier to controlling and eliminating the disease, as illustrated by bovine tuberculosis in badgers in the United Kingdom (Donnelly et al., 2003) and Guinea worm in feral dog populations (Callaway, 2016). This pathway demands implementation of new measures to prevent subsequent epidemics in humans: medical countermeasures and non‐pharmaceutical interventions cannot prevent recurring outbreaks, and authorities must monitor and prevent pathogen reintroduction using One Health strategies such as surveillance, vaccination, or population management of the new reservoir. Evidence for Pathway 2 requires demonstrating that a pathogen has established itself in a vertebrate population following human introduction and circulated in said population, before re‐emerging into humans. As yet, it is unclear how likely this phenomenon is. Importantly, pathogens that cause high mortality (fulfilling Pathway 1) may less often achieve sustained transmission (Blumberg & Lloyd‐Smith, 2013), so these two pathways may trade off (i.e. if a pathogen fulfils Pathway 1 it may be less likely to fulfil Pathway 2).
Crucially, these pathways represent only a fraction of possible transmission routes through a complex metapopulation of hosts; the sequence of events from original hosts through humans to the secondary reservoirs depicted in Figure Figure11 are a simplified narrative of directional transmission through time. Instead, transmission may occur regularly between hosts of different species, and each depicted host may in fact represent a community or reservoir complex. Nevertheless, considering these pathways as simplified routes from animal‐to‐human (and vice versa) allows us to critically assess the evidence for each stage of the process, which is especially important given that these narratives are commonly used in the literature when assessing the risk of human‐to‐wildlife transmission (Cerdà‐Cuéllar et al. 2019; Britton et al., 2019; Zachariah et al., 2017).
THE EVIDENCE FOR HUMAN‐TO‐WILDLIFE PATHOGEN TRANSMISSION
To synthesise existing knowledge on spillback, we developed an evidence base using a primary literature search (Table S1). As has often been done for animal‐to‐human transmission research, our goal was to develop the most stringent set of criteria possible, defined as the confirmed transmission of a pathogen from a human to a non‐domesticated animal host, either wild or in captivity (e.g. zoo, rehabilitation centre, etc). Owing to difficulties inferring directionality or temporality of transmission, we excluded certain lines of evidence (serology, phylogenetic reconstructions, or historical documents) and certain categories of related processes (pathogen life cycles with obligate vectors or arthropod hosts, transmission between domestic animals and wild animals, or infection of wildlife resulting from anthropogenic activities) (Text S1). We also excluded a small number of cases describing human‐to‐invertebrate transmission (e.g. shellfish).
We found 97 studies describing human‐to‐wildlife transmission of a wide range of pathogens (Table S1). This list represents a substantial advance from the 19 studies retained from a previous review of zooanthroponoses (Messenger et al., 2014). Overall, these studies suggest that zooanthroponotic events themselves are well‐documented and diverse, but the evidence for their potential ecological and public health‐related consequences is far sparser (Table S1).
Our review uncovered a phylogenetically skewed selection of 104 host species. Most notably, our literature search reaffirmed that published zooanthroponotic events disproportionately feature non‐human primates (hereafter, “primates”), comprising 57/97 (58.8%) of our studies. Animals that are more closely related to humans generally host more parasites that can infect humans (Albery et al., 2021; Olival et al., 2017), and our literature review supports the bidirectional nature of this relationship. That is, perhaps because they are our closest living relatives, primates are both more likely to act as sources for pathogens that can infect humans, and to become infected with pathogens of humans (Davies & Pedersen, 2008). Nevertheless, human pathogens were also reported in a diverse range of non‐primate hosts, including carnivores, birds, and bats (Table S1). Over a quarter (15/44; 34.1%) of non‐primate records involved Mycobacterium tuberculosis, with hosts including elephants, a number of different ungulates, captive birds, mesocarnivores, and a rodent (Table S1).
Although it is thus clear that spillback can happen in a wide range of hosts, the non‐primate species involved were largely long‐lived, charismatic, and from captive populations, which are generally subject to elevated study effort and increased exposure to human pathogens. Specifically, many records involved elephants, large carnivores in zoos, and long‐lived birds. There were relatively few ungulates (only 18 of 327 extant species), rodents (only 7 rodent species out of >2000), and fast‐lived animals in general. There were only 6 species of bat, despite the fact that bats comprise ~20% of mammal species and (because they are important zoonotic hosts) have been heavily sampled for pathogens (Banskar et al., 2016; Barr et al., 2015; Ge et al., 2012; Li et al., 2010; Wu et al., 2016). Although life history is an important predictor of zoonotic risk (Plourde et al., 2017), it seems unlikely that charismatic, long‐lived species are truly more susceptible to human‐infecting pathogens; instead, these species are probably known to host human pathogens because they more frequently live alongside humans and are therefore more often exposed, or because they have simply been more intensively monitored and sampled (Albery et al., 2021). This skew confirms that the observation of human pathogens in animal populations depends heavily on the attention humans pay to those populations, and future analyses of zooanthroponotic transmission may have to deal with this confounded exposure and observation effort.
Overall, data suggest that human pathogens from diverse taxonomic groups and transmission modes can be transmitted to wildlife, and primates are disproportionately represented as hosts within the available literature. Beyond primates, zooanthroponotic transmission events are widely distributed, but the collected evidence base is heavily skewed towards large, slow‐lived animals. These findings demonstrate a widely‐observed pattern that emerges from human proximity: animals under stricter surveillance have more often been observed with human pathogens, demonstrating that spillback inference is vulnerable to well‐documented sampling biases (Albery et al., 2021; Mollentze & Streicker, 2020; Olival et al., 2017). We next sought to identify whether these pathogens affected the health of the infected animals.
PATHWAY 1: THE POTENTIAL CONSERVATION THREAT OF SPILLBACK
We found strong evidence that human pathogens can cause morbidity and mortality in new wildlife hosts, thus presenting a potential cost to conservation. In fact, most documented zooanthroponotic events (61/97, or 62.9%) were associated with observed morbidity or mortality in the naive animal, across a wide range of hosts (Table S1). Unsurprisingly, a substantial proportion of these reports (50/61, or 82.0%) described captive populations or habituated primates subject to ecotourism (Table (Table1,1, Table S1), leaving only 11 (18%) in free‐ranging animals. These studies generally describe animals receiving frequent visits from humans, allowing for more effective health monitoring compared to their wild counterparts, but at the costs of increased exposure to human‐sourced pathogens and overrepresentation in the literature.
TABLE 1
Studies reviewed are summarised (Table 1a), and further stratified based on primate‐only studies (Table 1b) and non‐primate studies (Table 1c). Study counts for Table 1b and c do not equal the total count in Table 1a because four studies described pathogens present in both primate and non‐primate populations, and thus, were counted in both Table 1b and c. The fourth column, “Comparative study”, describes articles in which both free‐ranging and captive animals were sampled for human‐sourced pathogens
| (a) All studies describing spillback (n = 97) | |||||
|---|---|---|---|---|---|
| Captive | Free‐ranging | Habituated (primates) | Comparative study | Total | |
| Healthy (or not discussed) | 7 | 10 | 14 | 5 | 36 |
| Morbidity ± mortality | 37 | 11 | 10 | 3 | 61 |
| Total | 44 | 21 | 24 | 8 | 97 |
| (b) Studies describing spillback in primates (n = 57) | |||||
|---|---|---|---|---|---|
| Captive | Free‐ranging | Habituated | Comparative study | Total | |
| Healthy (or not discussed) | 4 | 1 | 14 | 2 | 21 |
| Morbidity ± mortality | 23 | 0 | 10 | 3 | 36 |
| Total | 27 | 1 | 24 | 5 | 57 |
| (c) Studies describing spillback in non‐primates (n = 44) | ||||
|---|---|---|---|---|
| Captive | Free‐ranging | Comparative study | Total | |
| Healthy (or not discussed) | 3 | 9 | 3 | 15 |
| Morbidity ± mortality | 16 | 11 | 2 | 29 |
| Total | 19 | 20 | 5 | 44 |
Interestingly, there were no reports from long‐term ecological studies. Worldwide, dozens of wild animal populations have been studied by human researchers for decades (Clutton‐Brock & Sheldon, 2010; Hayes & Schradin, 2017). These populations are generally well‐understood and individuals are often known by name; as such, oddly behaving, sick, or dying animals are noticed and sampled, with pathogens considered as a potential cause. For example, an avian pox epidemic in Britain was quickly detected in a long‐term study of great tits (Parus major) using standard non‐disease‐focussed sampling procedures (Lachish et al., 2012). When a disease‐related conservation crisis arises, researchers do sometimes consider spillback: for example, when >60% of the global population of Saiga antelope (Saiga tatarica tatarica) died in Kazakhstan in 2015, human pathogens were specifically considered and rejected as a potential cause (Kock et al., 2018). Nevertheless, despite their close proximity to humans and decades of high‐intensity observations, we found no incidental records of spillback into long‐term study populations. It is possible that individual animals are regularly infected with human pathogens and suffer undetected morbidity; however, this possibility itself implies limited clinical significance of the pathogens involved, because infections are more likely to be detected if they cause mortality. Therefore, either human pathogens have had relatively low health impacts, or these impacts go unreported in the primary literature because researchers are not motivated to disclose or publish their findings.
Despite the common assertion that captive animals are commonly kept in high‐risk environments for disease, there are surprisingly few reports of human pathogens in zoos or wildlife rehabilitation centres—only 37 examples emerged from the millions of zoo animals that exist under extreme scrutiny by veterinary professionals (Table (Table1).1). We interpret this to demonstrate that either spillback events (and their associated health impacts) happen relatively infrequently, or that they most often occur through events that are unlikely to arise within zoos, such as faecal‐oral transmission or violent interactions (Figure (Figure3).3). Alternatively, concerns about public perception may result in limited disclosure of human pathogen transmission in zoos and other similar establishments, as with wild study systems. Regardless of the cause, given the scarcity of reports in wild animals, it remains difficult to assess the conservation threat posed by human‐to‐wildlife pathogen transmission – particularly relative to other anthropogenic drivers like land use change.
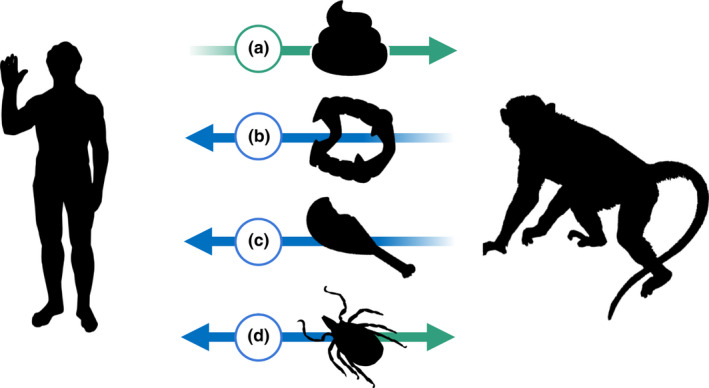
Asymmetry in transmission between humans and wildlife. Many studies that investigate animals’ susceptibility to human pathogens rest on the implicit assumption that interspecific pathogen transmission is symmetrical—that is, that pathogens go through the same series of hurdles in transmitting from humans to animals as they do in the reverse direction. One of the greatest unknowns concerning zooanthroponotic transmission is its symmetry: that is, do the same processes govern transmission from humans to animals as those governing animal‐to‐human transmission? A great many processes could create asymmetry in this relationship. For example, host immune cells often use cell surface proteins such as glycans to identify self from non‐self; when one species encounters a virus that has just budded off another species’ cells, its immune response may be able to more easily identify the glycans of the other species, and the propensity to identify other species’ glycans may not be equally effective in both directions. Several mechanisms act on humans specifically—most notably non‐pharmaceutical interventions (e.g. disinfectant, face masks, bednets, etc)—to lower incidence and transmission of infectious disease, both between humans and to/from animals. We suggest that the symmetry of transmission between humans and wildlife is likely to depend on the pathogen's transmission mode. For example, humans may be less likely to inhabit areas that involve concentrated animal waste, whereas a great many animal species are subjected to human sewage or runoff, exposing them more readily to human faecal‐oral pathogens (a). Similarly, it is unlikely that humans will bite wild animals, but relatively more likely that the opposite will happen; consequently humans are regularly exposed to rabies from animals, but the reverse is not true (b). Relatively few humans are eaten by animals, but many humans eat animals, which provides a well‐established spillover route for pathogen transmission during handling, slaughter, and consumption (c). However, there are some transmission modes that are likely to be more‐or‐less symmetrical—most notably vector‐borne transmission, provided the arthropod does not have narrow host‐feeding preferences, and feeds on both humans and non‐human vertebrates (d). Animal silhouettes are from phylopic.org
PATHWAY 2: THE SURPRISING SCARCITY OF SPILLBACK‐DRIVEN MAINTENANCE RESERVOIRS
In contrast to Pathway 1, documented instances of Pathway 2 (Figure 1d–e) appear extremely infrequently. In fact, we were unable to identify a verified example of novel maintenance of a human pathogen in any of our 97 studies. There were multiple examples of infected individuals that appeared healthy, and were therefore more likely to survive to transmit the pathogen (Table S1); however, all of these findings were the results of cross‐sectional surveillance and thus could have been produced by a recent transmission event rather than extended maintenance. Other examples of novel maintenance populations are marred by uncertainty or caveats, and some have been refuted: for example, although humans are thought to be the main source of leprosy (Mycobacterium leprae) for wild animals, leprosy was recently confirmed in disparate populations of chimpanzees with no history of prolonged contact with humans, implying the existence of an unknown reservoir (Hockings et al., 2021). This absence of evidence accentuates that future spillback‐related studies should aim to assess whether the focal pathogen is being maintained, and the time elapsed since the human‐to‐animal transmission event.
Despite our targeted search, the only documented reports of “secondary spillover” from a novel host back into humans (Figure (Figure1e)1e) were of SARS‐CoV‐2, from farmed mink into farm employees (Munnink et al., 2020) and from Syrian hamsters into pet shop visitors (Yen et al., 2022). Wildlife reservoirs like these could severely complicate elimination efforts, as is the case for dogs and dracunculiasis (“Guinea worm”) in sub‐Saharan Africa. Although dracunculiasis is nearing elimination, recent re‐emergence events have been traced to feral dog populations (Eberhard et al., 2014; McDonald et al., 2020), interfering with the disease's elimination (Hopkins et al., 2019). Note however that we do not consider this an example of secondary spillover because it is unclear whether dogs’ infections were sourced by humans recently, or whether they are a long‐standing maintenance reservoir for D. medinensis (McDonald et al., 2020).
The surprising scarcity of evidence for Pathway 2 suggests that transmission of a human‐sourced pathogen from wildlife back into human populations is not commonly observed. The dynamics observed with SARS‐CoV‐2 appear relatively unique, and contemporary concerns about secondary spillover are therefore mostly speculative. In cases like a multinational epidemic (e.g. Ebola virus in West Africa) or a pandemic (e.g. SARS‐CoV‐2), our data suggest that novel reservoirs represent a minor risk compared to the challenges of outbreak response and the possibility of unsuccessful containment in (or elimination from) humans themselves. Nevertheless, given that many important zoonotic pathogens have undergone historic human‐to‐wildlife transmission (Dean et al., 2018; Michalak et al., 1998; Palacios et al., 2011), our publication process appears to be failing to identify these events in their early prerequisite stages. Understanding the mechanisms underlying this failure will be important for remedying our evidence base in the future.
CHALLENGES TO OBSERVING HUMAN‐TO‐WILDLIFE TRANSMISSION EVENTS
Given that human‐origin pathogens have undoubtedly been transmitted from animals back to humans, why do these events appear so infrequently in the primary literature? This scarcity may be explained by the complexity of interspecific transmission, and the number of steps required for these events to occur and be observed, mediated by sampling and reporting biases. Interspecific pathogen transmission is a multi‐stage process, involving interspecific encounters, exposure to a novel pathogen, pathogen invasion, replication, and (possibly) transmission onwards (Alexander et al., 2018; Becker et al., 2019). Each of these processes requires the pathogen to pass a series of filters, such that the probability of progression decreases with each step (Plowright et al., 2017). It is relatively simple for Pathway 1 to manifest empirically: a human introduces a pathogen into a wildlife population (exposure and invasion) and the pathogen causes disease, involving some degree of pathogen survival and replication within the new host. In contrast, for Pathway 2 to manifest empirically, exposure and invasion must occur as in Pathway 1, and subsequently the pathogen must replicate to the point of onward transmission and attain an R0 of greater than 1 in the new animal population. This event is seemingly rare: only ~10% of zoonotic pathogens are known to have achieved this status in humans (Woolhouse & Brierley, 2018). Subsequently, the pathogen must then be transmitted again into human populations, following the exact same series of events but in the reverse direction, and all under the assumption that the pathogen does not cause rapid mortality (as it frequently will (Best & Kerr, 2000; Rothenburg & Brennan, 2020)), thereby diverting it towards Pathway 1 (Figure (Figure1c).1c). These processes may happen infrequently enough that they have very rarely occurred, particularly over the past few decades.
Assuming that this series of events (human‐to‐wildlife pathogen transmission, maintenance, and secondary spillover into human populations) does take place, our empirical inference must then contend with a series of sampling processes to be observed and published. For Pathway 1, sampling is fairly simple: human observers notice sick or dying animals, and identify the aetiological agent as being human in origin (e.g. SARS‐CoV‐2 in tigers at Bronx zoo (McAloose et al., 2020)). This process is facilitated by the fact that most of these scenarios involve captive, domestic, or habituated animals that are under close observation and in close proximity to humans—the same characteristics that drive their infection risk by elevating rates of exposure to human pathogens. This sampling process presumably contributes to the bias towards large charismatic animals that we detail above. For Pathway 2, because novel maintenance reservoirs are perhaps more likely to become established if the pathogen does not cause extreme pathology, they may be inherently more difficult to detect—potentially requiring active sampling rather than passive observation. If prior knowledge of the wild animal's pathogen community is limited before the zooanthroponotic transmission event, it may be impossible to reconcile the directionality of the observed shift – e.g. it remains plausible that palm civets were spillback hosts for SARS‐CoV in at least some circumstances (Tu et al., 2004; Wang et al., 2005). Moreover, if the maintenance population does source subsequent infections in human populations, these new human infections must then be traced back to the animal population of origin, which must then be verified as a novel host that was infected by a human (rather than a natural reservoir of a known multi‐host pathogen).
Even when a human‐to‐wildlife transmission event is observed, the observers must decide to report or publish it. It is possible that such events are commonly observed, but rarely reported – either because they are not considered novel or worth publishing, or because the individuals overseeing the facility in which transmission occurred are hesitant to draw attention to their operation. Motivating researchers and conservation practitioners to publish observations of human pathogens in wild and captive animals (while ensuring de‐identification of facility‐specific information to minimise endangering the validity of their operations) will therefore be important to remedy our skewed evidence base.
A perceived absence of secondary spillover events could also emerge from the bias towards surveillance of charismatic slow‐lived animals that we outline above. Fast‐lived animals are thought to host and source a disproportionate number of zoonoses as a result of their high abundance, lower immune investment, and proximity to humans (Albery & Becker, 2021); therefore, instances of pathogen transmission from humans into free‐ranging animal populations may be rare in the literature due to underrepresentation of fast‐lived animal species like rodents, and the use of pseudoabsences for unsampled species (Albery & Becker, 2021). Similarly, we found almost exclusively positive reports of human pathogens: only two studies reported negative findings for human pathogens in free‐ranging or habituated animal populations (Benavides et al., 2012; Bonnedahl et al., 2005). Although these studies were not included in our assessment and in the statistics we report (because they did not report a confirmed instance of human‐to‐wildlife transmission), confirming the absence of human pathogens in wild populations will provide an important baseline for future pathogen surveillance efforts, and for countering the observation bias evident in our dataset.
Due to the complexity of the processes involved and the limited relevant evidence, our ability to answer the question “how much of a threat do human pathogens pose to free‐ranging wildlife populations?” is currently severely limited. Nevertheless, it is vital that we attempt to answer this question to prevent a repetition of previous human‐to‐wildlife transmission events (Aguiló‐Gisbert et al., 2021; Ash et al., 2010; Britton et al., 2019), particularly as zoonotic pathogens continue to emerge in human populations at an accelerated rate.
HOW TO INFER AND UNDERSTAND HUMAN‐TO‐WILDLIFE TRANSMISSIONPOTENTIAL
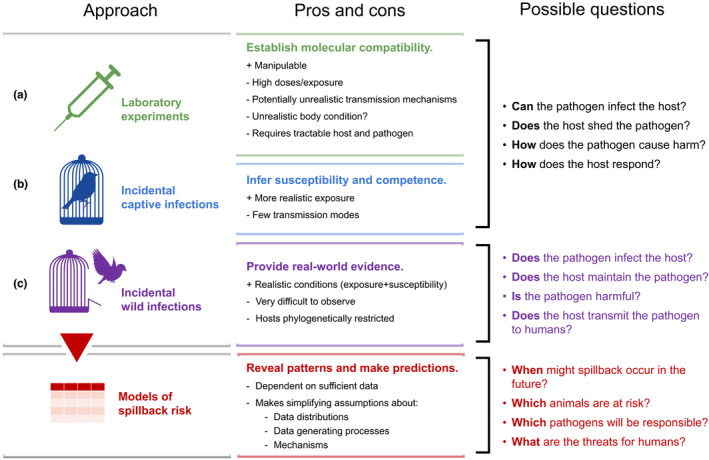
Given the difficulty of characterising high‐risk hosts and pathogens with the available evidence, researchers will likely have to continue relying on case‐by‐case assessment of human‐to‐wildlife transmission risk for individual pathogens (Figure (Figure2).2). Such understanding will come from three main sources: (A) laboratory experiments demonstrating host‐pathogen compatibility; (B) incidental transmission events in captive or managed animal populations; and (C) documented transmission events in free‐ranging animal populations. All of these scenarios possess different strengths and limitations in their ability to inform the complete ecological narrative, which we discuss here. To assess the risk of human‐to‐wildlife pathogen transmission, researchers should apply a combination of these approaches; in time, if the sampling biases evident in the literature are addressed, the evidence base may grow to the point that we can carry out meta‐analyses and build predictive models for zooanthroponotic transmission in the same way as others have for human infections sourced from wildlife populations (Olival et al., 2017; see below).
(A) Laboratory experiments. Because zoonotic pathogens are by definition able to infect multiple hosts, researchers often ask whether a particular animal could host the pathogen using in vivo and in vitro infection experiments. For example, in an effort to characterise potential sylvatic reservoirs for SARS‐CoV‐2 in the last year, researchers have successfully infected deer mice, tree shrews, ferrets, rabbits, bushy‐tailed woodrats, striped skunks, Egyptian fruit bats, white‐tailed deer, and raccoon dogs (Bosco‐Lauth et al., 2021; Chandler et al., 2021; Fagre et al., 2021; Freuling et al., 2020; Griffin et al., 2021; Hale et al., 2021; Mykytyn et al., 2021; Schlottau et al., 2020; Shi et al., 2020; Zhao et al., 2020). Equally importantly, researchers have identified wildlife species that are harder to infect (big brown bats, cottontail rabbits, fox squirrels, Wyoming ground squirrels, black‐tailed prairie dogs, house mice, and raccoons) (Bosco‐Lauth et al., 2021; Hall et al., 2020). These findings can establish (or exclude) new model animal systems, interrogate the cellular and molecular underpinnings of susceptibility, and provide clues about a given host's possible reservoir status.
Successfully infecting an animal in the lab is incontrovertible evidence that it could become infected in the wild through incidental human‐to‐wildlife pathogen transmission events. However, these approaches offer limited information about overall transmission risk in the wild for several reasons. First, they do not take into account the ecological nuances of contact rates or transmission routes between susceptible hosts: that is, while they approximate interspecific compatibility, they do not account for the role of opportunity in determining patterns of infection in nature. Further, experimental studies often use very high infectious doses or exposure techniques like intranasal inoculation (Gerdts et al., 2007), which are unlikely to occur in the wild. Laboratory animal populations may also differ immunologically from their wild counterparts in important ways. For example, recent work has shown that the immune systems and microbiota of laboratory house mice (Mus musculus) change when they are (re)introduced to wild environments, altering immune resistance (Bär et al., 2020; Leung et al., 2018). As such, mice could be interpreted as a potential sylvatic reservoir for a given pathogen based on results in laboratory populations, while wild counterparts are in fact minimally susceptible and therefore less vulnerable to zooanthroponotic transmission. Similarly, a recent deworming study in wild Peromyscus mice found that helminth coinfection reduces the prevalence of Sin Nombre Virus (Sweeny et al., 2020); lab‐reared Peromyscus do not host helminths, whereas wild animals are nearly all infected, and so their susceptibility in the wild is in fact lower than would be expected based on laboratory experiments. Finally, experimental approaches are limited to hosts that can be housed and maintained (which excludes many large animals and species of conservation concern), and pathogens that can be cultured and readily worked with (which eliminates several species of anaerobic bacteria, metazoan parasites with complex life cycles, certain vector‐borne pathogens, and many high‐consequence pathogens).
(B) Incidental captive infections. Incidental human‐to‐wildlife transmission events have been relatively commonly documented in captive animals, comprising 52/97(53.6%) of our records. While such transmission events have resulted in some morbidity or mortality (Crossley et al., 2012; McAloose et al., 2020; Munnink et al., 2020; Oh et al., 2002) and onward transmission of the pathogen (Li et al., 2014; McClure & Keeling, 1971; Oreshkova et al., 2020; Yen et al., 2022), these scenarios still do not accurately recapitulate the ecological conditions affecting free‐ranging populations for several reasons. For example, direct contact rates between humans and captive animals likely far surpass those in the wild, increasing the probability that zooanthroponotic transmission will occur in the first place. Captive animals may also be immunosuppressed compared to their free‐ranging counterparts due to inadequate housing conditions (Fischer & Romero, 2019; Seeber et al., 2020), which could lower their resistance and paint an exaggerated picture of their susceptibility. Further, following successful initial infection, confined housing may result in artificially high intraspecific contact and transmission rates, so that their ability to transmit and maintain the pathogen is similarly exaggerated compared to wild animals. As such, opportunity, compatibility, and reservoir competence are all potentially exaggerated in captive contexts, likely inflating perceived probability of pathogen transmissions from humans to truly free‐ranging wildlife for both Pathway 1 and 2.
(C) Incidental wild infections. Documented historical transmission events in wild animal populations are the most biologically relevant standard for human‐to‐wildlife pathogen transmission evidence, and comprised 45/97 (46.4%) of our records. Unfortunately, these examples come with their own set of inferential difficulties. Most importantly, directionality is difficult to determine in observational contexts in the absence of molecular analysis and epidemiological tracing (Nizeyi et al., 2001; Palacios et al., 2011). Verifying that humans transmitted a pathogen to an animal (rather than both receiving the pathogen from the same source) requires a combination of well‐understood transmission mechanisms, known local prevalence in both humans and animals, and time‐structured sampling of both populations. At macroscopic scales, time‐structuring or geographic expansions are occasionally sufficient to assume zooanthroponosis. For example, discovering newly emergent strains of Influenza A (H1N1) in striped skunks (Mephitis mephitis) at the height of flu season implied that humans were sourcing the disease (Britton et al., 2019). Similarly, Zika virus was discovered in New World primates following introduction of the virus to the Americas (Favoretto et al., 2019), implying that humans were responsible for transporting it.
Directionality becomes difficult to determine in observational scenarios when a pathogen is maintained in both human and animal populations. In the case of influenza A viruses (IAV), transmission from humans to swine has been documented, and is even considered responsible for a majority of viral diversity in farmed swine (Zhou et al., 1999), but directionality is difficult to infer without molecular tracing. While the decreased cost associated with next‐generation sequencing has accelerated these efforts, simultaneous circulation of IAVs in migratory birds, farmed swine, and human populations complicates analysis of spatiotemporal patterns and directionality (Lam et al., 2012; Roche et al., 2014). Similarly, Mycobacterium tuberculosis complex (MTC) bacteria have been transmitted from humans to cattle (Fritsche et al., 2004), primates (Coscolla et al., 2013), and elephants (Zachariah et al., 2017). Although they are ancestrally human pathogens, in many regions where MTC is both endemic and enzootic, epidemiological tracing becomes nearly impossible. A final obstacle comes in cases where the pathogen exhibits latent transmission through the environment, such that the lack of direct human‐animal contact makes causality and directionality even harder to infer. In this case, identifying contaminated areas or conducting thorough environmental sampling is important and can provide clues to the pathogen's source.
In summary, our evidence for human‐to‐wildlife pathogen transmission is based on several lines of inquiry (Figure (Figure2),2), none of which are sufficient alone. While laboratory experiments and captive infection reports are indicative of host‐pathogen compatibility, they occur in potentially non‐representative (high‐susceptibility and high‐exposure) environments that may be minimally relevant to the risk of zooanthroponotic transmission in situ. Meanwhile, observational reports of these incidents in wild populations are fraught with inferential difficulties inherent to ecological systems. Further, a lack of directed sampling for human pathogens in wild populations has produced an evidence base that is biased towards large, captive animals. Unfortunately, when observational reports are received, it is too late from a conservation and public health perspective: all they can tell us currently is whether or how often zooanthroponotic transmission has happened in the past. Therefore, when researchers are concerned about cross‐species transmission of an emerging pathogen (e.g., SARS‐CoV‐2), experiments are necessary to properly anticipate this risk.
Ideally, to overcome the shortcomings that we have outlined, we suggest that more experiments aimed at identifying risk of human‐to‐wildlife pathogen transmission be carried out in semi‐natural settings that more closely approximate the conditions experienced by wild populations. Such studies are already conducted in some contexts: for example, researchers explored the risk of SARS‐CoV‐2 transmission to Rousettus aegyptiacus bats by inoculating individuals and then observing as the virus was transmitted to co‐housed animals (Schlottau et al., 2020). Emulation of a natural environment is difficult, and answering these questions will require collaboration between virologists, immunologists, veterinarians, and field ecologists. Future investigations could also aim to surveil suspected high‐risk areas across a wide range of hosts to increase the observability of incidental wild infections when they do occur. Ultimately, the more evidence that we can accumulate, the easier it will be to build advanced models of zooanthroponotic transmission risk for use in prediction and prevention efforts.
THE FUTURE OF ZOOANTHROPONOSIS RESEARCH
A crucial step to understanding zooanthroponosis, and the logical end point of a sufficiently bolstered evidence base, is the construction of sophisticated predictive models. Using analogous models applied to other infectious disease processes, researchers have made enormous strides towards understanding host‐pathogen ecology, allowing reservoir host identification (Albery et al., 2020; Babayan et al., 2018), zoonotic risk prediction (Mollentze & Streicker, 2020), geographic sampling prioritisation (Han et al., 2016; Olival et al., 2017), and more. Although model results are not always well‐integrated into pandemic prevention efforts (Carlson, 2020; Holmes et al., 2018), they are crucial for understanding broad‐scale patterns of animal‐to‐human pathogen transmission—and the same will be true of human‐to‐wildlife transmission. As it stands, researchers have just produced a model‐based prediction of zooanthroponosis risk for the most intensively studied pathogen in recent years (SARS‐CoV‐2 (Fischhoff et al., 2021)). In this study, the authors used structural modelling of ACE2 (the binding receptor of SARS‐CoV‐2) and trait‐based analyses to examine species’ propensity to host the virus, with some success (Table (Table2).2). This study represents a significant step forward for a focal pathogen, and given enough data on broad‐scale zooanthroponotic transmission trends, we may be able to do the same more broadly in the near future.
TABLE 2
In addition to in vivo experiments to characterise susceptibility of novel hosts, analytical tools (e.g. comparative genomics, structural modelling for receptor binding, and trait‐based machine learning) are being used to predict the potential host range of SARS‐CoV‐2
| Species | Zoonotic compatibility score (Fischhoff) | Damas | Citation | |
|---|---|---|---|---|
| Experimental infection | Chinese hamster (Cricetulus (barabensis) griseus) | 0.756 | High | Bertzbach et al. (2021) |
| Black‐tailed prairie dog (Cynomys ludovicianus) | 0.200 | Not listed | Bosco‐Lauth et al. (2021) | |
| Red fox (Vulpes vulpes) | 0.880 | Low | Porter et al. (2022) | |
| Natural infection | Binturong (Arctictis binturong) | 0.796 | Not listed | Animal Plant and Health Inspection Service, United States Department of Agriculture (2021a) |
| Fishing cat (Prionailurus viverrinus) | 0.680 | Not listed | ||
| Spotted hyena (Crocuta crocuta) | 0.892 | Not listed | Animal Plant and Health Inspection Service, United States Department of Agriculture (2021d) | |
| Hippopotamus (Hippopotamus amphibius) | 0.442 | Medium | British Broadcasting Corporation (2021) | |
| Lynx (Lynx canadensis) | 0.223 | Medium | Animal Plant and Health Inspection Service, United States Department of Agriculture (2021b) | |
| Coati (Nasua nasua) | 0.384 | Not listed | Animal Plant and Health Inspection Service, United States Department of Agriculture (2021c) |
Featured is a list of wildlife species for which empirical evidence of susceptibility has been demonstrated, either through natural infection or experimental inoculation, with predicted susceptibilities as determined by structural analysis of ACE2 alone (Damas et al., 2020) and with the addition of ecological and life history traits as predictors (Fischhoff et al., 2021). Following a procedure described in Becker et al. for evaluating the performance of a classifier model when the validation data can only meaningfully be presence‐only (Becker et al., 2022), we evaluated the area under the training prevalence‐test sensitivity curve (AUC‐TPTSC). We found that the model performed slightly better than random (AUC = 0.637). Code to reproduce the analysis is available at github.com/viralemergence/fischhoff‐validation.
In principle, host‐pathogen models should be applicable to zooanthroponotic pathogen transmission, just as they are to animal‐to‐human spillover. Interspecific pathogen transmission is a bilateral process, and humans are merely a node in a diverse host‐pathogen network. For example, as outlined above, non‐human primates both host more zoonotic pathogens and comprise a majority of zooanthroponotic transmission reports (Engel et al., 2002; Heldstab et al., 1981; Terzian et al., 2018). Given this potential symmetry, it stands to reason that larger datasets on zooanthroponotic processes (and large‐scale analyses of host and pathogen traits) may facilitate prediction of bidirectional transmission between humans and animals. Doing so may require subtle changes in analytical framing. For example, models are often framed as “predicting the original reservoir host(s)” (Babayan et al., 2018; Becker et al., 2022; Brierley & Fowler, 2021), rather than “predicting potential future reservoir hosts.” This is also often true of laboratory infection studies (Botten et al., 2000; Cogswell‐Hawkinson et al., 2012; Jones et al., 2019; Richter et al., 2004), although work on SARS‐CoV‐2 is a notable exception given the concern over establishment of secondary sylvatic reservoirs (Fagre et al., 2021; Fischhoff et al., 2021; Griffin et al., 2021; Hall et al., 2020; Schlottau et al., 2020). Because of the different ways that we obtain and interpret human and animal infection data, it remains a near‐total unknown whether these approaches are equally valid, and whether spillover is generally symmetrical between humans and animals; nevertheless, this symmetry is sometimes implicitly assumed to be the case.
In Figure Figure3,3, we outline how asymmetry in transmission could manifest between humans and animals. Researchers may be able to construct models of human‐to‐animal transmission risk based on spillover‐related processes and then to test their predictions with datasets of animal‐to‐human transmission, or vice versa, in order to verify whether the symmetry of these processes (Table (Table2)2) (Fischhoff et al., 2021). Validating the generality of these interrelationships will further the perception of zoonotic pathogens as part of a complex metapopulation of hosts, rather than reinforcing a narrative of animal‐to‐human directionality. Importantly, while Fischhoff et al. (2021) succeeded in modelling species’ ability to become infected with SARS‐CoV‐2, researchers have yet to model species’ propensity to transmit it onwards (i.e., transmission competence); general multi‐host pathogen transmission prediction frameworks will have to incorporate elements of both.
While it is likely universally useful to understand and predict how pathogens are transmitted from animals to humans, it is possible that a smaller subset of efforts related to transmission risk of zooanthroponoses will be worthy of (urgent) consideration or investment in mitigation. For example, we are increasingly able to understand the genomic basis of zoonotic potential (Mollentze & Streicker, 2020), but considerable resources might be required to quantify a pathogen's ability to infect any given animal species, rendering this approach economically unviable for mitigating human‐to‐wildlife transmission. Regardless, identification of at‐risk species or groups could lead to researchers prioritising high‐risk groups for further identification, leading to a better understanding of the general drivers of infection in both humans and animals. Although experimental infections of animals with SARS‐CoV‐2 were partially motivated by concerns around novel maintenance reservoirs (Bosco‐Lauth et al., 2020; Cool et al., 2022; Fagre et al., 2021; Olival et al., 2020; Schlottau et al., 2020; Shi et al., 2020), their findings are likely to eventually inform the underlying physiological and ecological mechanisms responsible for mediating susceptibility. For example, the knowledge that big brown bats are relatively resistant to infection with SARS‐CoV‐2 may lead to further investigations into the immune mechanisms responsible for viral clearance (Hall et al., 2020).
Ultimately, integrating statistical models to predict bidirectional human‐animal pathogen transmission, and more accurately delineating the similarities and differences between the two processes, will draw necessary attention to the perception of humans as part of a complex metapopulation of hosts (Frutos et al., 2021; Viana et al., 2014). Researchers studying infectious diseases at the human‐animal interface could benefit from the renewed interest in zooanthroponotic pathogen transmission engendered by the SARS‐CoV‐2 pandemic, building on the established knowledge in this review. A fuller understanding of such processes, and the ecology of multi‐host systems in general, will reveal at‐risk hosts and associated pathogens, inform fundamental biological understanding of the symmetry of interspecific pathogen transmission, and improve human and animal health in the coming century. This kind of understanding is only likely to increase in importance: ongoing increases in human population density, epidemic and pandemic risk, human‐wildlife contact, and anthropogenic stressors on animal health stand to drive up the rate of human‐to‐wildlife pathogen transmission over the coming decades. Ensuring that our evidence base can track its progression is an important prerequisite to effective prevention.
AUTHORSHIP
ACF and GFA conceptualised and designed the study. ACF collected and analyzed papers. ACF, CJC and GFA wrote the first draft of the manuscript, and all authors contributed substantially to revisions.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1111/ele.14003.
Supporting information
Table S1
Supplementary Material
Notes
Fagre A.C., Cohen L.E., Eskew E.A., Farrell M., Glennon E. & Joseph M.B. et al. (2022) Assessing the risk of human‐to‐wildlife pathogen transmission for conservation and public health. Ecology Letters, 25, 1534–1549. Available from: 10.1111/ele.14003 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
Contributor Information
Anna C. Fagre, Email: [email protected].
Gregory F. Albery, Email: moc.liamg@yreblafg.
DATA AVAILABILITY STATEMENT
The data for this paper are available at zenodo.org: https://doi.org/10.5281/zenodo.6334167.
REFERENCES
- Aguiló‐Gisbert J., Padilla‐Blanco M., Lizana V., Maiques E., Muñoz‐Baquero M., Chillida‐Martínez E. et al. (2021) First description of SARS‐CoV‐2 infection in two feral American mink (Neovison vison) caught in the wild. Animals, 11, 1422. [Europe PMC free article] [Abstract] [Google Scholar]
- Aizenman N. (2020) Why the U.S. government stopped funding a research project on bats and coronaviruses. National Public Radio. Available at: https://www.npr.org/sections/goatsandsoda/2020/04/29/847948272/why‐the‐u‐s‐government‐stopped‐funding‐a‐research‐project‐on‐bats‐and‐coronaviru, Last accessed 18 March 2022. [Google Scholar]
- Albery G.F. & Becker D.J. (2021) Fast‐lived hosts and zoonotic risk. Trends in Parasitology, 37, 117–129. [Abstract] [Google Scholar]
- Albery G.F., Carlson C.J., Cohen L.E., Eskew E.A., Gibb R., Ryan S.J. et al. (2021). Urban‐adapted mammal species have more known pathogens. bioRxiv, 2021.01.02.425084. 10.1101/2021.01.02.425084 [Abstract] [CrossRef] [Google Scholar]
- Albery G.F., Eskew E.A., Ross N. & Olival K.J. (2020) Predicting the global mammalian viral sharing network using phylogeography. Nature Communications, 11, 2260. [Europe PMC free article] [Abstract] [Google Scholar]
- Alexander K.A., Carlson C.J., Lewis B.L., Getz W.M., Marathe M.V., Eubank S.G. et al. (2018). The ecology of pathogen spillover and disease emergence at the human‐wildlife‐environment interface. In: Hurst, C.J. , editor. (Ed.) The connections between ecology and infectious disease. Cham: Springer, pp. 267–298. [Google Scholar]
- Animal Plant and Health Inspection Service, United States Department of Agriculture (2021a). Confirmation of COVID‐19 in a Binturong and a Fishing Cat at an Illinois Zoo. Anim. Plant Health Insp. Serv. Newsroom. Available at: https://www.aphis.usda.gov/aphis/newsroom/stakeholder‐info/sa_by_date/sa‐2021/sa‐10/covid‐binturong‐fishing‐cat, Last accessed 5 February 2022
- Animal Plant and Health Inspection Service, United States Department of Agriculture (2021b). Confirmation of COVID‐19 in a Canada Lynx at an Pennsylvania Zoo. Anim. Plant Health Insp. Serv. Newsroom. Available at: https://www.aphis.usda.gov/aphis/newsroom/stakeholder‐info/sa_by_date/sa‐2021/sa‐12/covid‐lynx‐pa, Last accessed 5 February 2022
- Animal Plant and Health Inspection Service, United States Department of Agriculture (2021c). Confirmation of COVID‐19 in a Coatimundi at an Illinois Zoo. Anim. Plant Health Insp. Serv. Newsroom. Available at: https://www.aphis.usda.gov/aphis/newsroom/stakeholder‐info/sa_by_date/sa‐2021/sa‐10/covid‐coatimundi, Last accessed 5 February 2022
- Animal Plant and Health Inspection Service, United States Department of Agriculture (2021d). Confirmation of COVID‐19 in Hyenas at a Colorado Zoo. Anim. Plant Health Insp. Serv. Newsroom. Available at: https://www.aphis.usda.gov/aphis/newsroom/stakeholder‐info/sa_by_date/sa‐2021/sa‐11/covid‐hyenas, Last accessed 5 February 2022
- Ash A., Lymbery A., Lemon J., Vitali S. & Thompson R.C.A. (2010) Molecular epidemiology of Giardia duodenalis in an endangered carnivore – The African painted dog. Veterinary Parasitology, 174, 206–212. [Abstract] [Google Scholar]
- Babayan S.A., Orton R.J. & Streicker D.G. (2018) Predicting reservoir hosts and arthropod vectors from evolutionary signatures in RNA virus genomes. Science, 362, 577–580. [Europe PMC free article] [Abstract] [Google Scholar]
- Banerjee A., Mossman K. & Baker M.L. (2021) Zooanthroponotic potential of SARS‐CoV‐2 and implications of reintroduction into human populations. Cell Host & Microbe, 29, 160–164. [Europe PMC free article] [Abstract] [Google Scholar]
- Banskar S., Bhute S.S., Suryavanshi M.V., Punekar S. & Shouche Y.S. (2016) Microbiome analysis reveals the abundance of bacterial pathogens in Rousettus leschenaultii guano. Scientific Reports, 6, 1–13. [Europe PMC free article] [Abstract] [Google Scholar]
- Bär J., Leung J.M., Hansen C., Loke P., Hall A.R., Conour L. et al. (2020) Strong effects of lab‐to‐field environmental transitions on the bacterial intestinal microbiota of Mus musculus are modulated by Trichuris muris infection. FEMS Microbiology Ecology, 96(10), fiaa167. 10.1093/femsec/fiaa167 [Abstract] [CrossRef] [Google Scholar]
- Barlow J., Lennox G.D., Ferreira J., Berenguer E., Lees A.C., Nally R.M. et al. (2016) Anthropogenic disturbance in tropical forests can double biodiversity loss from deforestation. Nature, 535, 144–147. [Abstract] [Google Scholar]
- Barr J., Smith C., Smith I., de Jong C., Todd S., Melville D. et al. (2015) Isolation of multiple novel paramyxoviruses from pteropid bat urine. Journal of General Virology, 96, 24–29. [Abstract] [Google Scholar]
- Becker D.J., Albery G.F., Sjodin A.R., Poisot T., Bergner L.M., Chen B. et al. (2022) Optimising predictive models to prioritise viral discovery in zoonotic reservoirs. The Lancet Microbe, 10.1016/S2666-5247(21)00245-7 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Becker D.J., Washburne A.D., Faust C.L., Pulliam J.R.C., Mordecai E.A., Lloyd‐Smith J.O. et al. (2019) Dynamic and integrative approaches to understanding pathogen spillover. Philosophical Transactions of the Royal Society B: Biological Sciences, 374(1782), 20190014. 10.1098/rstb.2019.0014 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Benavides J.A., Godreuil S., Bodenham R., Ratiarison S., Devos C., Petretto M.‐O. et al. (2012) No evidence for transmission of antibiotic‐resistant Escherichia coli strains from humans to wild western lowland gorillas in Lopé National Park. Applied and Environmental Microbiology, 78, 4281–4287. [Europe PMC free article] [Abstract] [Google Scholar]
- Bertzbach L.D., Vladimirova D., Dietert K., Abdelgawad A., Gruber A.D., Osterrieder N. et al. (2021) SARS‐CoV‐2 infection of Chinese hamsters (Cricetulus griseus) reproduces COVID‐19 pneumonia in a well‐established small animal model. Transboundary and Emerging Diseases, 68, 1075–1079. [Europe PMC free article] [Abstract] [Google Scholar]
- Best S.M. & Kerr P.J. (2000) Coevolution of host and virus: the pathogenesis of virulent and attenuated strains of myxoma virus in resistant and susceptible European rabbits. Virology, 267, 36–48. [Abstract] [Google Scholar]
- Blumberg S. & Lloyd‐Smith J.O. (2013) Inference of R0 and transmission heterogeneity from the size distribution of stuttering chains. PLoS Computational Biology, 9, e1002993. [Europe PMC free article] [Abstract] [Google Scholar]
- Bonnedahl J., Broman T., Waldenström J., Palmgren H., Niskanen T. & Olsen B. (2005) In search of human‐associated bacterial pathogens in Antarctic wildlife: report from six penguin colonies regularly visited by tourists. AMBIO: A Journal of the Human Environment, 34, 430–432. [Abstract] [Google Scholar]
- Bosco‐Lauth A.M., Hartwig A.E., Porter S.M., Gordy P.W., Nehring M., Byas A.D. et al. (2020) Experimental infection of domestic dogs and cats with SARS‐CoV‐2: pathogenesis, transmission, and response to reexposure in cats. Proceedings of the National Academy of Sciences, 117, 26382–26388. [Europe PMC free article] [Abstract] [Google Scholar]
- Bosco‐Lauth A.M., Root J.J., Porter S.M., Walker A.E., Guilbert L., Hawvermale D. et al. (2021) Peridomestic mammal susceptibility to severe acute respiratory syndrome coronavirus 2 infection. Emerging Infectious Diseases, 27, 2073. [Europe PMC free article] [Abstract] [Google Scholar]
- Botten J., Mirowsky K., Kusewitt D., Bharadwaj M., Yee J., Ricci R. et al. (2000) Experimental infection model for Sin Nombre hantavirus in the deer mouse (Peromyscus maniculatus). Proceedings of the National Academy of Sciences, 97, 10578–10583. [Europe PMC free article] [Abstract] [Google Scholar]
- Brierley L. & Fowler A. (2021) Predicting the animal hosts of coronaviruses from compositional biases of spike protein and whole genome sequences through machine learning. PLoS Path, 17, e1009149. [Europe PMC free article] [Abstract] [Google Scholar]
- Briggs H. (2020) Coronavirus: Spain orders culling of almost 100,000 mink. Br. Broadcast. Corp.
- British Broadcasting Corporation (2021). Belgian zoo hippos test positive for Covid.
- Britton A.P., Trapp M., Sabaiduc S., Hsiao W., Joseph T. & Schwantje H. (2019) Probable reverse zoonosis of influenza A (H1N1) pdm 09 in a striped skunk (Mephitis mephitis). Zoonoses Public Health, 66, 422–427. [Abstract] [Google Scholar]
- Bryant J.E., Holmes E.C. & Barrett A.D.T. (2007) Out of Africa: a molecular perspective on the introduction of yellow fever virus into the Americas. PLoS Path, 3, e75. [Europe PMC free article] [Abstract] [Google Scholar]
- Callaway E. (2016) Dogs thwart effort to eradicate Guinea worm. Nature News, 529, 10. [Abstract] [Google Scholar]
- Carlson C.J. (2020) From PREDICT to prevention, one pandemic later. Lancet Microbe, 1, e6–e7. [Europe PMC free article] [Abstract] [Google Scholar]
- Cerdà‐Cuéllar M., Moré E., Ayats T., Aguilera M., Muñoz‐González S., Antilles N. et al. (2019) Do humans spread zoonotic enteric bacteria in Antarctica? The Science of the Total Environment, 654, 190–196. 10.1016/j.scitotenv.2018.10.272 [Abstract] [CrossRef] [Google Scholar]
- Chandler J.C., Bevins S.N., Ellis J.W., Linder T.J., Tell R.M., Jenkins‐Moore M. et al. (2021) SARS‐CoV‐2 exposure in wild white‐tailed deer (Odocoileus virginianus). Proceedings of the National Academy of Sciences of the United States of America, 118(47), e2114828118. [Europe PMC free article] [Abstract] [Google Scholar]
- Clutton‐Brock T. & Sheldon B.C. (2010) Individuals and populations: the role of long‐term, individual‐based studies of animals in ecology and evolutionary biology. Trends in Ecology & Evolution, 25, 562–573. [Abstract] [Google Scholar]
- Cogswell‐Hawkinson A., Bowen R., James S., Gardiner D., Calisher C.H., Adams R. et al. (2012) Tacaribe virus causes fatal infection of an ostensible reservoir host, the Jamaican fruit bat. Journal of Virology, 86, 5791–5799. [Europe PMC free article] [Abstract] [Google Scholar]
- Cook J.D., Grant E.H.C., Coleman J.T.H., Sleeman J.M. & Runge M.C. (2021) Risks posed by SARS‐CoV‐2 to North American bats during winter fieldwork. Conservation Science and Practice, 3(6), 10.1111/csp2.410 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Cool K., Gaudreault N.N., Morozov I., Trujillo J.D., Meekins D.A., McDowell C. et al. (2022) Infection and transmission of ancestral SARS‐CoV‐2 and its alpha variant in pregnant white‐tailed deer. Emerging Microbes & Infections, 11, 95–112. [Europe PMC free article] [Abstract] [Google Scholar]
- Coscolla M., Lewin A., Metzger S., Maetz‐Rennsing K., Calvignac‐Spencer S., Nitsche A. et al. (2013) Novel mycobacterium tuberculosis complex isolate from a wild chimpanzee. Emerging Infectious Diseases, 19, 969. [Europe PMC free article] [Abstract] [Google Scholar]
- Crossley B., Hietala S., Hunt T., Benjamin G., Martinez M., Darnell D. et al. (2012) Pandemic (H1N1) 2009 in captive cheetah. Emerging Infectious Diseases, 18, 315. [Europe PMC free article] [Abstract] [Google Scholar]
- Damas J., Hughes G.M., Keough K.C., Painter C.A., Persky N.S., Corbo M. et al. (2020) Broad host range of SARS‐CoV‐2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proceedings of the National Academy of Sciences, 117(36), 22311–22322. 10.1073/pnas.2010146117 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Davies T.J. & Pedersen A.B. (2008) Phylogeny and geography predict pathogen community similarity in wild primates and humans. Proceedings of the Royal Society B‐Biological Sciences, 275, 1695–1701. [Europe PMC free article] [Abstract] [Google Scholar]
- Dean K.R., Krauer F., Walløe L., Lingjærde O.C., Bramanti B., Stenseth N.C. et al. (2018) Human ectoparasites and the spread of plague in Europe during the Second Pandemic. Proceedings of the National Academy of Sciences, 115, 1304–1309. [Europe PMC free article] [Abstract] [Google Scholar]
- Dobson A.P., Pimm S.L., Hannah L., Kaufman L., Ahumada J.A., Ando A.W. et al. (2020) Ecology and economics for pandemic prevention. Science, 369, 379–381. [Abstract] [Google Scholar]
- Donahue M. (2020) U.S. advises suspending bat research over concerns coronavirus could infect North American species. Wash. Post.
- Donnelly C.A., Woodroffe R., Cox D.R., Bourne J., Gettinby G., Le Fevre A.M. et al. (2003) Impact of localized badger culling on tuberculosis incidence in British cattle. Nature, 426, 834–837. [Abstract] [Google Scholar]
- Eberhard M.L., Ruiz‐Tiben E., Hopkins D.R., Farrell C., Toe F., Weiss A. et al. (2014) The peculiar epidemiology of dracunculiasis in Chad. American Journal of Tropical Medicine and Hygiene, 90, 61–70. [Europe PMC free article] [Abstract] [Google Scholar]
- Edwards S.J.L. & Santini J.M. (2020) Anthroponotic risk of SARS‐CoV‐2, precautionary mitigation, and outbreak management. The Lancet Microbe, 1(5), e187–e188. 10.1016/S2666-5247(20)30086-0 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Engel G.A., Jones‐Engel L., Schillaci M.A., Suaryana K.G., Putra A., Fuentes A. et al. (2002) Human exposure to herpesvirus B–seropositive macaques, Bali, Indonesia. Emerging Infectious Diseases, 8, 789. [Europe PMC free article] [Abstract] [Google Scholar]
- Fagre A., Lewis J., Eckley M., Zhan S., Rocha S.M., Sexton N.R. et al. (2021) SARS‐CoV‐2 infection, neuropathogenesis and transmission among deer mice: implications for spillback to New World rodents. PLoS Path, 17, e1009585. [Europe PMC free article] [Abstract] [Google Scholar]
- Farrell M.J. & Davies T.J. (2019) Disease mortality in domesticated animals is predicted by host evolutionary relationships. Proceedings of the National Academy of Sciences, 116, 7911–7915. [Europe PMC free article] [Abstract] [Google Scholar]
- Favoretto S.R., Araujo D.B., Duarte N.F.H., Oliveira D.B.L., da Crus N.G., Mesquita F. et al. (2019) Zika Virus in peridomestic neotropical primates, Northeast Brazil. EcoHealth, 16, 61–69. [Abstract] [Google Scholar]
- Fischer C.P. & Romero L.M. (2019) Chronic captivity stress in wild animals is highly species‐specific. Conservation Physiology, 7, coz093. [Europe PMC free article] [Abstract] [Google Scholar]
- Fischhoff I.R., Castellanos A.A., Rodrigues J.P.G.L.M., Varsani A. & Han B.A. (2021) Predicting the zoonotic capacity of mammals to transmit SARS‐CoV‐2. Proceedings of the Royal Society B: Biological Sciences, 288, 20211651. https://royalsocietypublishing.org/doi/10.1098/rspb.2021.1651 [Europe PMC free article] [Abstract] [Google Scholar]
- Freuling C.M., Breithaupt A., Müller T., Sehl J., Balkema‐Buschmann A., Rissmann M. et al. (2020) Susceptibility of raccoon dogs for experimental SARS‐CoV‐2 infection. Emerging Infectious Diseases, 26, 2982–2985. [Europe PMC free article] [Abstract] [Google Scholar]
- Fritsche A., Engel R., Buhl D. & Zellweger J.P. (2004) Mycobacterium bovis tuberculosis: from animal to man and back. The International Journal of Tuberculosis and Lung Disease, 8, 903–904. [Abstract] [Google Scholar]
- Frutos R., Gavotte L. & Devaux C.A. (2021) Understanding the origin of COVID‐19 requires to change the paradigm on zoonotic emergence from the spillover model to the viral circulation model. Infection, Genetics and Evolution, 95, 104812. [Europe PMC free article] [Abstract] [Google Scholar]
- Garigliany M., Van Laere A.‐S., Clercx C., Giet D., Escriou N., Huon C. et al. (2020) SARS‐CoV‐2 natural transmission from human to cat, Belgium, March 2020. Emerging Infectious Diseases, 26(12), 3069–3071. 10.3201/eid2612.202223 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Ge X., Li Y., Yang X., Zhang H., Zhou P., Zhang Y. et al. (2012) Metagenomic analysis of viruses from bat fecal samples reveals many novel viruses in insectivorous bats in China. Journal of Virology, 86, 4620–4630. [Europe PMC free article] [Abstract] [Google Scholar]
- Gerdts V., Littel‐van den Hurk S.V.D., Griebel P.J. & Babiuk L.A. (2007) Use of animal models in the development of human vaccines. Future Microbiology, 2, 667–675. [Abstract] [Google Scholar]
- Gibbons A. (2021) Captive gorillas test positive for coronavirus. Science Magazine. Available at: https://www.science.org/content/article/captive‐gorillas‐test‐positive‐coronavirus, Last accessed 18 March 2022. [Google Scholar]
- Goldberg T.L., Gillespie T.R., Rwego I.B., Wheeler E., Estoff E.L. & Chapman C.A. (2007) Patterns of gastrointestinal bacterial exchange between chimpanzees and humans involved in research and tourism in western Uganda. Biological Conservation, 135, 511–517. [Google Scholar]
- Gorman J. (2020) Can humans give coronaviruses to bats, and other wildlife? N. Y. Times.
- Griffin B.D., Chan M., Tailor N., Mendoza E.J., Leung A., Warner B.M. et al. (2021) SARS‐CoV‐2 infection and transmission in the North American deer mouse. Nature Communications, 12, 3612. [Europe PMC free article] [Abstract] [Google Scholar]
- Hale V.L., Dennis P.M., McBride D.S., Nolting J.M., Madden C., Huey D. et al. (2021) SARS‐CoV‐2 infection in free‐ranging white‐tailed deer. Nature, 602, 481–486. [Europe PMC free article] [Abstract] [Google Scholar]
- Hall J.S., Knowles S., Nashold S.W., Ip H.S., Leon A.E., Rocke T. et al. (2020) Experimental challenge of a North American bat species, big brown bat (Eptesicus fuscus), with SARS‐CoV‐2. Transboundary and Emerging Diseases, 68(6), 3443–3452. [Abstract] [Google Scholar]
- Han B.A., Kramer A.M. & Drake J.M. (2016) Global patterns of zoonotic disease in mammals. Trends in Parasitology, 32, 565–577. [Europe PMC free article] [Abstract] [Google Scholar]
- Hayes L.D. & Schradin C. (2017) Long‐term field studies of mammals: what the short‐term study cannot tell us. Journal of Mammalogy, 98(3), 600–602. 10.1093/jmammal/gyx027 [CrossRef] [Google Scholar]
- Hedman H.D., Krawczyk E., Helmy Y.A., Zhang L. & Varga C. (2021) Host diversity and potential transmission pathways of SARS‐CoV‐2 at the human‐animal interface. Pathogens, 10, 180. [Europe PMC free article] [Abstract] [Google Scholar]
- Heldstab A., Rüedi D., Sonnabend W. & Deinhardt F. (1981) Spontaneous generalized Herpesvirus hominis infection of a lowland gorilla (Gorilla gorilla gorilla). Journal of Medical Primatology, 10, 129–135. [Abstract] [Google Scholar]
- Hockings K.J., Mubemba B., Avanzi C., Pleh K., Düx A., Bersacola E. et al. (2021) Leprosy in wild chimpanzees. Nature, 598, 652–656. [Europe PMC free article] [Abstract] [Google Scholar]
- Holmes E.C., Rambaut A. & Andersen K.G. (2018) Pandemics: spend on surveillance, not prediction. Nature, 558(7709), 180–182. 10.1038/d41586-018-05373-w [Abstract] [CrossRef] [Google Scholar]
- Hopkins D.R., Weiss A.J., Roy S.L., Zingeser J. & Guagliardo S.A.J. (2019) Progress toward global eradication of dracunculiasis ‐ January 2018‐June 2019. MMWR. Morbidity and Mortality Weekly Report, 68, 979–984. [Europe PMC free article] [Abstract] [Google Scholar]
- Jones B.A., Grace D., Kock R., Alonso S., Rushton J., Said M.Y. et al. (2013) Zoonosis emergence linked to agricultural intensification and environmental change. Proceedings of the National Academy of Sciences, 110, 8399–8404. [Europe PMC free article] [Abstract] [Google Scholar]
- Jones M.E., Amman B.R., Sealy T.K., Uebelhoer L.S., Schuh A.J., Flietstra T. et al. (2019) Clinical, histopathologic, and immunohistochemical characterization of experimental Marburg virus infection in a natural reservoir host, the Egyptian rousette bat (Rousettus aegyptiacus). Viruses, 11, 214. [Europe PMC free article] [Abstract] [Google Scholar]
- Kaur T., Singh J., Tong S., Humphrey C., Clevenger D., Tan W. et al. (2008) Descriptive epidemiology of fatal respiratory outbreaks and detection of a human‐related metapneumovirus in wild chimpanzees (Pan troglodytes) at Mahale Mountains National Park, Western Tanzania. American Journal of Primatology, 70, 755–765. [Europe PMC free article] [Abstract] [Google Scholar]
- Kock R.A., Orynbayev M., Robinson S., Zuther S., Singh N.J., Beauvais W. et al. (2018) Saigas on the brink: multidisciplinary analysis of the factors influencing mass mortality events. Science Advances, 4, eaao2314. [Europe PMC free article] [Abstract] [Google Scholar]
- Kuchipudi S.V., Surendran‐Nair M., Ruden R.M., Yon M., Nissly R.H., Nelli R.K., et al. (2021). Multiple spillovers and onward transmission of SARS‐CoV‐2 in free‐living and captive white‐tailed deer. [Europe PMC free article] [Abstract]
- Lachish S., Lawson B., Cunningham A.A. & Sheldon B.C. (2012) Epidemiology of the emergent disease Paridae pox in an intensively studied wild bird population. PLoS One, 7, e38316. [Europe PMC free article] [Abstract] [Google Scholar]
- Lam T.‐Y., Hon C.‐C., Lemey P., Pybus O.G., Shi M., Tun H.M. et al. (2012) Phylodynamics of H5N1 avian influenza virus in Indonesia. Molecular Ecology, 21, 3062–3077. [Abstract] [Google Scholar]
- Leung J.M., Budischak S.A., Chung The H., Hansen C., Bowcutt R., Neill R. et al. (2018) Rapid environmental effects on gut nematode susceptibility in rewilded mice. PLoS Biology, 16, e2004108. [Europe PMC free article] [Abstract] [Google Scholar]
- Li D., Zhu L., Cui H., Ling S., Fan S., Yu Z. et al. (2014) Influenza A(H1N1)pdm09 virus infection in giant pandas, China. Emerging Infectious Diseases, 20, 480–483. [Europe PMC free article] [Abstract] [Google Scholar]
- Li L., Victoria J.G., Wang C., Jones M., Fellers G.M., Kunz T.H. et al. (2010) Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. Journal of Virology, 84, 6955–6965. [Europe PMC free article] [Abstract] [Google Scholar]
- McAloose D., Laverack M., Wang L., Killian M.L., Caserta L.C., Yuan F., et al. (2020). From people to Panthera: Natural SARS‐CoV‐2 infection in tigers and lions at the Bronx Zoo. bioRxiv, 2020.07.22.213959. [Europe PMC free article] [Abstract] [Google Scholar]
- McClure H.M. & Keeling M.E. (1971) Viral diseases noted in the Yerkes Primate Center colony. Laboratory Animal Science, 21, 1002–1010. [Abstract] [Google Scholar]
- McDonald R.A., Wilson‐Aggarwal J.K., Swan G.J.F., Goodwin C.E.D., Moundai T., Sankara D. et al. (2020) Ecology of domestic dogs Canis familiaris as an emerging reservoir of Guinea worm Dracunculus medinensis infection. PLoS Neglected Tropical Diseases, 14, e0008170. [Europe PMC free article] [Abstract] [Google Scholar]
- McKinney L.A., Kick E.L. & Fulkerson G.M. (2010) World system, anthropogenic, and ecological threats to bird and mammal species: a structural equation analysis of biodiversity loss. Organization & Environment, 23, 3–31. [Google Scholar]
- Messenger A.M., Barnes A.N. & Gray G.C. (2014) Reverse zoonotic disease transmission (zooanthroponosis): a systematic review of seldom‐documented human biological threats to animals. PLoS One, 9, e89055. [Europe PMC free article] [Abstract] [Google Scholar]
- Michalak K., Austin C., Diesel S., Bacon M.J., Zimmerman P. & Maslow J.N. (1998) Mycobacterium tuberculosis infection as a zoonotic disease: transmission between humans and elephants. Emerging Infectious Diseases, 4, 283. [Europe PMC free article] [Abstract] [Google Scholar]
- Molenaar R.J., Vreman S., Hakze‐van der Honing R.W., Zwart R., de Rond J., Weesendorp E. et al. (2020) Clinical and pathological findings in SARS‐CoV‐2 disease outbreaks in farmed mink (Neovison vison). Veterinary Pathology, 57, 653–657. [Abstract] [Google Scholar]
- Mollentze N. & Streicker D.G. (2020) Viral zoonotic risk is homogenous among taxonomic orders of mammalian and avian reservoir hosts. Proceedings of the National Academy of Sciences, 117, 9423. [Europe PMC free article] [Abstract] [Google Scholar]
- Mubemba B., Chanove E., Mätz‐Rensing K., Gogarten J.F., Düx A., Merkel K. et al. (2020) Yaws Disease Caused by Treponema pallidum subspecies pertenue in Wild Chimpanzee, Guinea, 2019. Emerging Infectious Diseases, 26, 1283–1286. [Europe PMC free article] [Abstract] [Google Scholar]
- Munnink B.B.O., Sikkema R.S., Nieuwenhuijse D.F., Molenaar R.J., Munger E., Molenkamp R. et al. (2020) Transmission of SARS‐CoV‐2 on mink farms between humans and mink and back to humans. Science, 371(6525), 172–177. [Europe PMC free article] [Abstract] [Google Scholar]
- Mykytyn A.Z., Lamers M.M., Okba N.M.A., Breugem T.I., Schipper D., van den Doel P.B. et al. (2021) Susceptibility of rabbits to SARS‐CoV‐2. Emerging Microbes & Infections, 10, 1–7. [Europe PMC free article] [Abstract] [Google Scholar]
- Nizeyi J.B., Innocent R.B., Erume J., Kalema G.R., Cranfield M.R. & Graczyk T.K. (2001) Campylobacteriosis, salmonellosis, and shigellosis in free‐ranging human‐habituated mountain gorillas of Uganda. Journal of Wildlife Diseases, 37, 239–244. [Abstract] [Google Scholar]
- Obanda V., Poghon J., Yongo M., Mulei I., Ngotho M., Waititu K. et al. (2013) First reported case of fatal tuberculosis in a wild African elephant with past human–wildlife contact. Epidemiology and Infection, 141, 1476–1480. [Europe PMC free article] [Abstract] [Google Scholar]
- Oh P., Granich R., Scott J., Sun B., Joseph M., Stringfield C. et al. (2002) Human exposure following Mycobacterium tuberculosis infection of multiple animal species in a metropolitan zoo. Emerging Infectious Diseases, 8, 1290. [Europe PMC free article] [Abstract] [Google Scholar]
- OIE‐World Organisation for Animal Health (2020). Infection with SARS‐CoV‐2, Slovenia. (Immediate notification (Final Report)).
- Olival K.J., Cryan P.M., Amman B.R., Baric R.S., Blehert D.S., Brook C.E. et al. (2020) Possibility for reverse zoonotic transmission of SARS‐CoV‐2 to free‐ranging wildlife: a case study of bats. PLoS Path, 16, e1008758. [Europe PMC free article] [Abstract] [Google Scholar]
- Olival K.J., Hosseini P.R., Zambrana‐Torrelio C., Ross N., Bogich T.L. & Daszak P. (2017) Host and viral traits predict zoonotic spillover from mammals. Nature, 546, 646–650. [Europe PMC free article] [Abstract] [Google Scholar]
- Oreshkova N., Molenaar R.J., Vreman S., Harders F., Oude Munnink B.B., Hakze‐van der Honing R.W. et al. (2020) SARS‐CoV‐2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance, 25(23), 10.2807/1560-7917.ES.2020.25.23.2001005 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Osterhaus A., Rimmelzwaan G.F., Martina B.E.E., Bestebroer T.M. & Fouchier R.A.M. (2000) Influenza B virus in seals. Science, 288, 1051–1053. [Abstract] [Google Scholar]
- Palacios G., Lowenstine L.J., Cranfield M.R., Gilardi K.V.K., Spelman L., Lukasik‐Braum M. et al. (2011) Human metapneumovirus infection in wild mountain gorillas. Emerging Infectious Diseases, 17, 711. [Europe PMC free article] [Abstract] [Google Scholar]
- Patterson E.I., Elia G., Grassi A., Giordano A., Desario C., Medardo M. et al. (2020) Evidence of exposure to SARS‐CoV‐2 in cats and dogs from households in Italy. Nature Communications, 11, 1–5. [Europe PMC free article] [Abstract] [Google Scholar]
- Pedersen A.B., Jones K.E., Nunn C.L. & Altizer S. (2007) Infectious diseases and extinction risk in wild mammals. Conservation Biology, 21, 1269–1279. [Europe PMC free article] [Abstract] [Google Scholar]
- Plourde B.T., Burgess T.L., Eskew E.A., Roth T.M., Stephenson N. & Foley J.E. (2017) Are disease reservoirs special? Taxonomic and life history characteristics. PLoS One, 12, e0180716. [Europe PMC free article] [Abstract] [Google Scholar]
- Plowright R.K., Eby P., Hudson P.J., Smith I.L., Westcott D., Bryden W.L. et al. (2015) Ecological dynamics of emerging bat virus spillover. Proceedings of the Royal Society B‐Biological Sciences, 282, 20142124. [Europe PMC free article] [Abstract] [Google Scholar]
- Plowright R.K., Parrish C.R., McCallum H., Hudson P.J., Ko A.I., Graham A.L. et al. (2017) Pathways to zoonotic spillover. Nature Reviews Microbiology, 15, 502–510. [Europe PMC free article] [Abstract] [Google Scholar]
- Porter S.M., Hartwig A.E., Bielefeldt‐Ohmann H., Bosco‐Lauth A.M. & Root J.J. (2022) Susceptibility of wild canids to severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). bioRxiv. 10.1101/2022.01.27.478082 [CrossRef] [Google Scholar]
- Prince T., Smith S.L., Radford A.D., Solomon T., Hughes G.L. & Patterson E.I. (2021) SARS‐CoV‐2 infections in animals: reservoirs for reverse zoonosis and models for study. Viruses, 13, 494. [Europe PMC free article] [Abstract] [Google Scholar]
- Richter D., Klug B., Spielman A. & Matuschka F.‐R. (2004) Adaptation of diverse Lyme disease spirochetes in a natural rodent reservoir host. Infection and Immunity, 72, 2442–2444. [Europe PMC free article] [Abstract] [Google Scholar]
- Roche B., Drake J.M., Brown J., Stallknecht D.E., Bedford T. & Rohani P. (2014) Adaptive evolution and environmental durability jointly structure phylodynamic patterns in avian influenza viruses. PLoS Biology, 12, e1001931. [Europe PMC free article] [Abstract] [Google Scholar]
- Rothenburg S. & Brennan G. (2020) Species‐specific host–virus interactions: implications for viral host range and virulence. Trends in Microbiology, 28, 46–56. [Europe PMC free article] [Abstract] [Google Scholar]
- Rwego I.B., Isabirye‐basuta G., Gillespie T.R. & Goldberg T.L. (2008) Gastrointestinal bacterial transmission among humans, mountain gorillas, and livestock in Bwindi Impenetrable National Park, Uganda. Conservation Biology, 22, 1600–1607. [Abstract] [Google Scholar]
- Ryan S.J. & Walsh P.D. (2011) Consequences of Non‐Intervention for Infectious Disease in African Great Apes. PLoS One, 6, e29030. [Europe PMC free article] [Abstract] [Google Scholar]
- Sailleau C., Dumarest M., Vanhomwegen J., Delaplace M., Caro V., Kwasiborski A. et al. (2020) First detection and genome sequencing of SARS‐CoV‐2 in an infected cat in France. Transboundary and Emerging Diseases, 67(6), 2324–2328. 10.1111/tbed.13659 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Santini J.M. & Edwards S.J.L. (2020) Host range of SARS‐CoV‐2 and implications for public health. Lancet Microbe, 1, e141–e142. [Europe PMC free article] [Abstract] [Google Scholar]
- Schlottau K., Rissmann M., Graaf A., Schön J., Sehl J., Wylezich C. et al. (2020) SARS‐CoV‐2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. The Lancet Microbe, 1(5), e218–e225. 10.1016/S2666-5247(20)30089-6 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Seeber P.A., Morrison T., Ortega A., East M.L., Greenwood A.D. & Czirják G.Á. (2020) Immune differences in captive and free‐ranging zebras (Equus zebra and E. quagga). Mammalian Biology, 100(2), 155–164. 10.1007/s42991-020-00006-0 [CrossRef] [Google Scholar]
- Segalés J., Puig M., Rodon J., Avila‐Nieto C., Carrillo J., Cantero G. et al. (2020) Detection of SARS‐CoV‐2 in a cat owned by a COVID‐19− affected patient in Spain. Proceedings of the National Academy of Sciences, 117, 24790–24793. [Europe PMC free article] [Abstract] [Google Scholar]
- Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B. et al. (2020) Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science, 368, 1016–1020. [Europe PMC free article] [Abstract] [Google Scholar]
- Sit T.H.C., Brackman C.J., Ip S.M., Tam K.W.S., Law P.Y.T., To E.M.W. et al. (2020) Infection of dogs with SARS‐CoV‐2. Nature, 586(7831), 1–6. [Europe PMC free article] [Abstract] [Google Scholar]
- Sweeny A.R., Thomason C.A., Carbajal E.A., Hansen C.B., Graham A.L. & Pedersen A.B. (2020) Experimental parasite community perturbation reveals associations between Sin Nombre virus and gastrointestinal nematodes in a rodent reservoir host. Biology Letters, 16, 20200604. [Europe PMC free article] [Abstract] [Google Scholar]
- Terzian A.C.B., Zini N., Sacchetto L., Rocha R.F., Parra M.C.P., Del Sarto J.L. et al. (2018) Evidence of natural Zika virus infection in neotropical non‐human primates in Brazil. Scientific Reports, 8, 1–15. [Europe PMC free article] [Abstract] [Google Scholar]
- Tu C., Crameri G., Kong X., Chen J., Sun Y., Yu M. et al. (2004) Antibodies to SARS coronavirus in civets. Emerging Infectious Diseases, 10, 2244. [Europe PMC free article] [Abstract] [Google Scholar]
- Viana M., Mancy R., Biek R., Cleaveland S., Cross P.C., Lloyd‐Smith J.O. et al. (2014) Assembling evidence for identifying reservoirs of infection. Trends in Ecology & Evolution, 29, 270–279. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang L., Mitchell P.K., Calle P.P., Bartlett S.L., McAloose D., Killian M.L. et al. (2020) Complete genome sequence of SARS‐CoV‐2 in a tiger from a US zoological collection. Microbiology Resource Announcements, 9(22), e00468‐20. 10.1128/MRA.00468-20 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wang M., Yan M., Xu H., Liang W., Kan B., Zheng B. et al. (2005) SARS‐CoV infection in a restaurant from palm civet. Emerging Infectious Diseases, 11, 1860. [Europe PMC free article] [Abstract] [Google Scholar]
- Woolhouse M.E. & Brierley L. (2018) Epidemiological characteristics of human‐infective RNA viruses. Science Data, 5, 180017. [Europe PMC free article] [Abstract] [Google Scholar]
- Wu Z., Yang L.I., Ren X., He G., Zhang J., Yang J. et al. (2016) Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME Journal, 10, 609–620. [Europe PMC free article] [Abstract] [Google Scholar]
- Yen H.L., Sit T.H., Brackman C.J., Chuk S.S., Cheng S., Gu H., et al. (2022) Transmission of SARS‐CoV‐2 (Variant Delta) from pet hamsters to humans and onward human propagation of the adapted strain: a case study. SSRN Electronic Journal, 10.2139/ssrn.4017393 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Zachariah A., Pandiyan J., Madhavilatha G.K., Mundayoor S., Chandramohan B., Sajesh P.K. et al. (2017) Mycobacterium tuberculosis in wild Asian elephants, southern India. Emerging Infectious Diseases, 23, 504. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhao Y., Wang J., Kuang D., Xu J., Yang M., Ma C. et al. (2020) Susceptibility of tree shrew to SARS‐CoV‐2 infection. Scientific Reports, 10, 16007. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhou N.N., Senne D.A., Landgraf J.S., Swenson S.L., Erickson G., Rossow K. et al. (1999) Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. Journal of Virology, 73, 8851–8856. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhou P., Yang X.‐L., Wang X.‐G., Hu B., Zhang L., Zhang W. et al. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579, 270–273. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1111/ele.14003
Read article for free, from open access legal sources, via Unpaywall:
https://www.repository.cam.ac.uk/bitstream/1810/335377/2/ele.14003.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/125285431
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1111/ele.14003
Article citations
Beliefs, taboos, usages, health perceptions, and practices toward wildlife among different ethnicities in Tak and Mae Hong Son Provinces, Thailand.
BMC Public Health, 24(1):2432, 06 Sep 2024
Cited by: 0 articles | PMID: 39242509 | PMCID: PMC11380376
CRISPR-Based Biosensors for Medical Diagnosis: Readout from Detector-Dependence Detection Toward Naked Eye Detection.
Biosensors (Basel), 14(8):367, 28 Jul 2024
Cited by: 0 articles | PMID: 39194596 | PMCID: PMC11353133
Review Free full text in Europe PMC
A One Health framework for exploring zoonotic interactions demonstrated through a case study.
Nat Commun, 15(1):5650, 15 Jul 2024
Cited by: 2 articles | PMID: 39009576 | PMCID: PMC11250852
Modeling zoonotic and vector-borne viruses.
Curr Opin Virol, 67:101428, 22 Jul 2024
Cited by: 0 articles | PMID: 39047313
Review
The reverse zoonotic potential of SARS-CoV-2.
Heliyon, 10(12):e33040, 13 Jun 2024
Cited by: 0 articles | PMID: 38988520 | PMCID: PMC11234007
Review Free full text in Europe PMC
Go to all (30) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Transmission dynamics and susceptibility patterns of SARS-CoV-2 in domestic, farmed and wild animals: Sustainable One Health surveillance for conservation and public health to prevent future epidemics and pandemics.
Transbound Emerg Dis, 69(5):2523-2543, 09 Nov 2021
Cited by: 21 articles | PMID: 34694705 | PMCID: PMC8662162
Review Free full text in Europe PMC
Possibility for reverse zoonotic transmission of SARS-CoV-2 to free-ranging wildlife: A case study of bats.
PLoS Pathog, 16(9):e1008758, 03 Sep 2020
Cited by: 89 articles | PMID: 32881980 | PMCID: PMC7470399
Review Free full text in Europe PMC
Lack of Evidence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Spillover in Free-Living Neotropical Non-Human Primates, Brazil.
Viruses, 13(10):1933, 25 Sep 2021
Cited by: 7 articles | PMID: 34696363 | PMCID: PMC8540180
Spillover of SARS-CoV-2 into novel wild hosts in North America: A conceptual model for perpetuation of the pathogen.
Sci Total Environ, 733:139358, 12 May 2020
Cited by: 42 articles | PMID: 32416535 | PMCID: PMC7214292

 1
,
2
1
,
2